Submitted:
09 October 2023
Posted:
10 October 2023
You are already at the latest version
Abstract
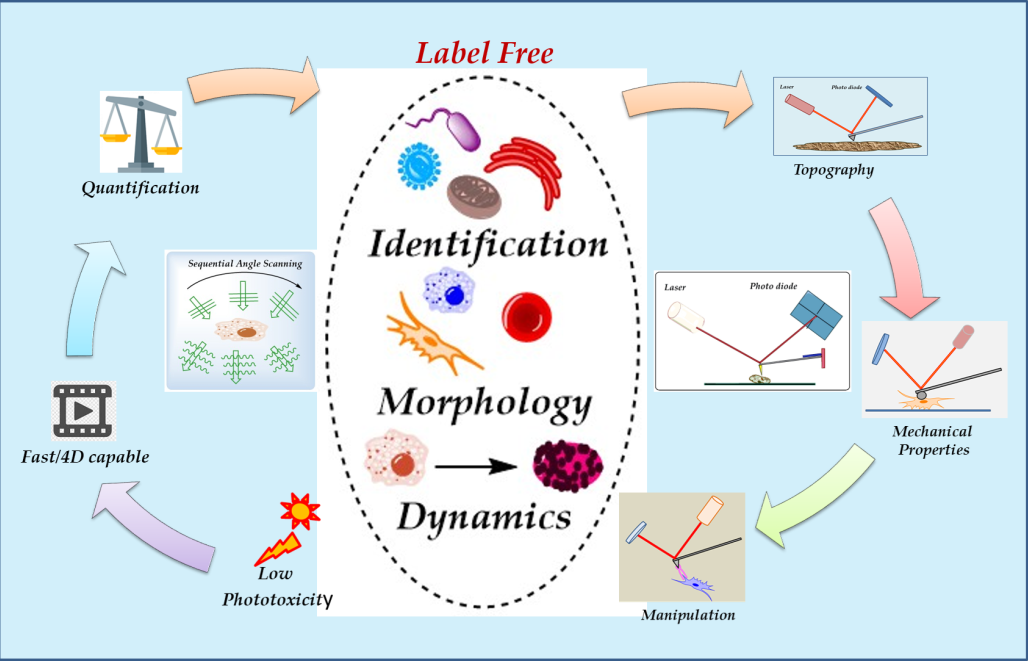
Keywords:
1. Introduction
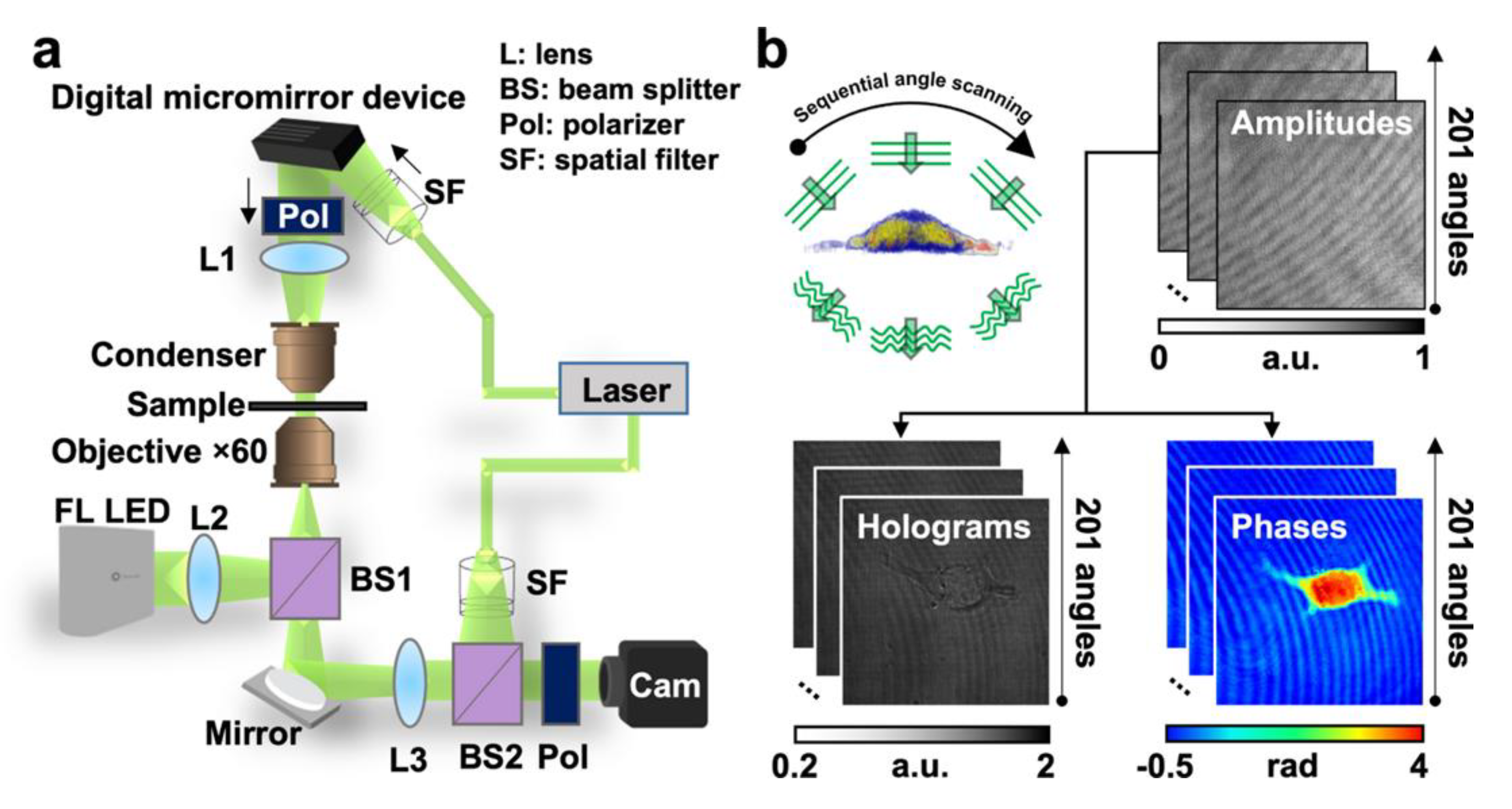
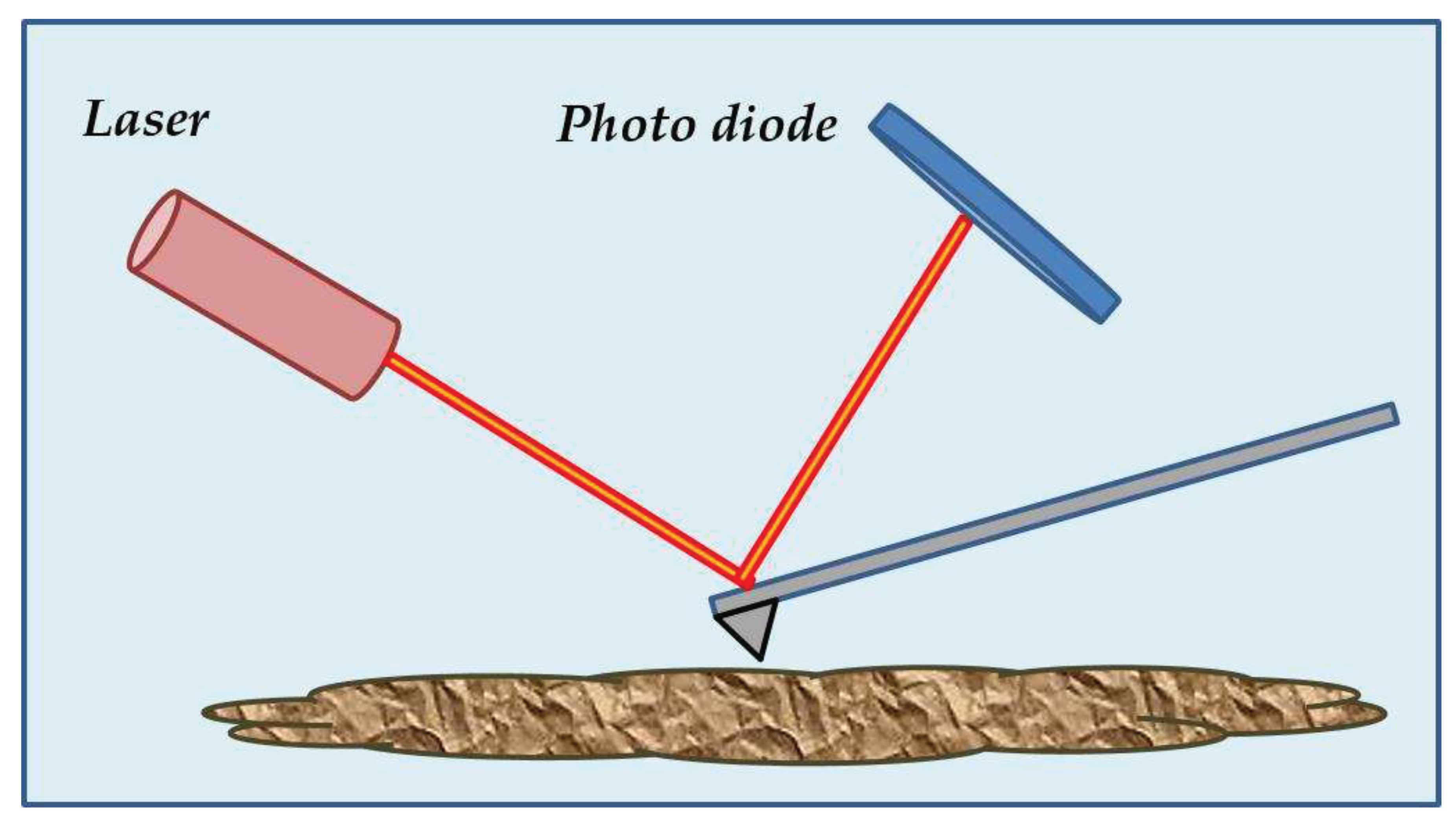
2. Holotomography Principle
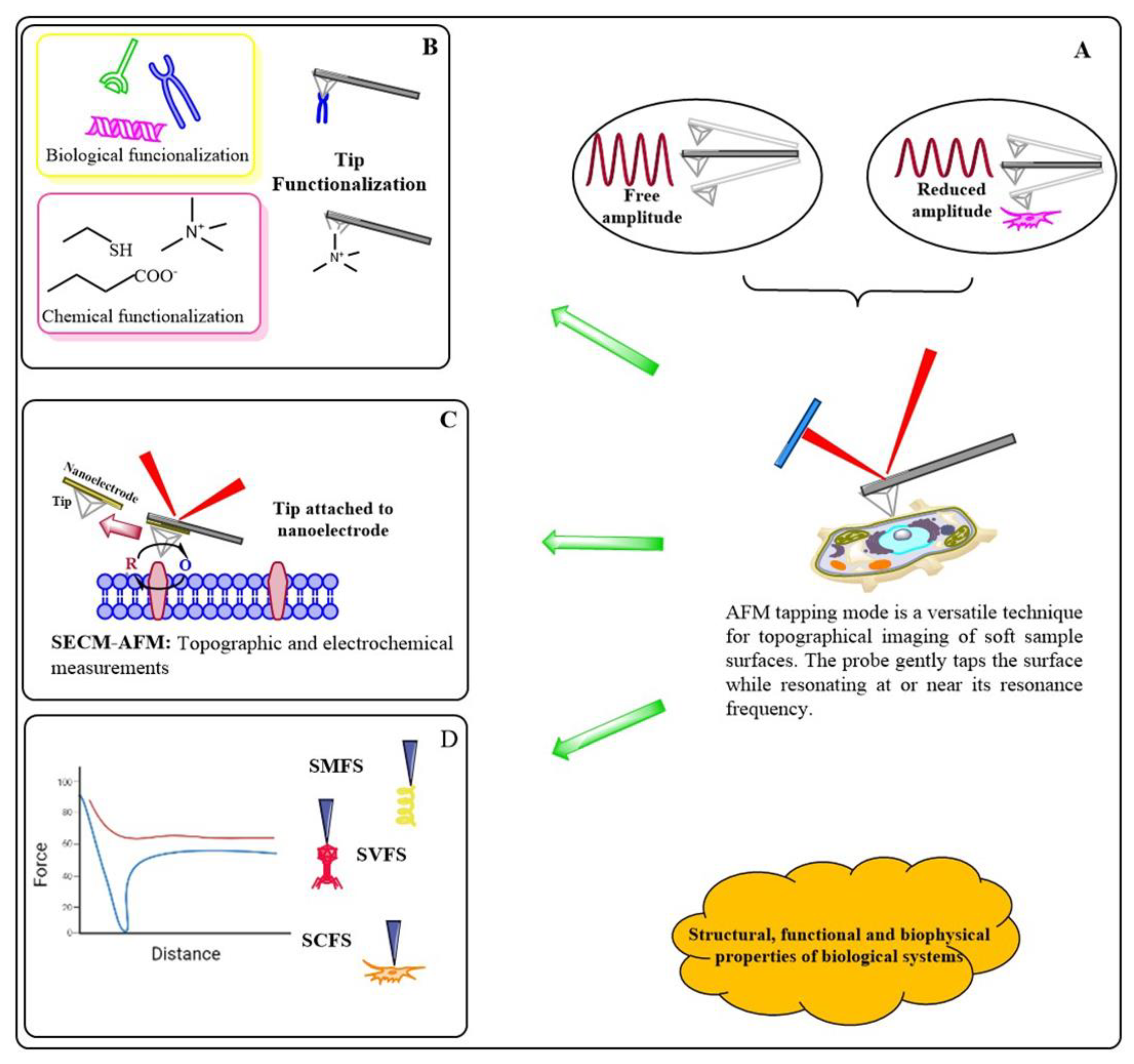
3. Atomic Force Microscopy Principle and Imaging Modes
- Application of AMF in biological research.
- High-speed AFM (HS-AFM).
- Fluid AFM and FluidFM (Fluidic force microscopy).
- IR-AFM
- Correlative Microscopy
4. Atomic Force and Holotomography Microscopy in Cancer, Microbiology, Nanotoxicology and Nanomedicine Research.
4.1. Cancer Research
4.2. Microbiology
4.3. Nanotoxicology and Nanomedicine
5. Conclusions and Prospects.
Author Contributions
Funding
Conflicts of Interest
References
- D. Kim, S. Lee, M. Lee, J. Oh, S.A. Yang, Y.K. Park, Holotomography: Refractive Index as an Intrinsic Imaging Contrast for 3-D Label-Free Live Cell Imaging, in: Adv. Exp. Med. Biol., 2021: pp. 211–238. https://doi.org/10.1007/978-981-33-6064-8_10. [CrossRef]
- S.Y. Kim, J.H. Lee, Y. Shin, T.K. Kim, J. won Lee, M.J. Pyo, A.R. Lee, C.G. Pack, Y.S. Cho, Label-free imaging and evaluation of characteristic properties of asthma-derived eosinophils using optical diffraction tomography, Biochem. Biophys. Res. Commun. 587 (2022) 42–48. [CrossRef]
- L.Y. D’Brant, H. Desta, T.C. Khoo, A. V. Sharikova, S.D. Mahajan, A. Khmaladze, Methamphetamine-induced apoptosis in glial cells examined under marker-free imaging modalities, J. Biomed. Opt. 24 (2019) 1–10. [CrossRef]
- Kim, Lee, Fujii, Kim, Pack, Physicochemical Properties of Nucleoli in Live Cells Analyzed by Label-Free Optical Diffraction Tomography, Cells. 8 (2019) 699. [CrossRef]
- R.A. Meyer, Light scattering from biological cells: dependence of backscatter radiation on membrane thickness and refractive index, Appl. Opt. 18 (1979) 585. [CrossRef]
- S. Chamot, E. Migacheva, O. Seydoux, P. Marquet, C. Depeursinge, Physical interpretation of the phase function related parameter γ studied with a fractal distribution of spherical scatterers, Opt. Express. 18 (2010) 23664. [CrossRef]
- Q. Zhang, L. Zhong, P. Tang, Y. Yuan, S. Liu, J. Tian, X. Lu, Quantitative refractive index distribution of single cell by combining phase-shifting interferometry and AFM imaging, Sci. Rep. 7 (2017) 2532. [CrossRef]
- P.Y. Liu, L.K. Chin, W. Ser, H.F. Chen, C.M. Hsieh, C.H. Lee, K.B. Sung, T.C. Ayi, P.H. Yap, B. Liedberg, K. Wang, T. Bourouina, Y. Leprince-Wang, Cell refractive index for cell biology and disease diagnosis: Past, present and future, Lab Chip. 16 (2016) 634–644. [CrossRef]
- P.A. Sandoz, C. Tremblay, S. Equis, S. Pop, L. Pollaro, Y. Cotte, F.G. van der Goot, M. Frechin, Label free 3D analysis of organelles in living cells by refractive index shows pre-mitotic organelle spinning in mammalian stem cells, BioRxiv. (2018) 407239.
- H. Moreno, L. Archetti, E. Gibbin, A.E. Grandchamp, M. Fréchin, Artificial Intelligence-Powered Automated Holotomographic Microscopy Enables Label-Free Quantitative Biology, Micros. Today. 29 (2021) 24–32. [CrossRef]
- P.A. Sandoz, C. Tremblay, F. Gisou van der Goot, M. Frechin, Image-based analysis of living mammalian cells using label-free 3D refractive index maps reveals new organelle dynamics and dry mass flux, PLoS Biol. 17 (2019) e3000553. [CrossRef]
- L. Pollaro, B. Dalla Piazza, Y. Cotte, Digital Staining: Microscopy of Live Cells Without Invasive Chemicals, Micros. Today. 23 (2015) 12–17. [CrossRef]
- Lambert, Live Cell Imaging with Holotomography and Fluorescence, Micros. Today. 28 (2020) 18–23. [CrossRef]
- S. Park, L.E. Lee, H. Kim, J.E. Kim, S.J. Lee, S. Yoon, S. Shin, H. Kang, Y.K. Park, J.J. Song, S. Lee, Detection of intracellular monosodium urate crystals in gout synovial fluid using optical diffraction tomography, Sci. Rep. 11 (2021) 1–8. [CrossRef]
- Y. He, N. Zhou, M. Ziemczonok, Y. Wang, L. Lei, L. Duan, R. Zhou, Standardizing image assessment in optical diffraction tomography, Opt. Lett. 48 (2023) 395–398. [CrossRef]
- S. Kang, R. Zhou, M. Brelen, H.K. Mak, P.T.C. So, Z. Yaqoob, Reflection-mode optical diffraction tomography for label-free imaging of thick biological specimens, (2022).
- D. Lee, M. Lee, H. Kwak, Y.S. Kim, J. Shim, J.H. Jung, W. Park, J.-H. Park, S. Lee, Y. Park, High-fidelity optical diffraction tomography of live organisms using iodixanol refractive index matching, Biomed. Opt. Express. 13 (2022) 6404. [CrossRef]
- Y.K. Park, C. Depeursinge, G. Popescu, Quantitative phase imaging in biomedicine, Nat. Photonics. 12 (2018) 578–589. [CrossRef]
- Y.J. Jo, H. Cho, S.Y. Lee, G. Choi, G. Kim, H.S. Min, Y.K. Park, Quantitative Phase Imaging and Artificial Intelligence: A Review, IEEE J. Sel. Top. Quantum Electron. 25 (2018) 1–14. [CrossRef]
- Shevkunov, M. Ziemczonok, M. Kujawińska, K. Egiazarian, Complex-domain SVD- and sparsity-based denoising for optical diffraction tomography, Opt. Lasers Eng. 159 (2022) 0–2. [CrossRef]
- M. Ziemczonok, A. Kuś, M. Kujawińska, Optical diffraction tomography meets metrology — Measurement accuracy on cellular and subcellular level, Meas. J. Int. Meas. Confed. 195 (2022). [CrossRef]
- D.H. Ryu, H. Nam, J.S. Jeon, Y.K. Park, Reagent- and actuator-free analysis of individual erythrocytes using three-dimensional quantitative phase imaging and capillary microfluidics, Sensors Actuators B Chem. 348 (2021) 130689. [CrossRef]
- Ali, Y. Abouleila, S. Amer, R. Furushima, S. Emara, S. Equis, Y. Cotte, T. Masujima, Quantitative live single-cell mass spectrometry with spatial evaluation by three-dimensional holographic and tomographic laser microscopy, Anal. Sci. 32 (2016) 125–127. [CrossRef]
- J. Zhao, A. Matlock, H. Zhu, Z. Song, J. Zhu, B. Wang, F. Chen, Y. Zhan, Z. Chen, Y. Xu, X. Lin, L. Tian, J.X. Cheng, Bond-selective intensity diffraction tomography, Nat. Commun. 13 (2022). [CrossRef]
- M. Baczewska, K. Eder, S. Ketelhut, B. Kemper, M. Kujawińska, Refractive Index Changes of Cells and Cellular Compartments Upon Paraformaldehyde Fixation Acquired by Tomographic Phase Microscopy, Cytom. Part A. 99 (2021) 388–398. [CrossRef]
- D. Park, D. Lee, Y. Kim, Y. Park, Y.J. Lee, J.E. Lee, M.K. Yeo, M.W. Kang, Y. Chong, S.J. Han, J. Choi, J.E. Park, Y. Koh, J. Lee, Y.K. Park, R. Kim, J.S. Lee, J. Choi, S.H. Lee, B. Ku, D.H. Kang, C. Chung, Cryobiopsy: A Breakthrough Strategy for Clinical Utilization of Lung Cancer Organoids, Cells. 12 (2023). [CrossRef]
- L. Pollaro, S. Equis, B. Dalla Piazza, Y. Cotte, Stain-free 3D Nanoscopy of Living Cells, Opt. Photonik. 11 (2016) 38–42. [CrossRef]
- H. Jiang, J. woo Kwon, S. Lee, Y.J. Jo, S. Namgoong, X. rui Yao, B. Yuan, J. bao Zhang, Y.K. Park, N.H. Kim, Reconstruction of bovine spermatozoa substances distribution and morphological differences between Holstein and Korean native cattle using three-dimensional refractive index tomography, Sci. Rep. 9 (2019) 8774. [CrossRef]
- L. Kreplak, Introduction to atomic force microscopy (AFM) in biology, Curr. Protoc. Protein Sci. 2016 (2016) 17.7.1-17.7.21. [CrossRef]
- Y.F. Dufrêne, T. Ando, R. Garcia, D. Alsteens, D. Martinez-Martin, A. Engel, C. Gerber, D.J. Müller, Imaging modes of atomic force microscopy for application in molecular and cell biology, Nat. Nanotechnol. 12 (2017) 295–307. [CrossRef]
- P. Parot, Y.F. Dufrêne, P. Hinterdorfer, C. Le Grimellec, D. Navajas, J.L. Pellequer, S. Scheuring, Past, present and future of atomic force microscopy in life sciences and medicine, J. Mol. Recognit. 20 (2007) 418–431. [CrossRef]
- Y.F. Dufrêne, Using nanotechniques to explore microbial surfaces, Nat. Rev. Microbiol. 2 (2004) 451–460. [CrossRef]
- Y.F. Dufrêne, Atomic force microscopy, a powerful tool in microbiology, J. Bacteriol. 184 (2002) 5205–5213. [CrossRef]
- C. Formosa-Dague, R.E. Duval, E. Dague, Cell biology of microbes and pharmacology of antimicrobial drugs explored by Atomic Force Microscopy, Semin. Cell Dev. Biol. 73 (2018) 165–176. [CrossRef]
- Y.C. Lin, C. Huang, H.C. Lai, Revealing the ultrastructure of the membrane pores of intact Serratia marcescens cells by atomic force microscopy, Heliyon. 5 (2019) e02636. [CrossRef]
- J.H. Martínez-Montelongo, I.E. Medina-Ramírez, Y. Romo-Lozano, J.A. Zapien, Development of a sustainable photocatalytic process for air purification., Chemosphere. 257 (2020). [CrossRef]
- I.E. Medina-Ramírez, A. Marroquin-Zamudio, J.H. Martínez-Montelongo, Y. Romo-Lozano, J.A. Zapien, A. Perez-Larios, Enhanced photocatalytic and antifungal activity of ZnO–Cu2+and Ag@ZnO–Cu2+ materials, Ceram. Int. 48 (2022) 12660–12674. [CrossRef]
- I.E. Medina-Ramírez, C.E. Díaz de León-Macias, G. Pedroza-Herrera, R. Gonzáles-Segovia, J.A. Zapien, J.L. Rodríguez-López, Evaluation of the biocompatibility and growth inhibition of bacterial biofilms by ZnO, Fe3O4 and ZnO@Fe3O4 photocatalytic magnetic materials, Ceram. Int. 46 (2020) 8979–8994. [CrossRef]
- I.E. Medina-Ramírez, M.A. Díaz de León Olmos, M.H. Muñoz Ortega, J.A. Zapien, I. Betancourt, N. Santoyo-Elvira, Development and Assessment of Nano-Technologies for Cancer Treatment: Cytotoxicity and Hyperthermia Laboratory Studies, Cancer Invest. 38 (2020) 61–84. [CrossRef]
- M. Krieg, G. Fläschner, D. Alsteens, B.M. Gaub, W.H. Roos, G.J.L. Wuite, H.E. Gaub, C. Gerber, Y.F. Dufrêne, D.J. Müller, Atomic force microscopy-based mechanobiology, Nat. Rev. Phys. 1 (2019) 41–57. [CrossRef]
- D.J. Müller, A.C. Dumitru, C. Lo Giudice, H.E. Gaub, P. Hinterdorfer, G. Hummer, J.J. De Yoreo, Y.F. Dufrêne, D. Alsteens, Atomic Force Microscopy-Based Force Spectroscopy and Multiparametric Imaging of Biomolecular and Cellular Systems, Chem. Rev. 121 (2021) 11701–11725. [CrossRef]
- T. Ando, High-speed atomic force microscopy and its future prospects, Biophys. Rev. 10 (2018) 285–292. [CrossRef]
- T. Ando, T. Uchihashi, S. Scheuring, Filming biomolecular processes by high-speed atomic force microscopy, Chem. Rev. 114 (2014) 3120–3188. [CrossRef]
- Y.F. Dufrêne, A. Viljoen, J. Mignolet, M. Mathelié-Guinlet, AFM in cellular and molecular microbiology, Cell. Microbiol. 23 (2021) 1–12. [CrossRef]
- K. Schoenwald, Z.C. Peng, D. Noga, S.R. Qiu, T. Sulchek, Integration of atomic force microscopy and a microfluidic liquid cell for aqueous imaging and force spectroscopy, Rev. Sci. Instrum. 81 (2010) 1–5. [CrossRef]
- Guillaume-Gentil, E. Potthoff, D. Ossola, C.M. Franz, T. Zambelli, J.A. Vorholt, Force-controlled manipulation of single cells: From AFM to FluidFM, Trends Biotechnol. 32 (2014) 381–388. [CrossRef]
- Dazzi, C.B. Prater, AFM-IR: Technology and applications in nanoscale infrared spectroscopy and chemical imaging, Chem. Rev. 117 (2017) 5146–5173. [CrossRef]
- J. Mathurin, A. Deniset-Besseau, D. Bazin, E. Dartois, M. Wagner, A. Dazzi, Photothermal AFM-IR spectroscopy and imaging: Status, challenges, and trends, J. Appl. Phys. 131 (2022). [CrossRef]
- L. Zhou, M. Cai, T. Tong, H. Wang, Progress in the correlative atomic force microscopy and optical microscopy, Sensors (Switzerland). 17 (2017). [CrossRef]
- N.A. Geisse, AFM and combined optical techniques, Mater. Today. 12 (2009) 40–45. [CrossRef]
- F. Colombo, E.G. Norton, E. Cocucci, Microscopy approaches to study extracellular vesicles, Biochim. Biophys. Acta - Gen. Subj. 1865 (2021) 129752. [CrossRef]
- M. Cascione, V. de Matteis, R. Rinaldi, S. Leporatti, Atomic force microscopy combined with optical microscopy for cells investigation, Microsc. Res. Tech. 80 (2017) 109–123. [CrossRef]
- J.R. Staunton, B.L. Doss, S. Lindsay, R. Ros, Correlating confocal microscopy and atomic force indentation reveals metastatic cancer cells stiffen during invasion into collagen i matrices, Sci. Rep. 6 (2016) 1–15. [CrossRef]
- A.R. Bagheri, N. Aramesh, M. Bilal, J. Xiao, H.W. Kim, B. Yan, Carbon nanomaterials as emerging nanotherapeutic platforms to tackle the rising tide of cancer – A review, Bioorganic Med. Chem. 51 (2021) 116493. [CrossRef]
- X. Deng, F. Xiong, X. Li, B. Xiang, Z. Li, X. Wu, C. Guo, X. Li, Y. Li, G. Li, W. Xiong, Z. Zeng, Application of atomic force microscopy in cancer research, J. Nanobiotechnology. 16 (2018) 1–16. [CrossRef]
- Stylianou, M. Lekka, T. Stylianopoulos, AFM assessing of nanomechanical fingerprints for cancer early diagnosis and classification: From single cell to tissue level, Nanoscale. 10 (2018) 20930–20945. [CrossRef]
- R. Di Santo, S. Romanò, A. Mazzini, S. Jovanović, G. Nocca, G. Campi, M. Papi, M. De Spirito, F. Di Giacinto, G. Ciasca, Recent advances in the label-free characterization of exosomes for cancer liquid biopsy: From scattering and spectroscopy to nanoindentation and nanodevices, Nanomaterials. 11 (2021). [CrossRef]
- Kubiak, T. Zieliński, J. Pabijan, M. Lekka, Nanomechanics in monitoring the effectiveness of drugs targeting the cancer cell cytoskeleton, Int. J. Mol. Sci. 21 (2020) 1–15. [CrossRef]
- H. Zhang, L. Xiao, Q. Li, X. Qi, A. Zhou, Microfluidic chip for non-invasive analysis of tumor cells interaction with anti-cancer drug doxorubicin by AFM and Raman spectroscopy, Biomicrofluidics. 12 (2018) 1–13. [CrossRef]
- L. Andrei, S. Kasas, I. Ochoa Garrido, T. Stanković, M. Suárez Korsnes, R. Vaclavikova, Y.G. Assaraf, M. Pešić, Advanced technological tools to study multidrug resistance in cancer, Drug Resist. Updat. 48 (2020) 100658. [CrossRef]
- W. Szlasa, S. Supplitt, M. Drąg-Zalesińska, D. Przystupski, K. Kotowski, A. Szewczyk, P. Kasperkiewicz, J. Saczko, J. Kulbacka, Effects of curcumin based PDT on the viability and the organization of actin in melanotic (A375) and amelanotic melanoma (C32)– in vitro studies, Biomed. Pharmacother. 132 (2020). [CrossRef]
- Z. Łapińska, M. Dębiński, A. Szewczyk, A. Choromańska, J. Kulbacka, J. Saczko, Electrochemotherapy with calcium chloride and 17β-estradiol modulated viability and apoptosis pathway in human ovarian cancer, Pharmaceutics. 13 (2021) 1–17. [CrossRef]
- Y. Xiao, Y. Cheng, P. He, X. Wu, Z. Li, New insights into external layers of cyanobacteria and microalgae based on multiscale analysis of AFM force-distance curves, Sci. Total Environ. 774 (2021) 145680. [CrossRef]
- S. Salucci, M. Battistelli, S. Burattini, F. Sbrana, E. Falcieri, Holotomographic microscopy: A new approach to detect apoptotic cell features, Microsc. Res. Tech. 83 (2020) 1464–1470. [CrossRef]
- S.K. Paidi, V. Shah, P. Raj, K. Glunde, R. Pandey, I. Barman, Coarse Raman and optical diffraction tomographic imaging enable label-free phenotyping of isogenic breast cancer cells of varying metastatic potential, Biosens. Bioelectron. 175 (2021) 112863. [CrossRef]
- N. Nissim, M. Dudaie, I. Barnea, N.T. Shaked, Real-Time Stain-Free Classification of Cancer Cells and Blood Cells Using Interferometric Phase Microscopy and Machine Learning, Cytom. Part A. 99 (2021) 511–523. [CrossRef]
- A.L. Palacios-Acedo, S. Mezouar, D. Mège, L. Crescence, C. Dubois, L. Panicot-Dubois, P2RY12-Inhibitors Reduce Cancer-Associated Thrombosis and Tumor Growth in Pancreatic Cancers, Front. Oncol. 11 (2021) 1–15. [CrossRef]
- W. Szlasa, A. Kiełbik, A. Szewczyk, N. Rembiałkowska, V. Novickij, M. Tarek, J. Saczko, J. Kulbacka, Oxidative Effects during Irreversible Electroporation of Melanoma Cells-In Vitro Study, Molecules. 26 (2021). [CrossRef]
- K.P. Lee, S. Baek, M.S. Yoon, J.S. Park, B.S. Hong, S.J. Lee, S.J. Oh, S.H. Kwon, R. Lee, D.H. Lee, K.S. Park, B.S. Moon, Potential anticancer effect of aspirin and 2’-hydroxy-2,3,5’-trimethoxychalcone-linked polymeric micelles against cervical cancer through apoptosis, Oncol. Lett. 23 (2022) 1–8. [CrossRef]
- X. Zhu, H. Shen, X. Yin, L. Long, C. Xie, Y. Liu, L. Hui, X. Lin, Y. Fang, Y. Cao, Y. Xu, M. Li, W. Xu, Y. Li, MiR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin, Oncogene. 35 (2016) 323–332. [CrossRef]
- L. Xin, W. Xiao, L. Che, J. Liu, L. Miccio, V. Bianco, P. Memmolo, P. Ferraro, X. Li, F. Pan, Label-Free Assessment of the Drug Resistance of Epithelial Ovarian Cancer Cells in a Microfluidic Holographic Flow Cytometer Boosted through Machine Learning, ACS Omega. 6 (2021) 31046–31057. [CrossRef]
- M.B.D. Aldonza, R.D.D. Reyes, Y.S. Kim, J. Ku, A.M. Barsallo, J.Y. Hong, S.K. Lee, H.S. Ryu, Y.K. Park, J.Y. Cho, Y. Kim, Chemotherapy confers a conserved secondary tolerance to EGFR inhibition via AXL-mediated signaling bypass, Sci. Rep. 11 (2021). [CrossRef]
- H. Yamashita, A. Taoka, T. Uchihashi, T. Asano, T. Ando, Y. Fukumori, Single-molecule imaging on living bacterial cell surface by high-speed AFM, J. Mol. Biol. 422 (2012) 300–309. [CrossRef]
- T. Uchihashi, H. Watanabe, S. Fukuda, M. Shibata, T. Ando, Functional extension of high-speed AFM for wider biological applications, Ultramicroscopy. 160 (2016) 182–196. [CrossRef]
- Y. Jiang, Z. Yuan, J. Huang, Substituted hydroxyapatite: a recent development, Mater. Technol. 35 (2020) 785–796. [CrossRef]
- E.H. Backes, L.D.N. Pires, C.A.G. Beatrice, L.C. Costa, F.R. Passador, L.A. Pessan, Fabrication of Biocompatible Composites of Poly(lactic acid)/Hydroxyapatite Envisioning Medical Applications, Polym. Eng. Sci. 60 (2020) 636–644. [CrossRef]
- E. Ungureanu, A. Vladescu, A.C. Parau, V. Mitran, A. Cimpean, M. Tarcolea, D.M. Vranceanu, C.M. Cotrut, In Vitro Evaluation of Ag- and Sr-Doped Hydroxyapatite Coatings for Medical Applications, Materials (Basel). 16 (2023). [CrossRef]
- R. Hernandez, A. Jimenez-Chávez, A. De Vizcaya, J.A. Lozano-Alvarez, K. Esquivel, I.E. Medina-Ramírez, Synthesis of TiO2-Cu2+/CuI Nanocomposites and Evaluation of Antifungal and Cytotoxic Activity, Nanomaterials. 13 (2023). [CrossRef]
- J. Oh, J.S. Ryu, M. Lee, J. Jung, S. Han, H.J. Chung, Y. Park, Three-dimensional label-free observation of individual bacteria upon antibiotic treatment using optical diffraction tomography, Biomed. Opt. Express. 11 (2020) 1257. [CrossRef]
- L.S. Zhao, H.N. Su, K. Li, B. Bin Xie, L.N. Liu, X.Y. Zhang, X.L. Chen, F. Huang, B.C. Zhou, Y.Z. Zhang, Supramolecular architecture of photosynthetic membrane in red algae in response to nitrogen starvation, Biochim. Biophys. Acta - Bioenerg. 1857 (2016) 1751–1758. [CrossRef]
- H. Atomic, F. Microscopy, K. Kobayashi, N. Kodera, T. Kasai, Y.O. Tahara, T. Toyonaga, Movements of Mycoplasma mobile Gliding Machinery Detected, MBio. 12 (2021) 1–14.
- Y. Kikuchi, N. Obana, M. Toyofuku, N. Kodera, T. Soma, T. Ando, Y. Fukumori, N. Nomura, A. Taoka, Diversity of physical properties of bacterial extracellular membrane vesicles revealed through atomic force microscopy phase imaging, Nanoscale. 12 (2020) 7950–7959. [CrossRef]
- Demir-Yilmaz, P. Guiraud, C. Formosa-Dague, The contribution of Atomic Force Microscopy (AFM) in microalgae studies: A review, Algal Res. 60 (2021) 102506. [CrossRef]
- J. Mignolet, Y.F. Dufre, AFM force-clamp spectroscopy captures the nanomechanics of the Tad pilus retraction, Nanoscale Horizons. 6 (2021) 489–496. [CrossRef]
- Chantraine, M. Mathelié-Guinlet, G. Pietrocola, P. Speziale, Y.F. Dufrêne, AFM Identifies a Protein Complex Involved in Pathogen Adhesion Which Ruptures at Three Nanonewtons, Nano Lett. 21 (2021) 7595–7601. [CrossRef]
- F. Viela, M.J. Alfeo, G. Pietrocola, P. Speziale, M. Mathelie, Single-Molecule Analysis Demonstrates Stress-Enhanced Binding between Staphylococcus aureus Surface Protein IsdB and Host Cell Integrins ́, Nano Lett. 20 (2020) 8919–8925. [CrossRef]
- G. Kim, D. Ahn, M. Kang, Y. Jo, D. Ryu, H. Kim, J. Song, J.S. Ryu, G. Choi, H.J. Chung, K. Kim, D.R. Chung, I.Y. Yoo, H.J. Huh, H. Min, N.Y. Lee, Y. Park, Rapid and label-free identification of individual bacterial pathogens exploiting three-dimensional quantitative phase imaging and deep learning, BioRxiv. (2019) 596486. [CrossRef]
- M. Kim, Y. Cheon, D. Shin, J. Choi, J.E. Nielsen, M.S. Jeong, H.Y. Nam, S.H. Kim, R. Lund, H. Jenssen, A.E. Barron, S. Lee, J. Seo, Real-Time Monitoring of Multitarget Antimicrobial Mechanisms of Peptoids Using Label-Free Imaging with Optical Diffraction Tomography, Adv. Sci. 10 (2023). [CrossRef]
- Y. Jo, S. Park, J. Jung, J. Yoon, H. Joo, M. Kim, S.-J. Kang, M.C. Choi, S.Y. Lee, Y. Park, Holographic deep learning for rapid optical screening of anthrax spores, Sci. Adv. 3 (2017) e1700606.
- Larrazabal, L.M.R. Silva, C. Hermosilla, A. Taubert, Ezetimibe blocks Toxoplasma gondii-, Neospora caninum- And Besnoitia besnoiti-tachyzoite infectivity and replication in primary bovine endothelial host cells, Parasitology. 148 (2021) 1107–1115. [CrossRef]
- S. Lopez-Osorio, Z.D. Velasquez, I. Conejeros, A. Taubert, C. Hermosilla, Morphometric analysis of aerobic Eimeria bovis sporogony using live cell 3D holotomographic microscopy imaging, Parasitol. Res. (2021). [CrossRef]
- S. López-Osorio, L.M.R. Silva, J.J. Chaparro-Gutierréz, Z.D. Velásquez, A. Taubert, C. Hermosilla, Optimized excystation protocol for ruminant Eimeria bovis- and Eimeria arloingi-sporulated oocysts and first 3D holotomographic microscopy analysis of differing sporozoite egress, Parasitol. Int. 76 (2020) 102068. [CrossRef]
- E.R. Firdaus, J.H. Park, S.K. Lee, Y.K. Park, G.H. Cha, E.T. Han, 3D morphological and biophysical changes in a single tachyzoite and its infected cells using three-dimensional quantitative phase imaging, J. Biophotonics. 13 (2020) e202000055. [CrossRef]
- Z.D. Velásquez, S. Lopez-Osorio, L. Pervizaj-Oruqaj, S. Herold, C. Hermosilla, A. Taubert, Besnoitia besnoiti–driven endothelial host cell cycle alteration, Parasitol. Res. 119 (2020) 2563–2577. [CrossRef]
- Z. Koutsogiannis, J.G.M. Mina, R. Suman, P.W. Denny, Assessment of Toxoplasma gondii lytic cycle and the impact of a gene deletion using 3D label-free optical diffraction holotomography, Front. Cell. Infect. Microbiol. 13 (2023) 1–6. [CrossRef]
- Zhou, L.M.R. Silva, I. Conejeros, Z.D. Velásquez, M. Hirz, U. Gärtner, P. Jacquiet, A. Taubert, C. Hermosilla, Besnoitia besnoiti bradyzoite stages induce suicidal- and rapid vital-NETosis, Parasitology. 147 (2020) 401–409. [CrossRef]
- L.M.R. Silva, D. Lütjohann, P. Hamid, Z.D. Velasquez, K. Kerner, C. Larrazabal, K. Failing, C. Hermosilla, A. Taubert, Besnoitia besnoiti infection alters both endogenous cholesterol de novo synthesis and exogenous LDL uptake in host endothelial cells, Sci. Rep. 9 (2019) 6650. [CrossRef]
- N. Hammoudeh, C. Soukkarieh, D.J. Murphy, A. Hanano, Involvement of hepatic lipid droplets and their associated proteins in the detoxification of aflatoxin B1 in aflatoxin-resistance BALB/C mouse, Toxicol. Reports. 7 (2020) 795–804. [CrossRef]
- S.Y. Lee, K. Kim, A. Mubarok, A. Panduwirawan, K.R. Lee, S. Lee, H.J. Park, Y.K. Park, High-resolution 3-D refractive index tomography and 2-D synthetic aperture imaging of live phytoplankton, J. Opt. Soc. Korea. 18 (2014) 691–697. [CrossRef]
- K. Umemura, Y. Matsukawa, Y. Ide, S. Mayama, Label-free imaging and analysis of subcellular parts of a living diatom cylindrotheca sp. using optical diffraction tomography, MethodsX. 7 (2020) 100889. [CrossRef]
- J.H. Jung, S.J. Hong, H.B. Kim, G. Kim, M. Lee, S. Shin, S.Y. Lee, D.J. Kim, C.G. Lee, Y.K. Park, Label-free non-invasive quantitative measurement of lipid contents in individual microalgal cells using refractive index tomography, Sci. Rep. 8 (2018) 6524. [CrossRef]
- A.V. Singh, P. Laux, A. Luch, C. Sudrik, S. Wiehr, A.M. Wild, G. Santomauro, J. Bill, M. Sitti, Review of emerging concepts in nanotoxicology: opportunities and challenges for safer nanomaterial design, Toxicol. Mech. Methods. 29 (2019) 378–387. [CrossRef]
- Bondarenko, M. Mortimer, A. Kahru, N. Feliu, I. Javed, A. Kakinen, S. Lin, T. Xia, Y. Song, T.P. Davis, I. Lynch, W.J. Parak, D.T. Leong, P.C. Ke, C. Chen, Y. Zhao, Nanotoxicology and nanomedicine: The Yin and Yang of nano-bio interactions for the new decade, Nano Today. 39 (2021) 101184. [CrossRef]
- Egbuna, V.K. Parmar, J. Jeevanandam, S.M. Ezzat, K.C. Patrick-Iwuanyanwu, C.O. Adetunji, J. Khan, E.N. Onyeike, C.Z. Uche, M. Akram, M.S. Ibrahim, N.M. El Mahdy, C.G. Awuchi, K. Saravanan, H. Tijjani, U.E. Odoh, M. Messaoudi, J.C. Ifemeje, M.C. Olisah, N.J. Ezeofor, C.J. Chikwendu, C.G. Ibeabuchi, Toxicity of Nanoparticles in Biomedical Application: Nanotoxicology, J. Toxicol. 2021 (2021). [CrossRef]
- G. Maiorano, S. Sabella, B. Sorce, V. Brunetti, M.A. Malvindi, R. Cingolani, P.P. Pompa, Effects of cell culture media on the dynamic formation of protein-nanoparticle complexes and influence on the cellular response, ACS Nano. 4 (2010) 7481–7491. [CrossRef]
- A.M. Alkilany, N.N. Mahmoud, F. Hashemi, M.J. Hajipour, F. Farvadi, M. Mahmoudi, Misinterpretation in Nanotoxicology: A Personal Perspective, Chem. Res. Toxicol. 29 (2016) 943–948. [CrossRef]
- A.L. Taka, C.M. Tata, M.J. Klink, X.Y. Mbianda, F.M. Mtunzi, E.B. Naidoo, A review on conventional and advanced methods for nanotoxicology evaluation of engineered nanomaterials, Molecules. 26 (2021). [CrossRef]
- R.P. Friedrich, E. Schreiber, R. Tietze, H. Yang, C. Pilarsky, C. Alexiou, Intracellular quantification and localization of label-free iron oxide nanoparticles by holotomographic microscopy, Nanotechnol. Sci. Appl. 13 (2020) 119–130. [CrossRef]
- L.S. Franqui, M.A. De Farias, R. V Portugal, C.A.R. Costa, R.R. Domingues, A.G. Souza Filho, V.R. Coluci, A.F.P. Leme, D.S.T. Martinez, Interaction of graphene oxide with cell culture medium: Evaluating the fetal bovine serum protein corona formation towards in vitro nanotoxicity assessment and nanobiointeractions, Mater. Sci. Eng. C. 100 (2019) 363–377. [CrossRef]
- P. Zhou, M. Guo, X. Cui, Effect of food on orally-ingested titanium dioxide and zinc oxide nanoparticle behaviors in simulated digestive tract, Chemosphere. 268 (2021) 128843. [CrossRef]
- S. Batasheva, G. Fakhrullina, F. Akhatova, R. Fakhrullin, Caenorhabditis elegans nematode: A versatile model to evaluate the toxicity of nanomaterials in vivo, in: Nanotechnol. Charact. Tools Environ. Heal. Saf., 2019: pp. 323–345. https://doi.org/10.1007/978-3-662-59600-5_11. [CrossRef]
- X. Jiang, C. Lu, M. Tang, Z. Yang, W. Jia, Y. Ma, P. Jia, D. Pei, H. Wang, Nanotoxicity of Silver Nanoparticles on HEK293T Cells: A Combined Study Using Biomechanical and Biological Techniques, ACS Omega. 3 (2018) 6770–6778. [CrossRef]
- Géloën, K. Isaieva, M. Isaiev, O. Levinson, E. Berger, V. Lysenko, Intracellular detection and localization of nanoparticles by refractive index measurement, Sensors. 21 (2021). [CrossRef]
- G. Liu, J. Gao, H. Ai, X. Chen, Applications and potential toxicity of magnetic iron oxide nanoparticles, Small. 9 (2013) 1533–1545. [CrossRef]
- Khanal, F. Zhang, Y. Song, H. Hau, A. Gautam, S. Yamaguchi, J. Uertz, S. Mills, A. Kondyurin, J.C. Knowles, G. Georgiou, I. Ramzan, W. Cai, K.W. Ng, W. Chrzanowski, Biological impact of nanodiamond particles–label free, high-resolution methods for nanotoxicity assessment, Nanotoxicology. 13 (2019) 1210–1226. [CrossRef]
- Roshanzadeh, S. Park, S.E. Ganjbakhsh, J. Park, D.H. Lee, S. Lee, E.S. Kim, Surface Charge-Dependent Cytotoxicity of Plastic Nanoparticles in Alveolar Cells under Cyclic Stretches, Nano Lett. 20 (2020) 7168–7176. [CrossRef]
- L. Zapor, L. Chojnacka-Puchta, D. Sawicka, K. Miranowicz-Dzierzawska, J. Skowron, Cytotoxic and pro - in fl ammatory e ff ects of molybdenum and tungsten disulphide on human bronchial cells, Nanotechnol. Rev. 11 (2022) 1263–1272.
- Y. Suematsu, Y.A. Tsai, S. Takeoka, C.M. Franz, S. Arai, T. Fujie, Ultra-thin, transparent, porous substrates as 3D culture scaffolds for engineering ASC spheroids for high-magnification imaging, J. Mater. Chem. B. 8 (2020) 6999–7008. [CrossRef]
- S. Kim, S.H. Kang, S.H. Byun, H.J. Kim, I.K. Park, H. Hirschberg, S.J. Hong, Intercellular bioimaging and biodistribution of gold nanoparticle-loaded macrophages for targeted drug delivery, Electron. 9 (2020) 1–12. https://doi.org/10.3390/electronics9071105.
- S.H. Kang, Y.S. Shin, D.H. Lee, I.S. Park, S.K. Kim, D. Ryu, Y. Park, S.H. Byun, J.H. Choi, S.J. Hong, Interactions of Nanoparticles with Macrophages and Feasibility of Drug Delivery for Asthma, Int. J. Mol. Sci. 23 (2022). [CrossRef]
- H. Sardarabadi, D.E. Chafai, F. Gheybi, P. Sasanpour, H. Rafii-Tabar, M. Cifra, Enhancement of the biological autoluminescence by mito-liposomal gold nanoparticle nanocarriers, J. Photochem. Photobiol. B Biol. 204 (2020) 111812. https://doi.org/10.1016/j.jphotobiol.2020.111812. [CrossRef]
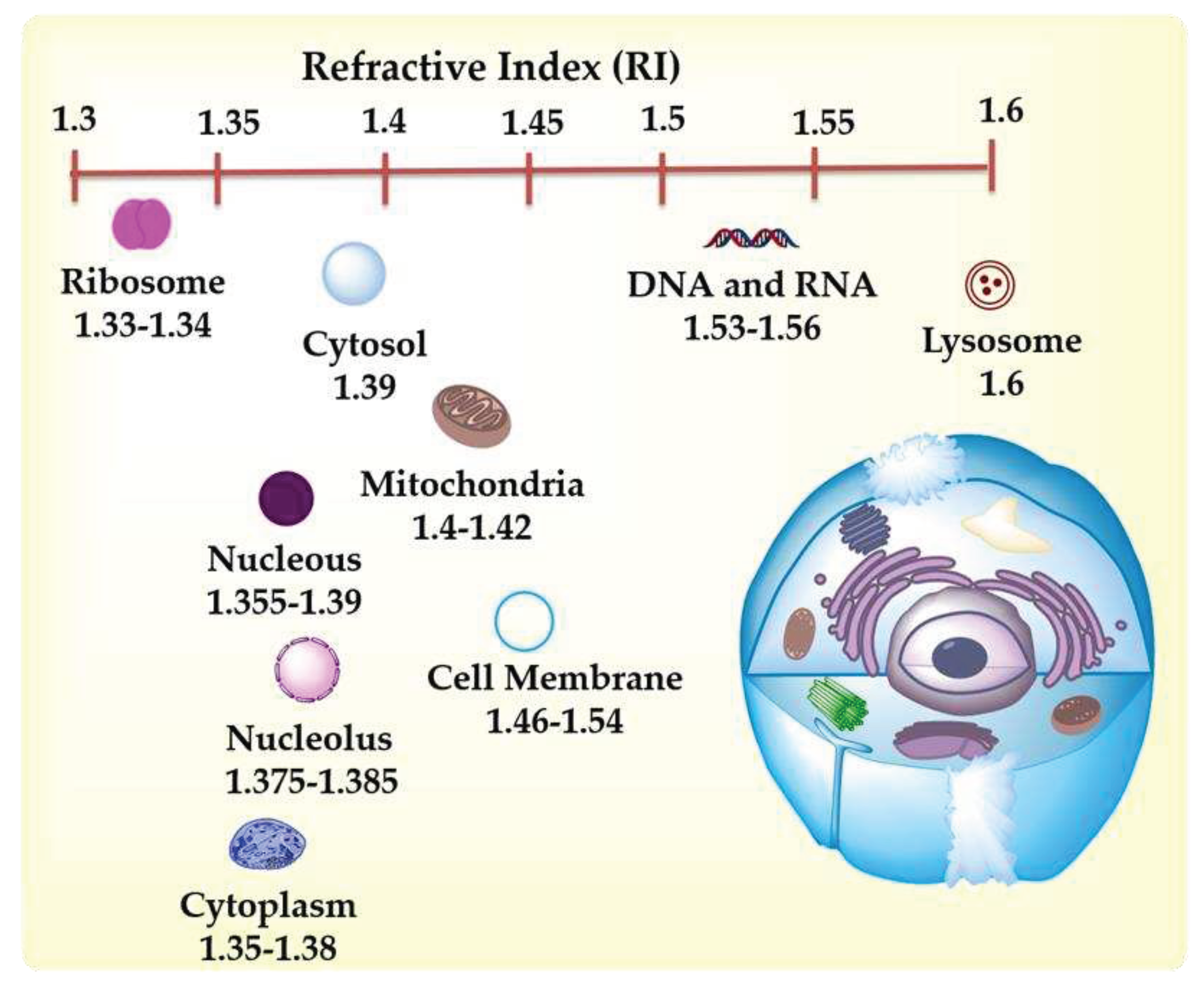
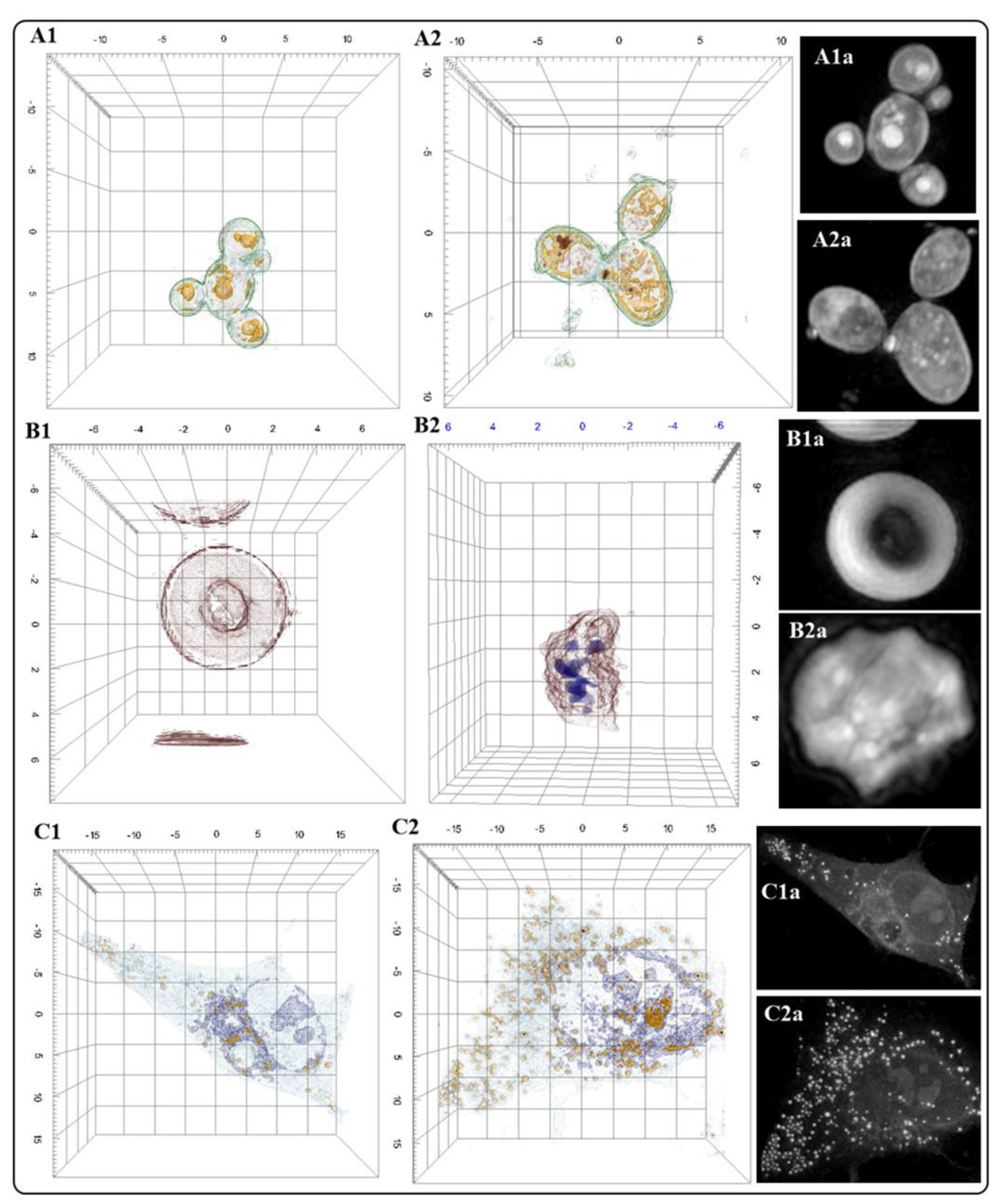
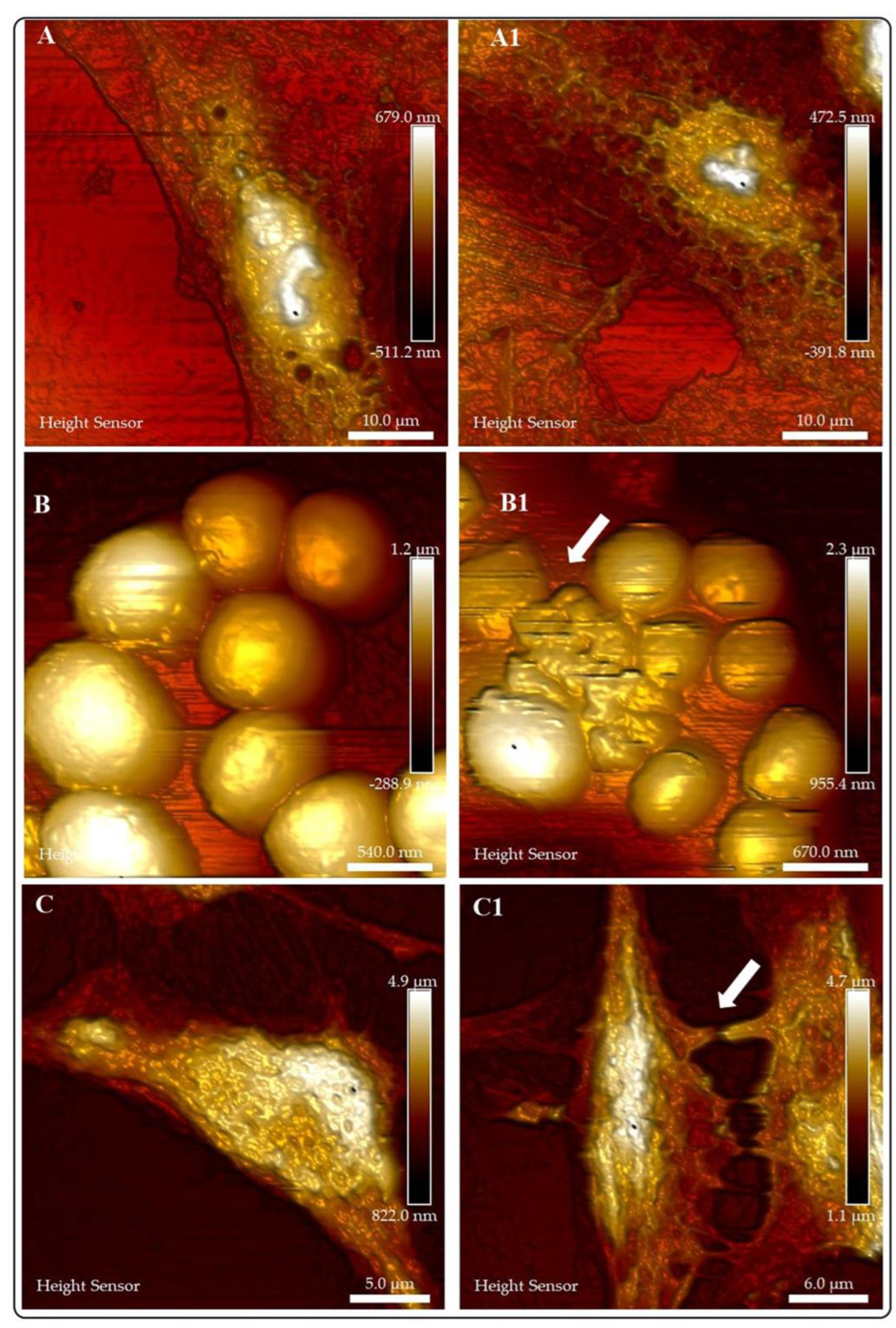
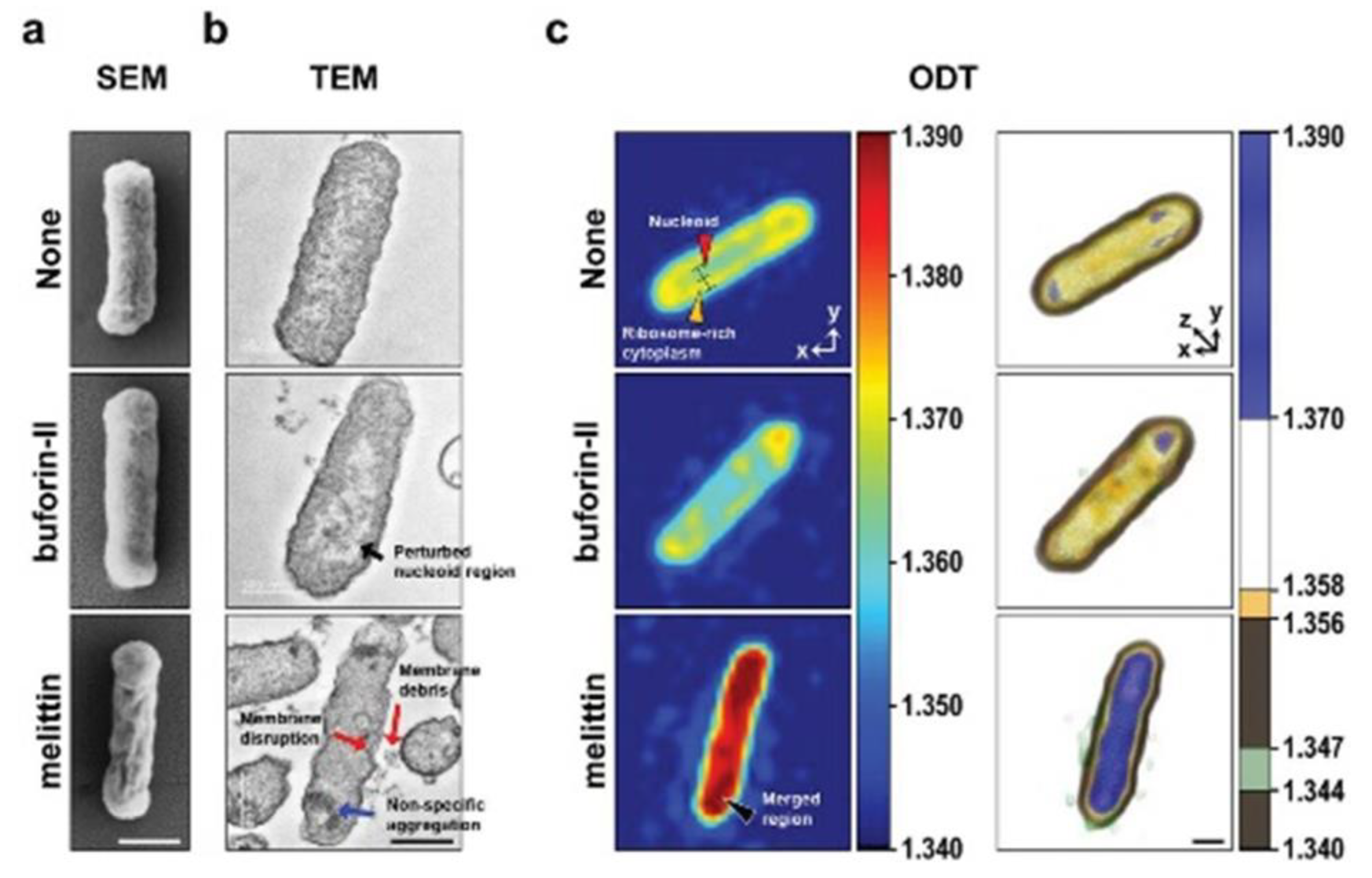
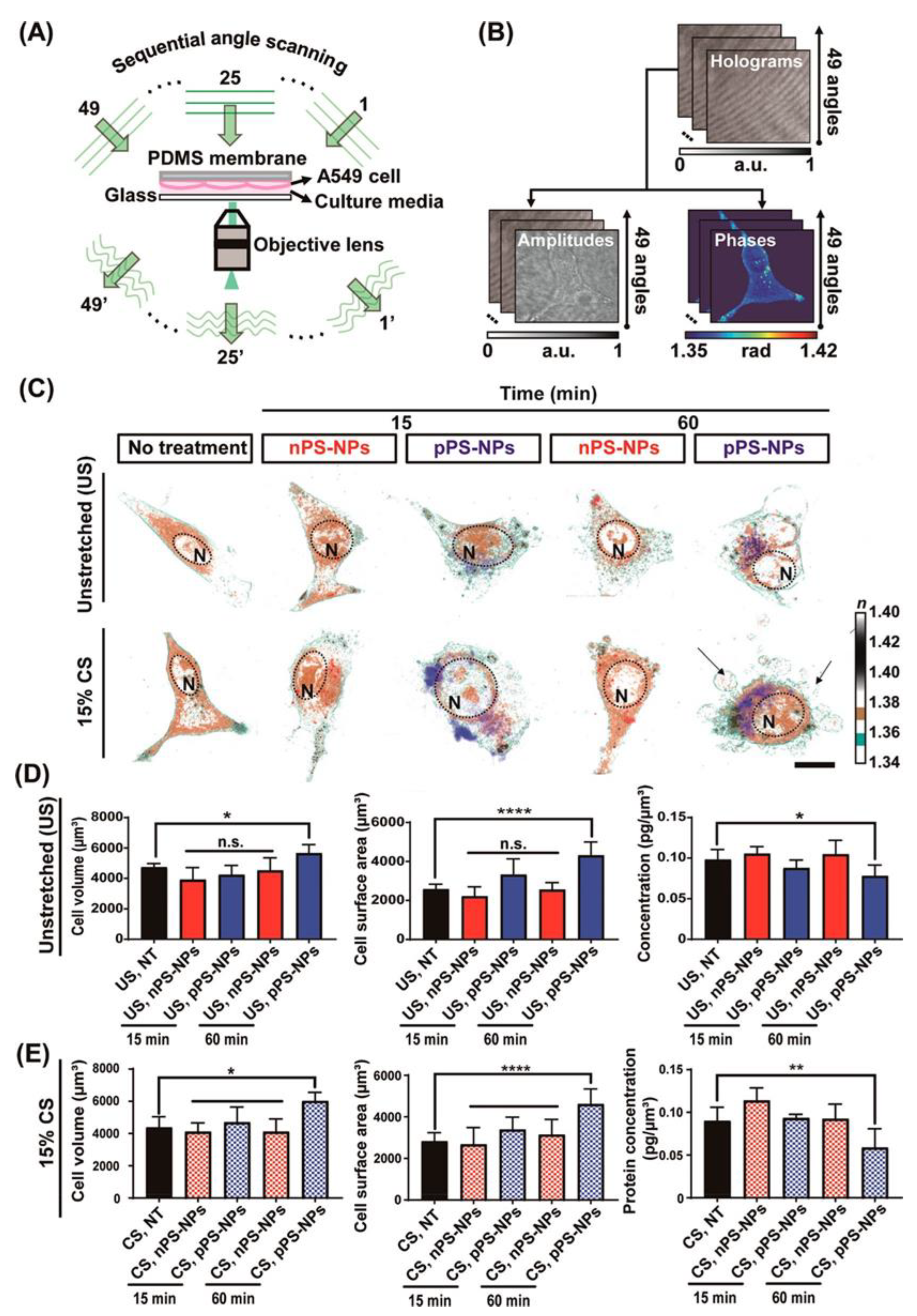
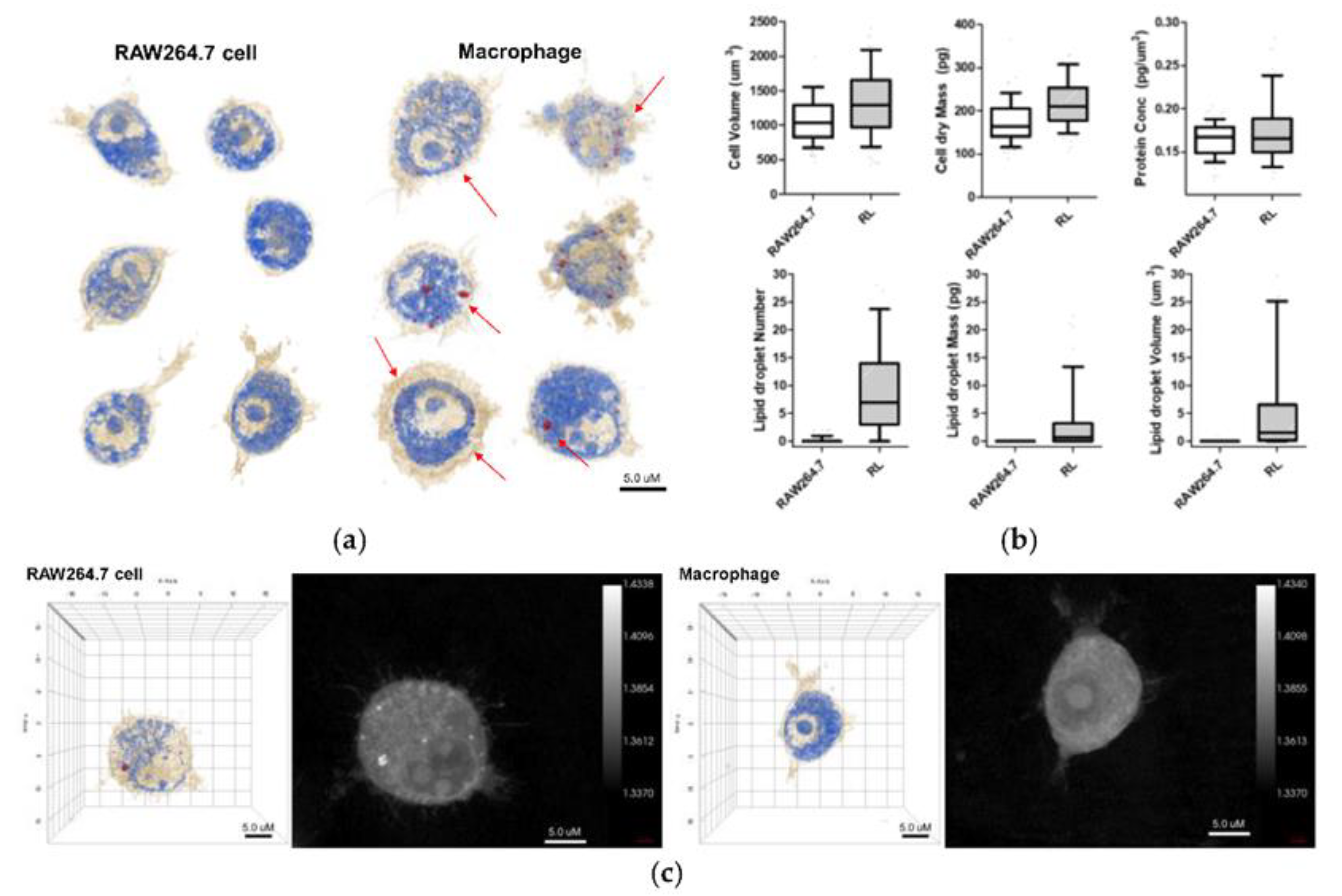
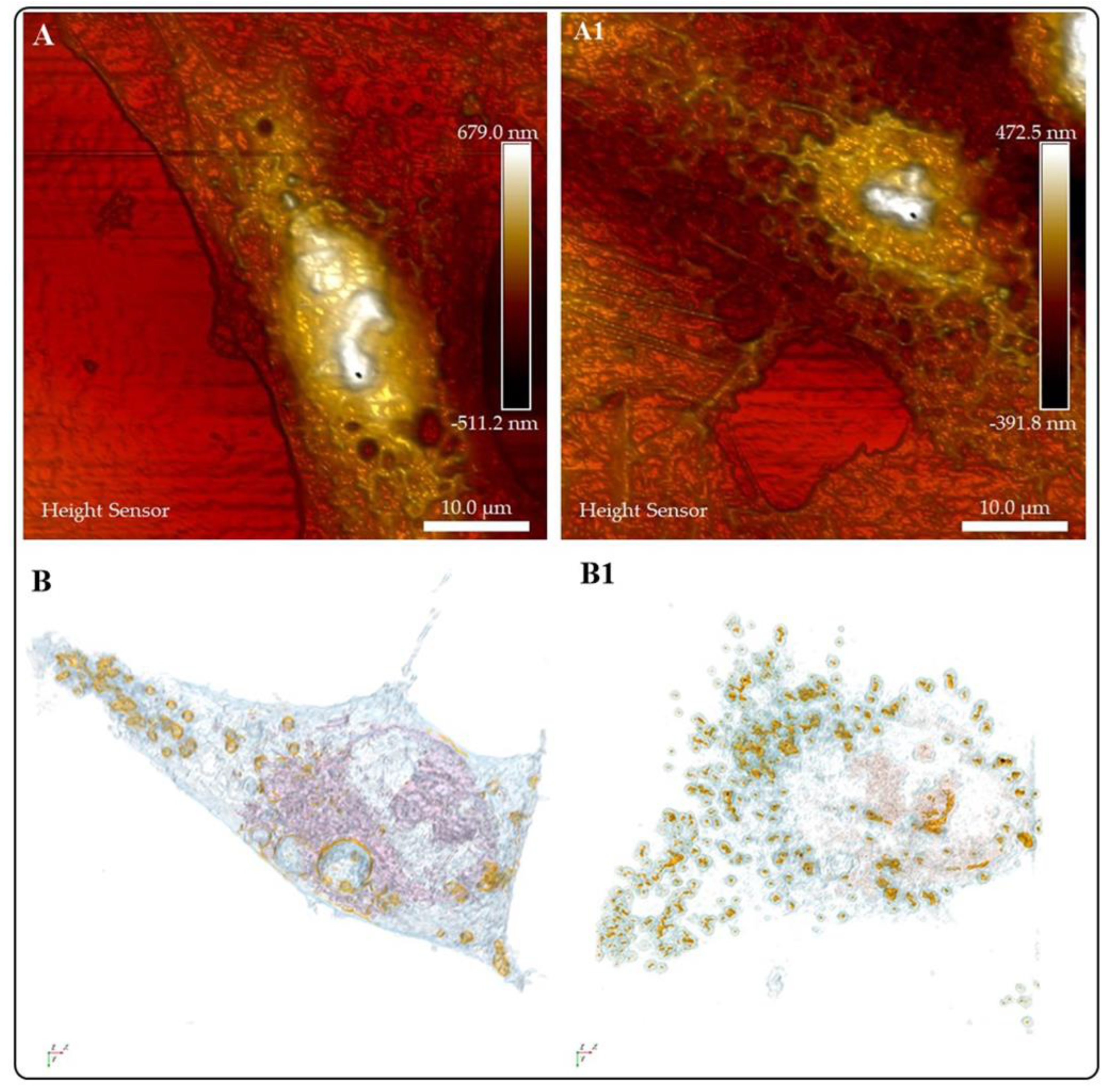
| Organelle | RI value | Ref |
|---|---|---|
| Cell membrane | 1.46-1.54 | [5] |
| Cytoplasm | 1.36-1.38 | [6,7] |
| Nuclei Nucleolus | 1.375-1.385 | [8] |
| Nucleus | 1.355-1.39 | [6,8] |
| Extracelullalr fluids | 1.35 | [6] |
| Mitochondria | 1.40-1.42 | [6,8] |
| Cytosol | 1.360-1.390 | [8] |
| Lysosome | 1.6 | [8] |
| DNA, RNA and ribonucleoprotein | 1.530-1.560 | [7] |
| Ribosome | 1.330-1.340 | [7] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





