Submitted:
06 October 2023
Posted:
09 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
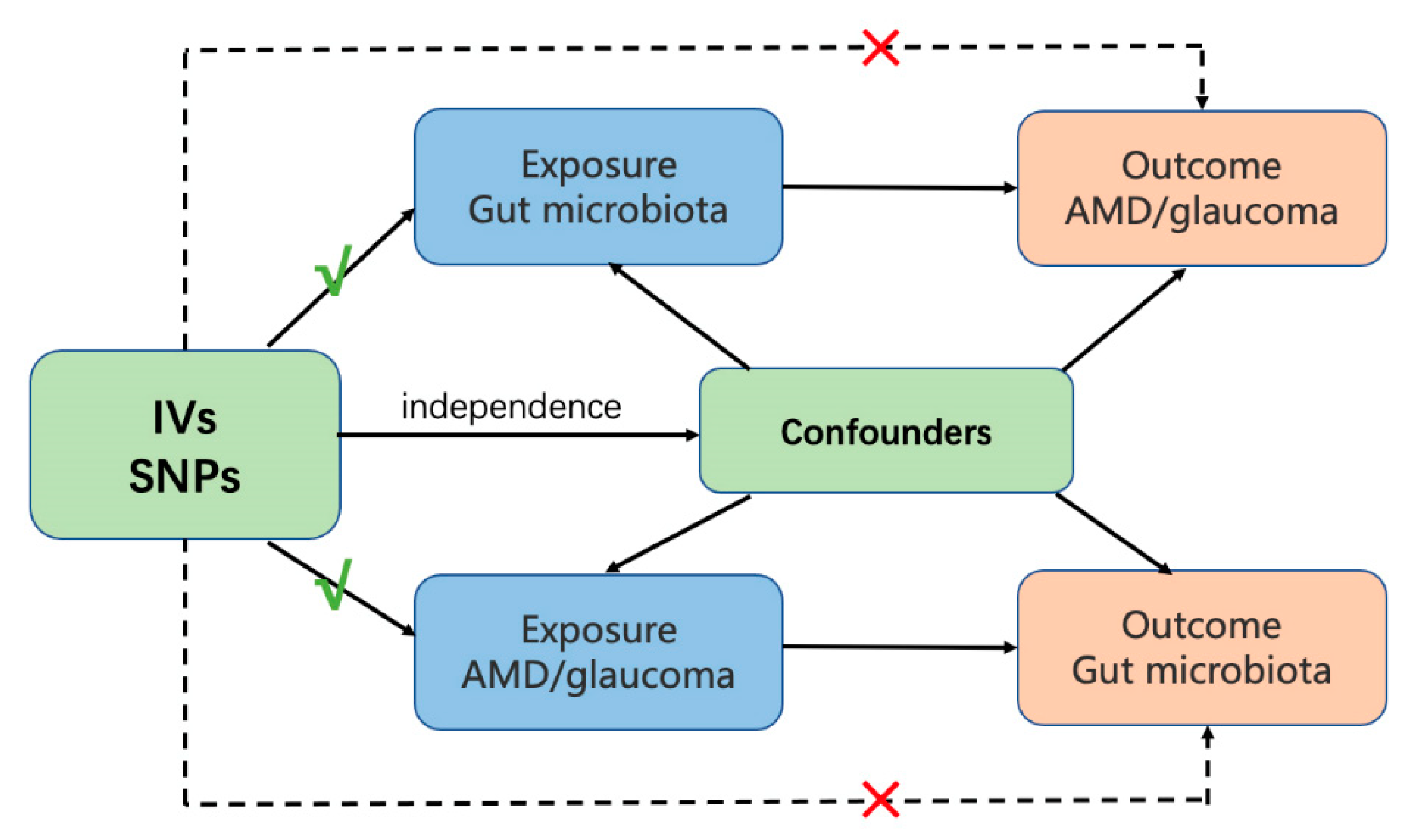
2. Materials and Methods
2.1. Data Sources
2.1.1. Gut Microbiota
2.1.2. AMD
2.1.3. Glaucoma
2.1.4. IVs
2.2. Statistical Analysis
3. Results
| Bacterial taxa (Exposures) | Methods | SNPs | OR | 95%CI | P |
|---|---|---|---|---|---|
| Dorea | MR Egger | 5 | 1.64 | 0.64-4.22 | 0.382 |
| Weighted median | 5 | 1.50 | 1.08-2.08 | 0.016 | |
| IVW | 5 | 1.46 | 1.15-1.85 | 0.002 | |
| Simple mode | 5 | 1.55 | 1.01-2.39 | 0.116 | |
| Weighted mode | 5 | 1.55 | 1.04-2.33 | 0.099 | |
| Eubacterium (oxidoreducens group) | MR Egger | 4 | 1.13 | 0.67-1.90 | 0.687 |
| Weighted median | 4 | 0.89 | 0.72-1.11 | 0.318 | |
| IVW | 4 | 0.84 | 0.70-1.00 | 0.049 | |
| Simple mode | 4 | 0.91 | 0.68-1.22 | 0.572 | |
| Weighted mode | 4 | 0.91 | 0.70-1.18 | 0.547 | |
| Eubacterium (ventriosum group) | MR Egger | 8 | 0.82 | 0.41-1.66 | 0.602 |
| Weighted median | 8 | 1.19 | 0.92-1.54 | 0.175 | |
| IVW | 8 | 1.23 | 1.01-1.50 | 0.038 | |
| Simple mode | 8 | 1.20 | 0.82-1.76 | 0.380 | |
| Weighted mode | 8 | 1.20 | 0.84-1.73 | 0.355 | |
| Lachnospiraceae (NK4A136 group) | MR Egger | 7 | 0.84 | 0.63-1.11 | 0.277 |
| Weighted median | 7 | 0.81 | 0.66-0.99 | 0.041 | |
| IVW | 7 | 0.84 | 0.71-0.98 | 0.031 | |
| Simple mode | 7 | 0.79 | 0.61-1.04 | 0.143 | |
| Weighted mode | 7 | 0.79 | 0.63-1.00 | 0.093 | |
| Parabacteroides | MR Egger | 3 | 0.84 | 0.10-6.75 | 0.896 |
| Weighted median | 3 | 0.71 | 0.48-1.04 | 0.080 | |
| IVW | 3 | 0.70 | 0.51-0.96 | 0.025 | |
| Simple mode | 3 | 0.72 | 0.45-1.13 | 0.290 | |
| Weighted mode | 3 | 0.72 | 0.47-1.11 | 0.280 | |
| Ruminococcaceae (UCG009) | MR Egger | 6 | 0.77 | 0.21-2.80 | 0.709 |
| Weighted median | 6 | 0.76 | 0.62-0.94 | 0.011 | |
| IVW | 6 | 0.83 | 0.70-0.99 | 0.036 | |
| Simple mode | 6 | 0.72 | 0.53-0.98 | 0.093 | |
| Weighted mode | 6 | 0.72 | 0.52-0.99 | 0.101 |
| Bacterial taxa (Exposures) | Methods | SNPs | OR | 95%CI | P |
|---|---|---|---|---|---|
| Eubacterium (nodatum group) | MR Egger | 3 | 3.37 | 0.70-16.2 | 0.371 |
| Weighted median | 3 | 1.16 | 1.01-1.35 | 0.041 | |
| IVW | 3 | 1.13 | 0.95-1.34 | 0.173 | |
| Simple mode | 3 | 1.21 | 1.01-1.45 | 0.176 | |
| Weighted mode | 3 | 1.21 | 1.01-1.45 | 0.179 | |
| Lachnospiraceae (NC2004 group) | MR Egger | 3 | 0.20 | 0.03-1.19 | 0.328 |
| Weighted median | 3 | 1.24 | 1.03-1.51 | 0.026 | |
| IVW | 3 | 1.12 | 0.84-1.50 | 0.427 | |
| Simple mode | 3 | 1.31 | 1.01-1.68 | 0.175 | |
| Weighted mode | 3 | 1.31 | 1.02-1.68 | 0.172 | |
| Roseburia | MR Egger | 6 | 0.99 | 0.41-2.41 | 0.984 |
| Weighted median | 6 | 1.28 | 1.03-1.59 | 0.028 | |
| IVW | 6 | 1.18 | 0.96-1.45 | 0.112 | |
| Simple mode | 6 | 1.41 | 0.98-2.03 | 0.124 | |
| Weighted mode | 6 | 1.40 | 0.95-2.05 | 0.146 | |
| Ruminococcaceae (UCG004) | MR Egger | 3 | 1.86 | 0.93-3.72 | 0.328 |
| Weighted median | 3 | 1.17 | 0.94-1.45 | 0.161 | |
| IVW | 3 | 1.21 | 1.02-1.43 | 0.029 | |
| Simple mode | 3 | 1.14 | 0.88-1.47 | 0.482 |
| Bacterial taxa (Exposures) | Methods | SNPs | OR | 95%CI | P |
|---|---|---|---|---|---|
| Dorea | MR Egger | 8 | 0.96 | 0.89-1.03 | 0.274 |
| Weighted median | 8 | 0.96 | 0.92-1.01 | 0.152 | |
| IVW | 8 | 0.96 | 0.92-1.01 | 0.098 | |
| Simple mode | 8 | 0.96 | 0.88-1.04 | 0.319 | |
| Weighted mode | 8 | 0.96 | 0.92-1.01 | 0.191 | |
| Eubacterium (oxidoreducens group) | MR Egger | 8 | 1.10 | 0.96-1.26 | 0.209 |
| Weighted median | 8 | 1.02 | 0.93-1.11 | 0.713 | |
| IVW | 8 | 1.02 | 0.95-1.10 | 0.579 | |
| Simple mode | 8 | 1.05 | 0.91-1.23 | 0.518 | |
| Weighted mode | 8 | 1.03 | 0.95-1.13 | 0.471 | |
| Eubacterium (ventriosum group) | MR Egger | 8 | 1.03 | 0.95-1.12 | 0.467 |
| Weighted median | 8 | 1.03 | 0.97-1.09 | 0.330 | |
| IVW | 8 | 1.03 | 0.99-1.08 | 0.163 | |
| Simple mode | 8 | 0.99 | 0.90-1.08 | 0.807 | |
| Weighted mode | 8 | 1.02 | 0.97-1.08 | 0.488 | |
| Lachnospiraceae (NK4A136 group) | MR Egger | 8 | 0.93 | 0.87-1.01 | 0.124 |
| Weighted median | 8 | 0.96 | 0.91-1.01 | 0.114 | |
| IVW | 8 | 0.96 | 0.92-1.01 | 0.086 | |
| Simple mode | 8 | 1.00 | 0.92-1.09 | 0.970 | |
| Weighted mode | 8 | 0.95 | 0.91-1.00 | 0.103 | |
| Parabacteroides | MR Egger | 8 | 0.94 | 0.85-1.04 | 0.302 |
| Weighted median | 8 | 0.98 | 0.94-1.03 | 0.483 | |
| IVW | 8 | 1.00 | 0.94-1.06 | 0.990 | |
| Simple mode | 8 | 0.96 | 0.88-1.04 | 0.307 | |
| Weighted mode | 8 | 0.98 | 0.93-1.03 | 0.419 | |
| Ruminococcaceae (UCG009) | MR Egger | 8 | 1.00 | 0.97-1.16 | 0.964 |
| Weighted median | 8 | 1.03 | 0.95-1.12 | 0.486 | |
| IVW | 8 | 1.04 | 0.96-1.12 | 0.308 | |
| Simple mode | 8 | 0.95 | 0.83-1.08 | 0.430 |
| Bacterial taxa (Exposures) | Methods | SNPs | OR | 95%CI | P |
|---|---|---|---|---|---|
| Eubacterium (nodatum group) | MR Egger | 81 | 1.06 | 0.87-1.28 | 0.578 |
| Weighted median | 81 | 1.15 | 1.04-1.28 | 0.005 | |
| IVW | 81 | 1.07 | 1.00-1.14 | 0.052 | |
| Simple mode | 81 | 1.27 | 0.97-1.67 | 0.086 | |
| Weighted mode | 81 | 1.24 | 0.99-1.55 | 0.071 | |
| Lachnospiraceae (NC2004group) | MR Egger | 75 | 0.89 | 0.77-1.03 | 0.113 |
| Weighted median | 75 | 0.95 | 0.88-1.03 | 0.217 | |
| IVW | 75 | 1.01 | 0.95-1.06 | 0.845 | |
| Simple mode | 75 | 0.93 | 0.78-1.11 | 0.433 | |
| Weighted mode | 75 | 0.92 | 0.81-1.06 | 0.263 | |
| Roseburia | MR Egger | 84 | 0.96 | 0.89-1.04 | 0.309 |
| Weighted median | 84 | 1.00 | 0.96-1.05 | 0.906 | |
| IVW | 84 | 1.00 | 0.97-1.03 | 0.996 | |
| Simple mode | 84 | 0.95 | 0.87-1.03 | 0.236 | |
| Weighted mode | 84 | 0.96 | 0.90-1.03 | 0.293 | |
| Ruminococcaceae (UCG004) | MR Egger | 83 | 0.99 | 0.89-1.11 | 0.906 |
| Weighted median | 83 | 1.05 | 0.99-1.12 | 0.113 | |
| IVW | 83 | 1.00 | 0.96-1.04 | 0.886 | |
| Simple mode | 83 | 1.08 | 0.93-1.25 | 0.322 |
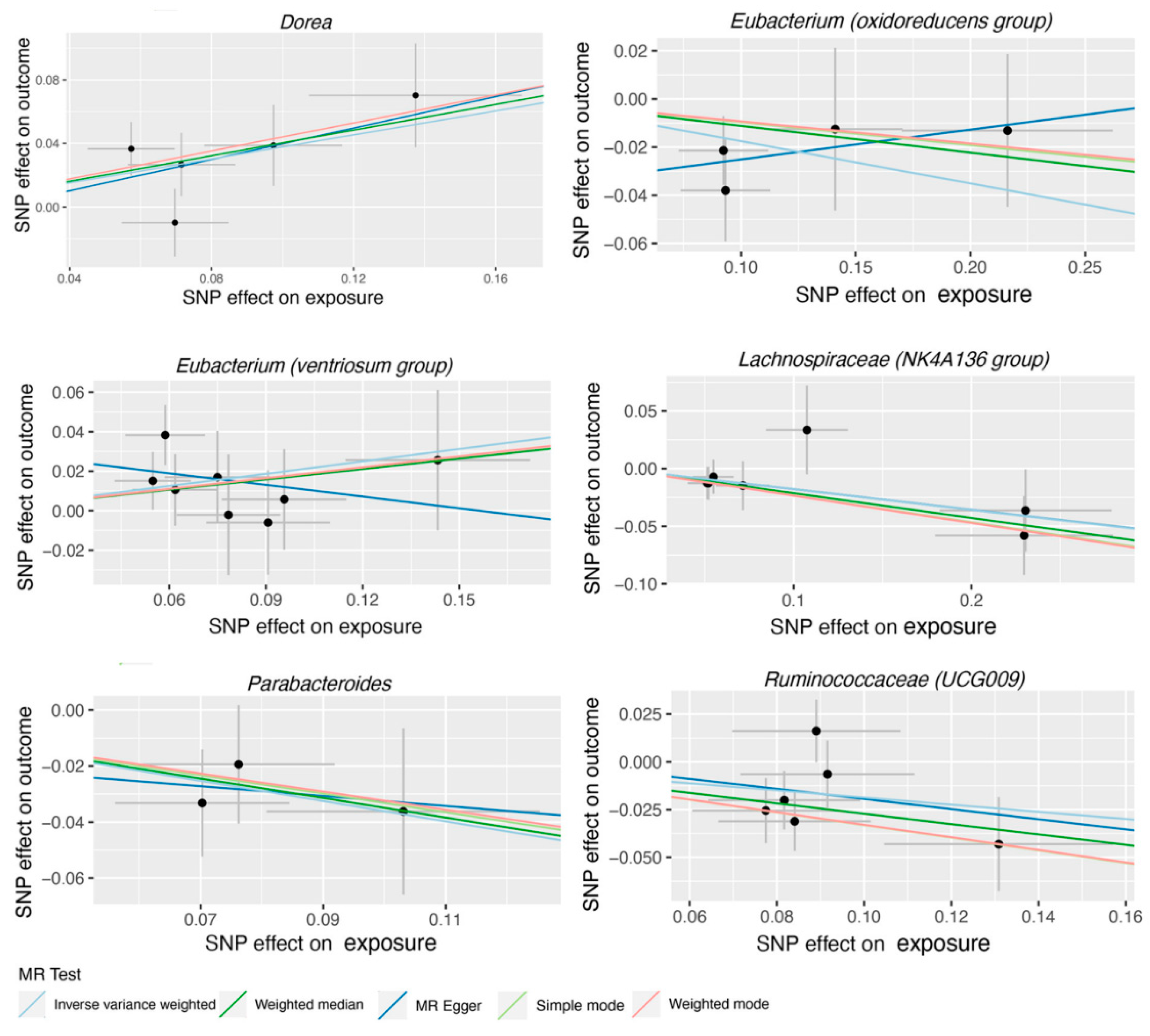
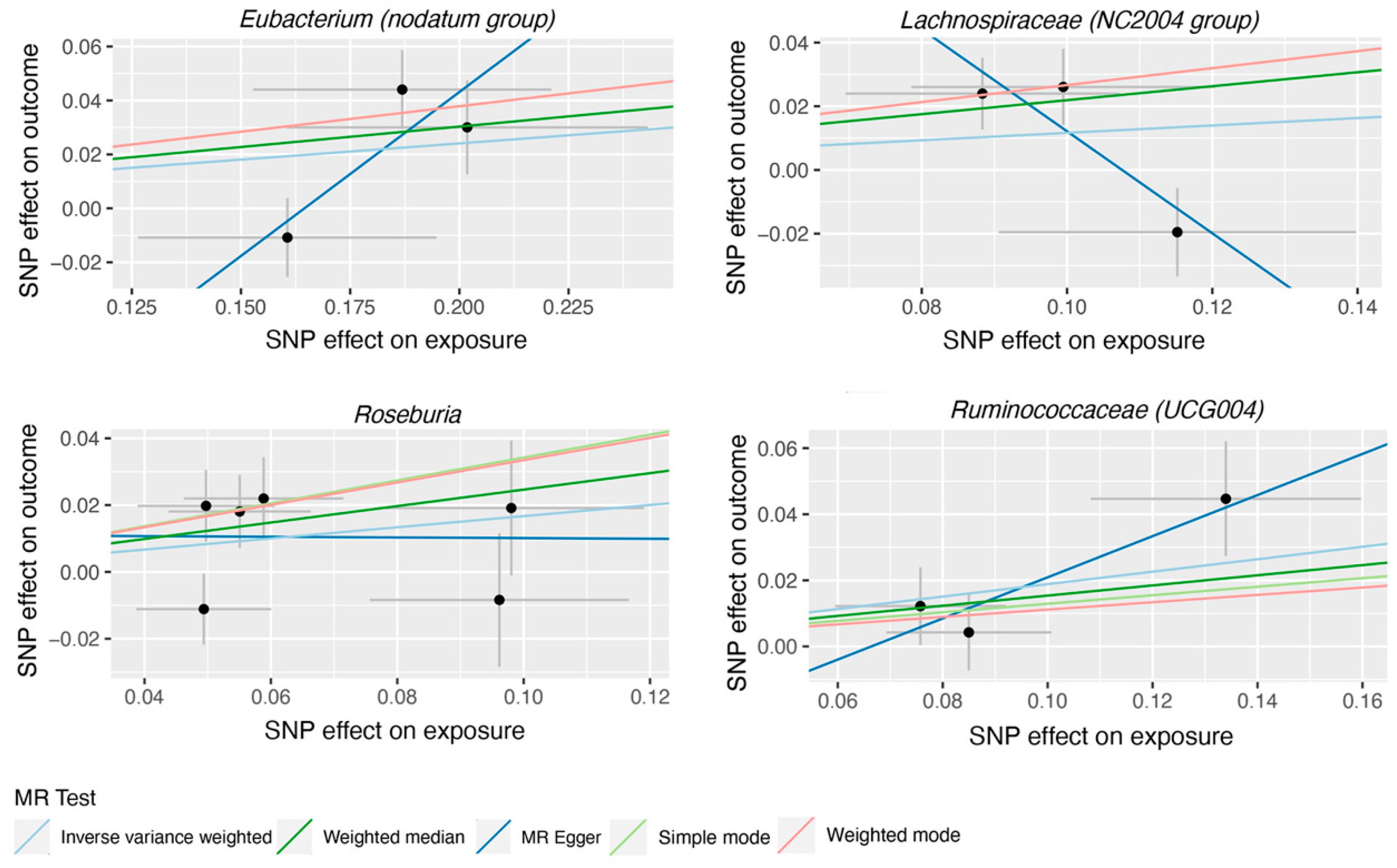
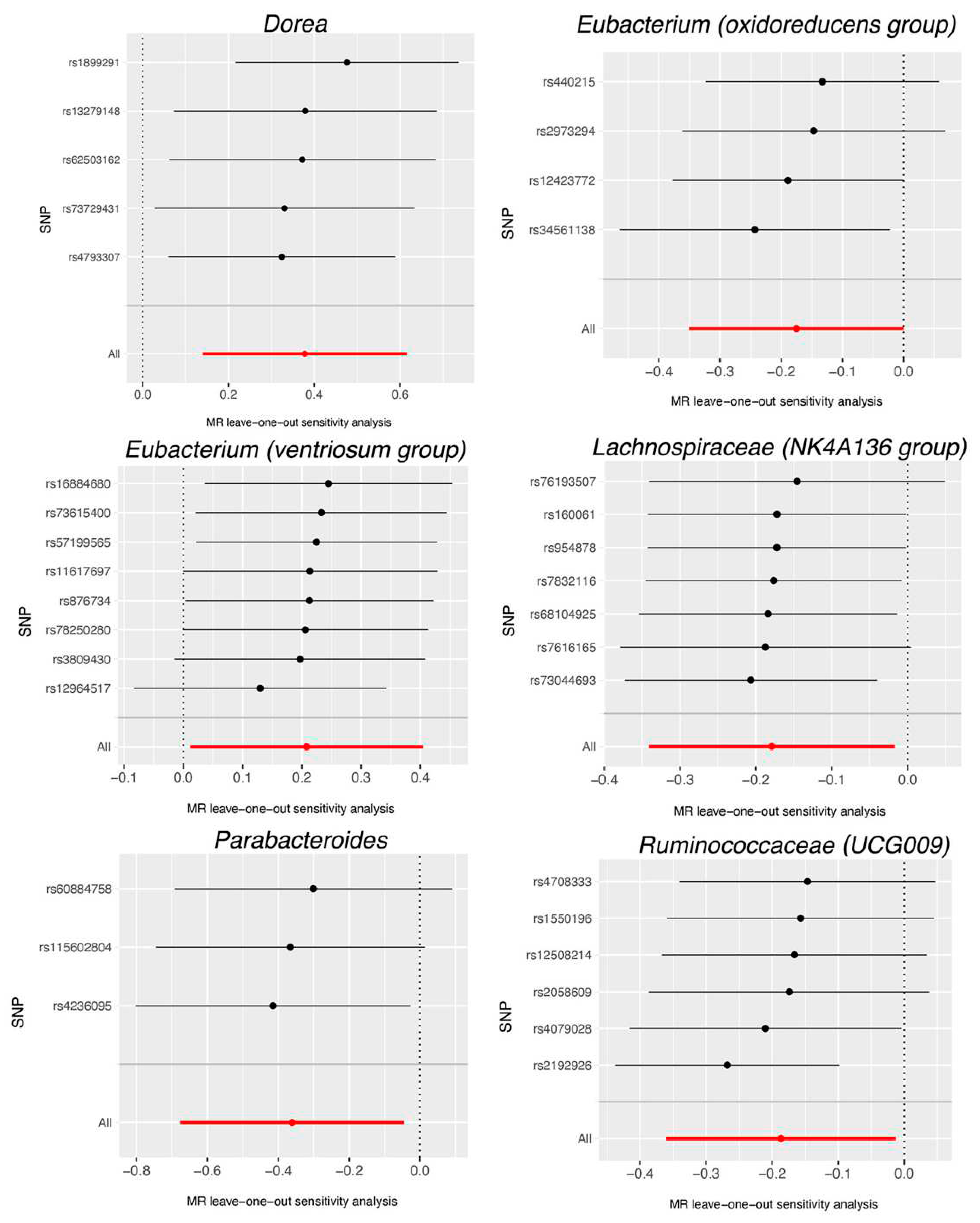
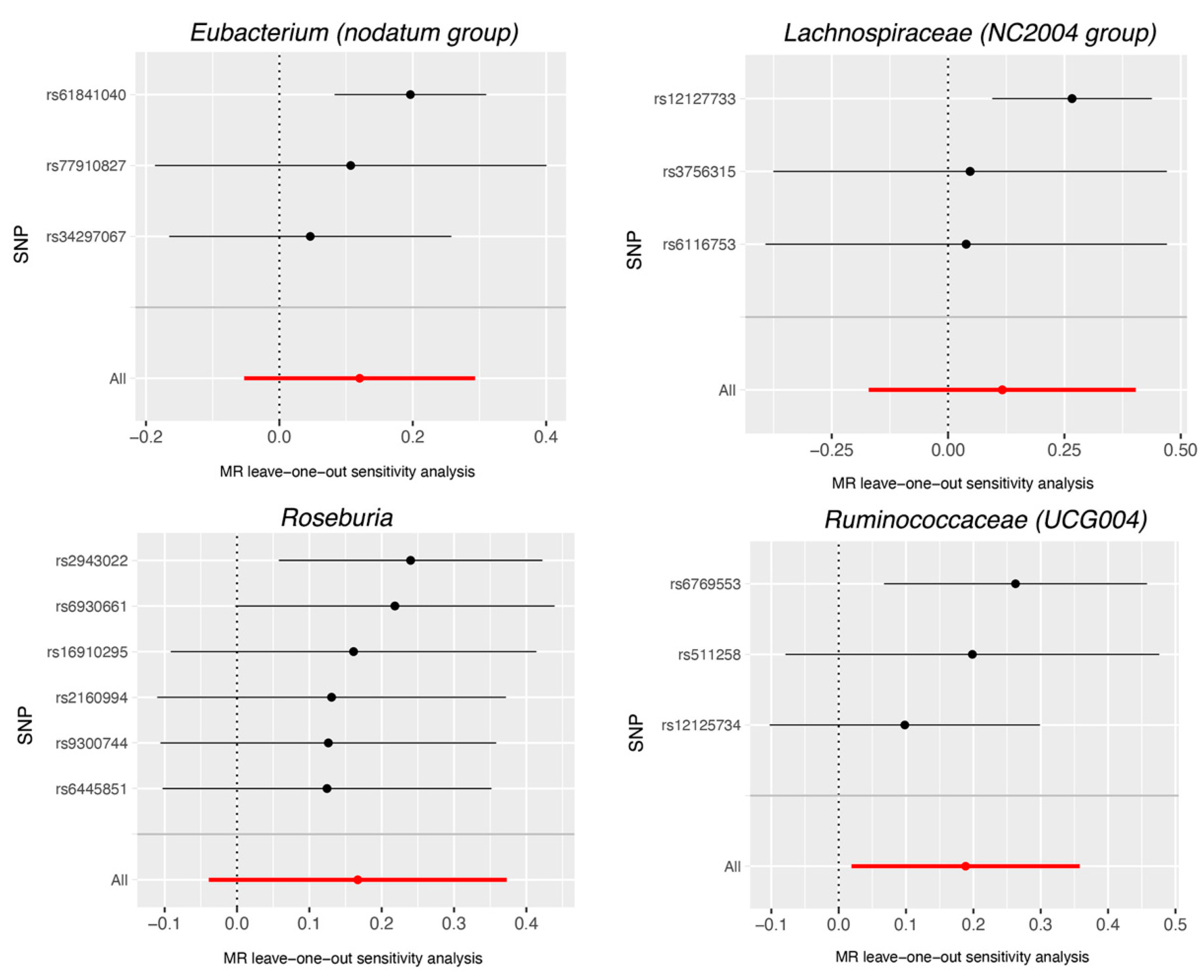
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ambati, J.; Atkinson, J.P.; Gelfand, B.D. Immunology of age-related macular degeneration. Nat Rev Immunol 2013, 13, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.; Liew, G.; Gopinath, B.; et al. Age-related macular degeneration. Lancet 2018, 392, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Tham, Y.C.; Li, X.; Wong, T.Y.; et al. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology 2014, 121, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Vujkovic-Cvijin, I.; Sklar, J.; Jiang, L.; et al. Host variables confound gut microbiota studies of human disease. Nature 2020, 587, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Rowan, S.; Taylor, A. The Role of Microbiota in Retinal Disease. Adv Exp Med Biol 2018, 1074, 429–435. [Google Scholar] [CrossRef]
- Zysset-Burri, D.C.; Keller, I.; Berger, L.E.; et al. Associations of the intestinal microbiome with the complement system in neovascular age-related macular degeneration. NPJ Genom Med 2020, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Lima-Fontes, M.; Meira, L.; Barata, P.; et al. Gut microbiota and age-related macular degeneration: A growing partnership. Surv Ophthalmol 2022, 67, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Zhang, S.; Li, Q.; et al. Gut microbiota compositional profile and serum metabolic phenotype in patients with primary open-angle glaucoma. Exp Eye Res 2020, 191, 107921. [Google Scholar] [CrossRef]
- Gong, H.; Zeng, R.; Li, Q.; et al. The profile of gut microbiota and central carbon-related metabolites in primary angle-closure glaucoma patients. Int Ophthalmol 2022, 42, 1927–1938. [Google Scholar] [CrossRef]
- Greenland, S. An introduction to instrumental variables for epidemiologists. Int J Epidemiol 2000, 29, 722–729. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Mendelian randomization: methods for causal inference using genetic variants: CRC Press.2021.
- Liu, K.; Zou, J.; Fan, H.; et al. Causal effects of gut microbiota on diabetic retinopathy: A Mendelian randomization study. Front Immunol 2022, 13, 930318. [Google Scholar] [CrossRef] [PubMed]
- Nusinovici, S.; Li, H.; Thakur, S.; et al. High-Density Lipoprotein 3 Cholesterol and Primary Open-Angle Glaucoma: Metabolomics and Mendelian Randomization Analyses. Ophthalmology 2022, 129, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wu, P.; Zou, J.; et al. Mendelian randomization analysis reveals causal relationships between gut microbiome and optic neuritis. Hum Genet 2022, 28. [Google Scholar] [CrossRef] [PubMed]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet 2021, 53, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Winkler, T.W.; Grassmann, F.; Brandl, C.; et al. Genome-wide association meta-analysis for early age-related macular degeneration highlights novel loci and insights for advanced disease. BMC Med Genomics 2020, 13, 120. [Google Scholar] [CrossRef] [PubMed]
- Craig, J.E.; Han, X.; Qassim, A.; et al. Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat Genet 2020, 52, 160–166. [Google Scholar] [CrossRef]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemio. 2015, 44, 512–525. [Google Scholar] [CrossRef]
- Pocock, S.J.; Simon, R. Sequential treatment assignment with balancing for prognostic factors in the controlled clinical trial. Biometrics 1975, 31, 103–115. [Google Scholar] [CrossRef]
- Burgess, S.; Dudbridge, F.; Thompson, S.G. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med 2016, 35, 1880–1906. [Google Scholar] [CrossRef]
- Scott, N.W.; McPherson, G.C.; Ramsay, C.R.; et al. The method of minimization for allocation to clinical trials. a review. Control Clin Trials 2002, 23, 662–674. [Google Scholar] [CrossRef]
- Hemani, G.; Tilling, K.; Davey Smith, G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet 2017, 13, e1007081. [Google Scholar] [CrossRef]
- Verbanck, M.; Chen, C.Y.; Neale, B.; et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Mele, M.C.; Merendino, N.; et al. The Role of Diet, Micronutrients and the Gut Microbiota in Age-Related Macular Degeneration: New Perspectives from the Gut⁻Retina Axis. Nutrients 2018, 10, 1677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, X.; Lu, Y. Gut microbiota and derived metabolomic profiling in glaucoma with progressive neurodegeneration. Front Cell Infect Microbiol 2022, 12, 968992. [Google Scholar] [CrossRef]
- Chaput, N.; Lepage, P.; Coutzac, C.; et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol 2017, 28, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Kalyana Chakravarthy, S.; Jayasudha, R.; et al. Alterations in the gut bacterial microbiome in fungal Keratitis patients. PLoS One 2018, 13, e0199640. [Google Scholar] [CrossRef]
- Moon, J.; Choi, S.H.; Yoon, C.H.; et al. Gut dysbiosis is prevailing in Sjögren's syndrome and is related to dry eye severity. PLoS One 2020, 15, e0229029. [Google Scholar] [CrossRef]
- Wang, J.; Qie, J.; Zhu, D.; et al. The landscape in the gut microbiome of long-lived families reveals new insights on longevity and aging - relevant neural and immune function. Gut Microbes 2022, 14, 2107288. [Google Scholar] [CrossRef]
- Jayasudha, R.; Chakravarthy, S.K.; Prashanthi, G.S.; et al. Alterations in gut bacterial and fungal microbiomes are associated with bacterial Keratitis, an inflammatory disease of the human eye. J Biosci 2018, 43, 835–856. [Google Scholar] [CrossRef]
- Kalyana Chakravarthy, S.; Jayasudha, R.; Sai Prashanthi, G.; et al. Dysbiosis in the Gut Bacterial Microbiome of Patients with Uveitis, an Inflammatory Disease of the Eye. Indian J Microbiol 2018, 58, 457–469. [Google Scholar] [CrossRef]
- Low, L.; Suleiman, K.; Shamdas, M.; et al. Gut Dysbiosis in Ocular Mucous Membrane Pemphigoid. Front Cell Infect Microbiol 2022, 12, 780354. [Google Scholar] [CrossRef] [PubMed]
- de Paiva, C.S.; Jones, D.B.; Stern, M.E.; et al. Altered Mucosal Microbiome Diversity and Disease Severity in Sjögren Syndrome. Sci Rep 2016, 6, 23561. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fan, H.; Shao, Y.; et al. Gut microbiota dysbiosis associated with different types of demyelinating optic neuritis in patients. Mult Scler Relat Disord 2023, 72, 104619. [Google Scholar] [CrossRef] [PubMed]
- Zinkernagel, M.S.; Zysset-Burri, D.C.; Keller, I.; et al. Association of the Intestinal Microbiome with the Development of Neovascular Age-Related Macular Degeneration. Sci Rep 2017, 7, 40826. [Google Scholar] [CrossRef]
- Lin, P. Importance of the intestinal microbiota in ocular inflammatory diseases: A review. Clin Exp Ophthalmol 2019, 47, 418–422. [Google Scholar] [CrossRef]
- Burgess, S.; Davies, N.M.; Thompson, S.G. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 2016, 40, 597–608. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




