Submitted:
04 October 2023
Posted:
05 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Method description
2.1. Grammatical evolution
- N is the set of the non-terminal symbols. Every symbol in N has a series of production rules, used to produce terminal symbols.
- Tis the set of terminal symbols.
- S denotes the start symbol of the grammar, with .
- P is the set of production rules, to create terminal symbols non - terminal symbols. These rules are in the form or .
- Get the next element V from the current chromosome.
- Select the next production rule according to: Rule = V mod , where is the total number of production rules for the current non – terminal symbol.
- NOTHING. This command means that no action takes place.
- EXPAND. With this command, the corresponding end of the value field is extended by 50% of the width of the field.
- DIVIDE. With this command, the corresponding end of the value field is shrunk by 50% of the width of the field.
2.2. The first phase of the proposed method
- Set as the number of chromosomes for the Grammatical Evolution.
- Set as H the number of weights for the neural network.
- Set the maximum number of allowed generations.
- Set as the selection rate , with .
- Set as the mutation rate, with .
- Set as the number of randomly created neural networks, that will be used in the fitness calculation.
- Initialize randomly the chromosomes. Every chromosome is a set of integer numbers used to produce valid programs through Grammatical Evolution and the associated grammar of Figure 1.
- Set , the best discovered fitness. For this algorithm, we consider the fitness function of any given chromosome g as an interval
- Set iter=0.
-
For do
- (a)
- Createfor the chromosome the corresponding program using the grammar of Figure 1.
- (b)
- Applythe program to in order to produce the bounds .
- (c)
- Set
- (d)
-
Fordo
- Create randomly as a set for the parameters of neural network.
- Calculate the associated training error
- If then
- If then
- (e)
- EndFor
- (f)
- Set as the fitness value for the chromosome .
- EndFor
-
Apply the selection procedure. Firstly, the chromosomes are sorted with correspondence to their fitness values. Since fitness is considered an interval, a fitness comparison function is required. For this reason the operator to compare two fitness values and as follows:In practice this means that the fitness value is considered smaller than if . The first chromosomes with the lowest fitness values are copied to the next generation. The remaining chromosomes are substituted by chromosomes produced by the crossover procedure. During the selection process, for every new offspring, two chromosomes are selected as parents from the population using well - known procedure of tournament selection.
- Apply the crossover procedure. For each pair of parents, two new chromosomes and are created using the one - point crossover, graphically shown in Figure 2.
- Apply the mutation procedure. For each element of every chromosome alter the corresponding element with probability .
- Set iter=iter+1
- If goto step 10.
2.3. The second phase of the proposed method
-
Initialization Step
- (a)
- Set as the number of chromosomes that participate in the genetic algorithm.
- (b)
- Set the maximum number of allowed iterations.
- (c)
- Set H the number of weights for the neural network.
- (d)
- Get the best interval S from the previous step of Subsection 2.2.
- (e)
- Initialize using uniform distribution the chromosomes in S.
- (f)
- Set as the selection rate, with .
- (g)
- Set as the mutation rate, with .
- (h)
- Set iter=0.
-
Fitness calculation Step
- (a)
-
Fordo
- Calculate the fitness of chromosome as
- (b)
- EndFor
-
Genetic operations step
- (a)
- Selection procedure. Initially, the chromosomes are sorted according to their fitness values. The first chromosomes with the lowest fitness values are copied to the next generation. The remaining chromosomes are substituted by chromosomes produced by the crossover procedure. During the selection process, for every new offspring, two chromosomes are selected as parents from the population using well - known procedure of tournament selection.
- (b)
-
Crossover procedure: For each pair of selected parents, two chromosomes and are constructed using the following equations:Where is random number with the property [64].
- (c)
- Mutation procedure: For each element of every chromosome alter the corresponding element with probability .
-
Termination Check Step
- (a)
- Set
- (b)
- If goto step 2. else apply a local search procedure to the best chromosome of the population. In the current work the BFGS variant of Powell [65] was used.
3. Experiments
- UCI dataset repository, https://archive.ics.uci.edu/ml/index.php [66]
- Keel repository, https://sci2s.ugr.es/keel/datasets.php [67].
- The Statlib URL ftp://lib.stat.cmu.edu/datasets/index.html.
3.1. Experimental datasets
- Appendictis a medical purpose dataset, suggested in [68].
- Australian dataset [69], a dataset related to credit card transactions.
- Balance dataset [70], related to psychological states.
- Dermatology dataset [73], a medical dataset related to erythemato-squamous diseases.
- Heart dataset [74], a medical dataset related to heart diseases.
- Hayes roth dataset [75].
- HouseVotes dataset [76], related to votes in the U.S. House of Representatives Congressmen.
- Liverdisorder dataset [79], a medical dataset related to liver disorders.
- Mammographic dataset [80], used to identify breast tumors.
- Parkinsons dataset, a medical dataset related to Parkinson’s disease (PD) [81].
- Pima dataset [82], used to detect the presence of diabetes.
- Popfailures dataset [83], a dataset related to climate measurements.
- Regions2 dataset, medical dataset related to hepatitis C [84].
- Saheart dataset [85], a medical dataset related to heart diseases.
- Segment dataset [86], an image processing dataset.
- Wdbc dataset [87], a medical dataset related to breast tumors.
- Eeg datasets, a medical dataset related to EEG measurements [90]. There are 3 different cases from this dataset used here denoted as Z_F_S, ZO_NF_S, ZONF_S.
- Zoo dataset [91], used to classify animals.
- Abalone dataset [93], used to to predict the age of abalone from physical measurements.
- Airfoildataset, derived from NASA [94].
- Baseball dataset, used to estimate the salary of baseball players.
- BK dataset [95], used to predict the points scored in a basketball game.
- BL dataset, an electrical engineering dataset.
- Concrete dataset [96].
- Dee dataset, used to predict the price of electricity.
- Diabetes dataset, a medical dataset.
- Housing dataset, provided in [97].
- FA dataset, used to predict the fit body fat.
- MB dataset, available from from Smoothing Methods in Statistics [98].
- MORTGAGE dataset, related to Economic data from USA.
- PYdataset, (Pyrimidines problem) [99].
- Quake dataset, used to predict the strength of a earthquakes.
- Treasure dataset, related to Economic data from USA.
- Wankara dataset, a dataset related to weather.
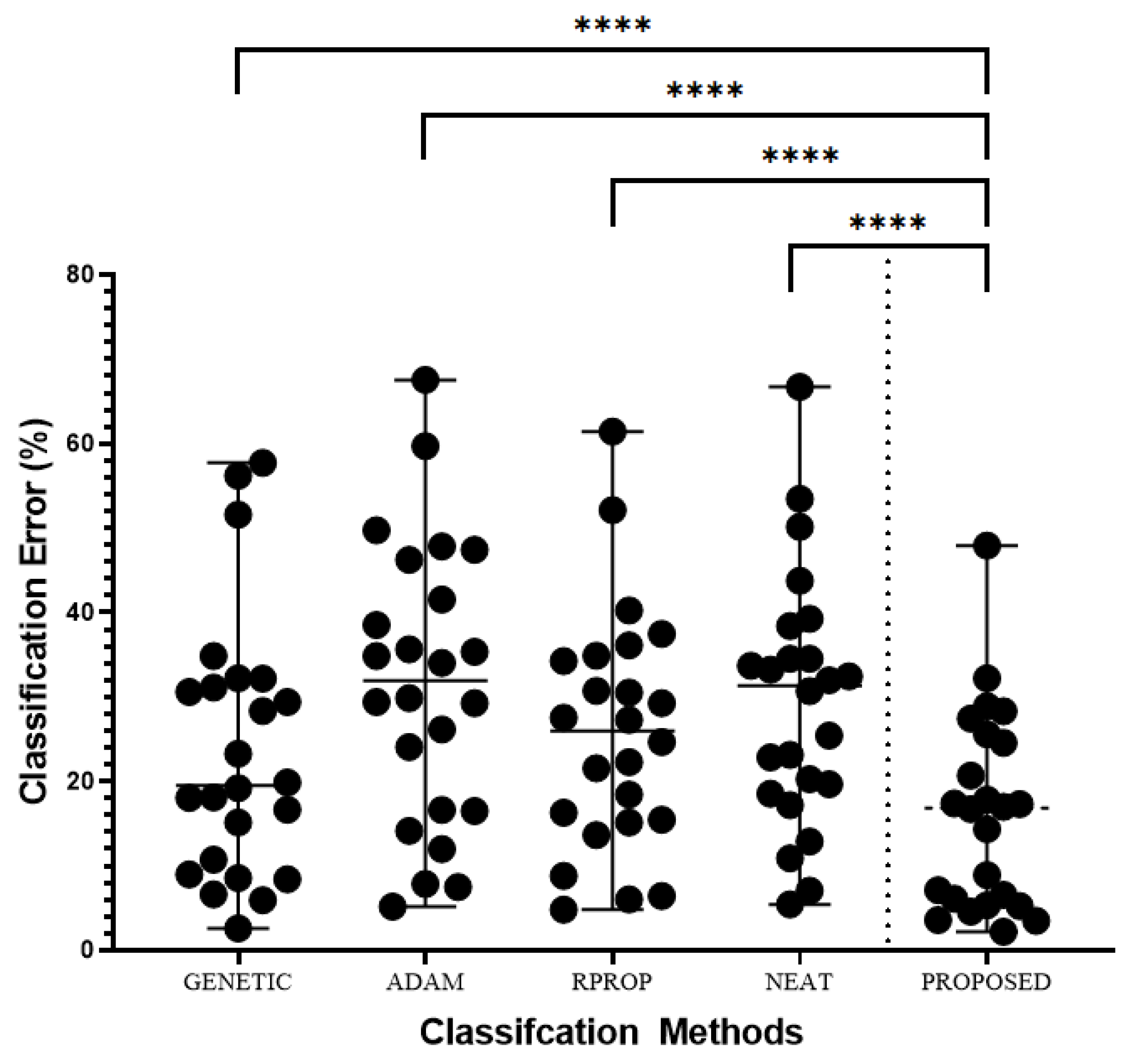
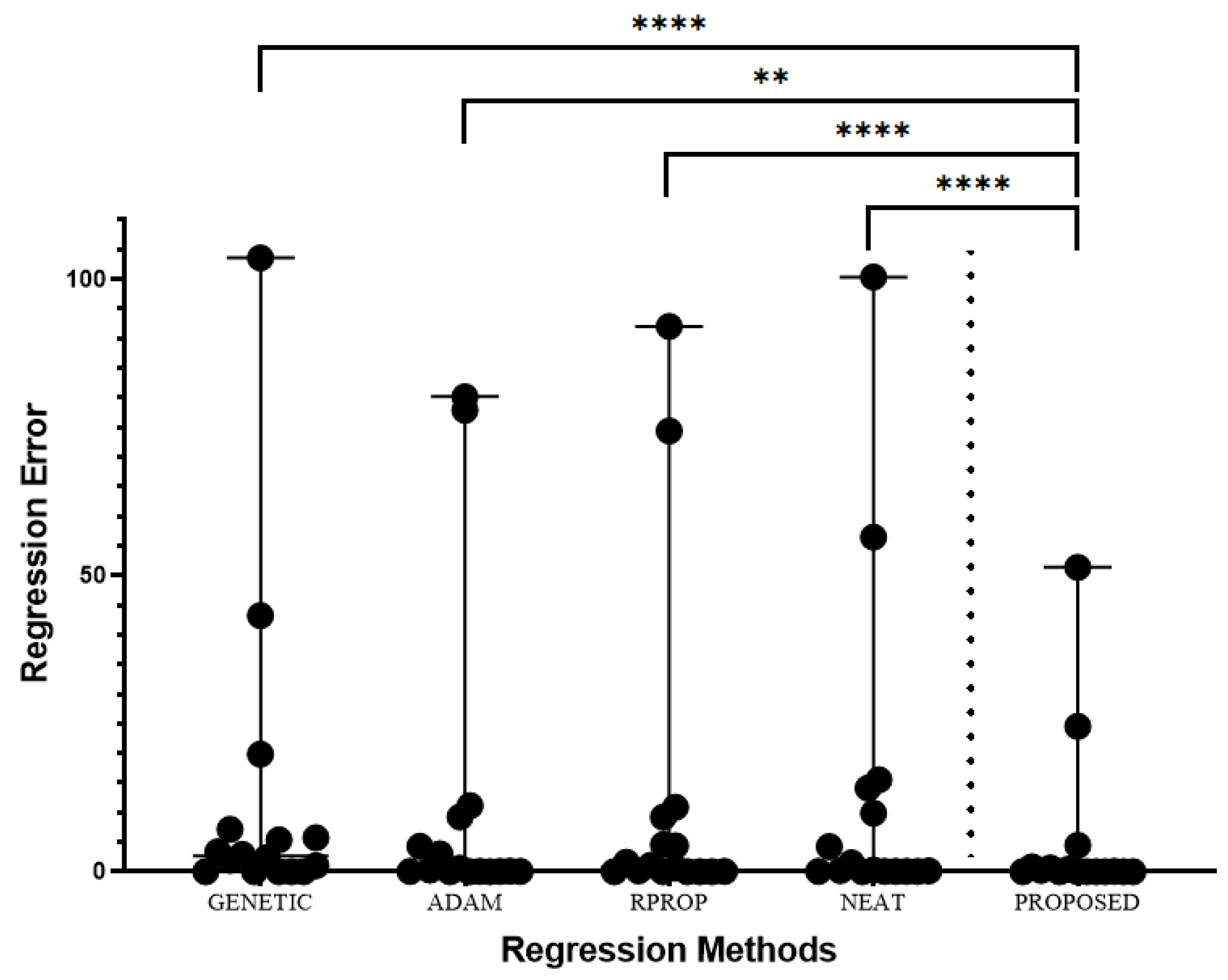
3.2. Experimental results
- A genetic algorithm where the parameters have the values of Table 2 used to train a neural network with H hidden nodes. The results in the experimental tables are denoted by the label GENETIC.
- The Adam optimization method is used to train a neural network with H hidden nodes. The column ADAM denotes the results for this method.
- The RPROP method is used to train a neural network with H hidden nodes. The corresponding results are denoted by RPROP in the relevant tables.
- The NEAT method (NeuroEvolution of Augmenting Topologies ) [100], where the maximum number of allowed generations is the same as in the case of the Genetic algorithm.
- The proposed method (denoted as PROPOSED) was used with the experimental settings and are shown in Table 2.
- An extra line was also added to the experimental tables under the title AVERAGE. This line represents the average classification or regression error for all datasets.
4. Conclusions
- There is a need for more efficient techniques for initializing the value space for artificial neural network parameters. In the present work, the optimal result from the execution of a limited number of steps by a genetic algorithm was used as an initial estimate of the value interval.
- In the present work, the same techniques as in any whole-chromosome genetic algorithm were used to perform the crossover and mutation operations. Research could be done at this point to find more focused crossover and mutation techniques for this particular problem.
- The present technique consists of two phases, in each of which a problem-adapted genetic algorithm is executed. This means that significant computational time is required to complete the algorithm. However, since genetic algorithms are inherently parallelizable, modern parallel programming techniques could be used here.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- C. Bishop, Neural Networks for Pattern Recognition, Oxford University Press, 1995.
- G. Cybenko, Approximation by superpositions of a sigmoidal function, Mathematics of Control Signals and Systems 2, pp. 303-314, 1989. https://doi.org/10.1007/BF02551274. [CrossRef]
- P. Baldi, K. Cranmer, T. Faucett et al, Parameterized neural networks for high-energy physics, Eur. Phys. J. C 76, 2016. https://doi.org/10.1140/epjc/s10052-016-4099-4. [CrossRef]
- J. J. Valdas and G. Bonham-Carter, Time dependent neural network models for detecting changes of state in complex processes: Applications in earth sciences and astronomy, Neural Networks 19, pp. 196-207, 2006. https://doi.org/10.1016/j.neunet.2006.01.006. [CrossRef]
- G. Carleo,M. Troyer, Solving the quantum many-body problem with artificial neural networks, Science 355, pp. 602-606, 2017. https://doi.org/10.1126/science.aag2302. [CrossRef]
- Y. Shirvany, M. Hayati, R. Moradian, Multilayer perceptron neural networks with novel unsupervised training method for numerical solution of the partial differential equations, Applied Soft Computing 9, pp. 20-29, 2009. https://doi.org/10.1016/j.asoc.2008.02.003. [CrossRef]
- A. Malek, R. Shekari Beidokhti, Numerical solution for high order differential equations using a hybrid neural network—Optimization method, Applied Mathematics and Computation 183, pp. 260-271, 2006. https://doi.org/10.1016/j.amc.2006.05.068. [CrossRef]
- A. Topuz, Predicting moisture content of agricultural products using artificial neural networks, Advances in Engineering Software 41, pp. 464-470, 2010. https://doi.org/10.1109/ICNN.1993.298623. [CrossRef]
- A. Escamilla-García, G.M. Soto-Zarazúa, M. Toledano-Ayala, E. Rivas-Araiza, A. Gastélum-Barrios, Abraham,Applications of Artificial Neural Networks in Greenhouse Technology and Overview for Smart Agriculture Development, Applied Sciences 10, Article number 3835, 2020. https://doi.org/10.3390/app10113835. [CrossRef]
- Lin Shen, Jingheng Wu, and Weitao Yang, Multiscale Quantum Mechanics/Molecular Mechanics Simulations with Neural Networks, Journal of Chemical Theory and Computation 12, pp. 4934-4946, 2016. https://doi.org/10.1021/acs.jctc.6b00663. [CrossRef]
- Sergei Manzhos, Richard Dawes, Tucker Carrington, Neural network-based approaches for building high dimensional and quantum dynamics-friendly potential energy surfaces, Int. J. Quantum Chem. 115, pp. 1012-1020, 2015. https://doi.org/10.1002/qua.24795. [CrossRef]
- Jennifer N. Wei, David Duvenaud, and Alán Aspuru-Guzik, Neural Networks for the Prediction of Organic Chemistry Reactions, ACS Central Science 2, pp. 725-732, 2016. https://doi.org/10.1021/acscentsci.6b00219. [CrossRef]
- Lukas Falat and Lucia Pancikova, Quantitative Modelling in Economics with Advanced Artificial Neural Networks, Procedia Economics and Finance 34, pp. 194-201, 2015. https://doi.org/10.1016/S2212-5671(15)01619-6. [CrossRef]
- Mohammad Namazi, Ahmad Shokrolahi, Mohammad Sadeghzadeh Maharluie, Detecting and ranking cash flow risk factors via artificial neural networks technique, Journal of Business Research 69, pp. 1801-1806, 2016. https://doi.org/10.1016/j.jbusres.2015.10.059. [CrossRef]
- G. Tkacz, Neural network forecasting of Canadian GDP growth, International Journal of Forecasting 17, pp. 57-69, 2001. https://doi.org/10.1016/S0169-2070(00)00063-7. [CrossRef]
- Igor I. Baskin, David Winkler and Igor V. Tetko, A renaissance of neural networks in drug discovery, Expert Opinion on Drug Discovery 11, pp. 785-795, 2016. https://doi.org/10.1080/17460441.2016.1201262. [CrossRef]
- Ronadl Bartzatt, Prediction of Novel Anti-Ebola Virus Compounds Utilizing Artificial Neural Network (ANN), Chemistry Faculty Publications 49, pp. 16-34, 2018. https://doi.org/10.20959/wjpr201813-12749. [CrossRef]
- I.G. Tsoulos, D. Gavrilis, E. Glavas, Neural network construction and training using grammatical evolution, Neurocomputing 72, pp. 269-277, 2008. https://doi.org/10.1016/j.neucom.2008.01.017. [CrossRef]
- D.E. Rumelhart, G.E. Hinton and R.J. Williams, Learning representations by back-propagating errors, Nature 323, pp. 533 - 536 , 1986. https://doi.org/10.1038/323533a0. [CrossRef]
- T. Chen and S. Zhong, Privacy-Preserving Backpropagation Neural Network Learning, IEEE Transactions on Neural Networks 20, , pp. 1554-1564, 2009. https://doi.org/10.1109/TNN.2009.2026902. [CrossRef]
- M. Riedmiller and H. Braun, A Direct Adaptive Method for Faster Backpropagation Learning: The RPROP algorithm, Proc. of the IEEE Intl. Conf. on Neural Networks, San Francisco, CA, pp. 586–591, 1993. https://doi.org/10.1109/ICNN.1993.298623. [CrossRef]
- T. Pajchrowski, K. Zawirski and K. Nowopolski, Neural Speed Controller Trained Online by Means of Modified RPROP Algorithm, IEEE Transactions on Industrial Informatics 11, pp. 560-568, 2015. https://doi.org/10.1109/TII.2014.2359620. [CrossRef]
- Rinda Parama Satya Hermanto, Suharjito, Diana, Ariadi Nugroho, Waiting-Time Estimation in Bank Customer Queues using RPROP Neural Networks, Procedia Computer Science 135, pp. 35-42, 2018. https://doi.org/10.1016/j.procs.2018.08.147. [CrossRef]
- D. P. Kingma, J. L. Ba, ADAM: a method for stochastic optimization, in: Proceedings of the 3rd International Conference on Learning Representations (ICLR 2015), pp. 1–15, 2015. https://doi.org/10.48550/arXiv.1412.6980. [CrossRef]
- Y. Xue, Y. Tong, F. Neri, An ensemble of differential evolution and Adam for training feed-forward neural networks. Information Sciences 608, pp. 453-471, 2022. https://doi.org/10.1016/j.ins.2022.06.036. [CrossRef]
- B. Robitaille and B. Marcos and M. Veillette and G. Payre, Modified quasi-Newton methods for training neural networks, Computers & Chemical Engineering 20, pp. 1133-1140, 1996. https://doi.org/10.1016/0098-1354(95)00228-6. [CrossRef]
- Q. Liu, J. Liu, R. Sang, J. Li, T. Zhang and Q. Zhang, Fast Neural Network Training on FPGA Using Quasi-Newton Optimization Method,IEEE Transactions on Very Large Scale Integration (VLSI) Systems 26, pp. 1575-1579, 2018. https://doi.org/10.1109/TVLSI.2018.2820016. [CrossRef]
- R.S. Sexton, B. Alidaee, R.E. Dorsey, J.D. Johnson, Global optimization for artificial neural networks: A tabu search application. European Journal of Operational Research 106, pp. 570-584, 1998. https://doi.org/10.1016/S0377-2217(97)00292-0. [CrossRef]
- A. Yamazaki, M. C. P. de Souto,T. B. Ludermir, Optimization of neural network weights and architectures for odor recognition using simulated annealing, In: Proceedings of the 2002 International Joint Conference on Neural Networks. IJCNN’02 1, pp. 547-552 , 2002. https://doi.org/10.1109/IJCNN.2002.1005531. [CrossRef]
- Y. Da, G. Xiurun, An improved PSO-based ANN with simulated annealing technique, Neurocomputing 63, pp. 527-533, 2005. https://doi.org/10.1016/j.neucom.2004.07.002. [CrossRef]
- F. H. F. Leung, H. K. Lam, S. H. Ling and P. K. S. Tam, Tuning of the structure and parameters of a neural network using an improved genetic algorithm, IEEE Transactions on Neural Networks 14, pp. 79-88, 2003. https://doi.org/10.1109/TNN.2002.804317. [CrossRef]
- X. Yao, Evolving artificial neural networks, Proceedings of the IEEE, 87(9), pp. 1423-1447, 1999. https://doi.org/10.1109/5.784219. [CrossRef]
- C. Zhang, H. Shao and Y. Li, Particle swarm optimisation for evolving artificial neural network, IEEE International Conference on Systems, Man, and Cybernetics, , pp. 2487-2490, 2000. https://doi.org/10.1109/ICSMC.2000.884366. [CrossRef]
- Jianbo Yu, Shijin Wang, Lifeng Xi, Evolving artificial neural networks using an improved PSO and DPSO 71, pp. 1054-1060, 2008. https://doi.org/10.1016/j.neucom.2007.10.013. [CrossRef]
- J. lonen, J.K. Kamarainen, J. Lampinen, Differential Evolution Training Algorithm for Feed-Forward Neural Networks, Neural Processing Letters 17, pp. 93–105, 2003. https://doi.org/10.1023/A:1022995128597. [CrossRef]
- M. Baioletti, G. Di Bari, A. Milani, V. Poggioni, Differential Evolution for Neural Networks Optimization, Mathematics 8, 2020. https://doi.org/10.3390/math8010069. [CrossRef]
- K.M. Salama, A.M. Abdelbar, Learning neural network structures with ant colony algorithms, Swarm Intell 9, pp. 229–265, 2015. https://doi.org/10.1007/s11721-015-0112-z. [CrossRef]
- J.R. Zhang, J. Zhang, T.M. Lok, M.R. Lyu, A hybrid particle swarm optimization–back-propagation algorithm for feedforward neural network training, Applied Mathematics and Computation 185, pp. 1026-1037, 2007. https://doi.org/10.1016/j.amc.2006.07.025. [CrossRef]
- S. Mishra, S. K. Patra, Short Term Load Forecasting Using Neural Network Trained with Genetic Algorithm & Particle Swarm Optimization, In: 2008 First International Conference on Emerging Trends in Engineering and Technology, Nagpur, India, 2008, pp. 606-611. https://doi.org/10.1109/ICETET.2008.94. [CrossRef]
- S. Mirjalili, S.Z. Mohd Hashim, H. Moradian Sardroudi, Training feedforward neural networks using hybrid particle swarm optimization and gravitational search algorithm, Applied Mathematics and Computation 218, pp. 11125-11137, 2012. https://doi.org/10.1016/j.amc.2012.04.069. [CrossRef]
- A. Kobrunov, I. Priezzhev, Hybrid combination genetic algorithm and controlled gradient method to train a neural network, Geophysics 81, pp,35–43, 2016. https://doi.org/10.1190/geo2015-0297.1. [CrossRef]
- I. Ivanova, M. Kubat, Initialization of neural networks by means of decision trees, Knowledge-Based Systems 8, pp. 333-344, 1995. https://doi.org/10.1016/0950-7051(96)81917-4. [CrossRef]
- J.Y.F. Yam, T.W.S. Chow, A weight initialization method for improving training speed in feedforward neural network, Neurocomputing 30, pp. 219-232, 2000. https://doi.org/10.1016/S0925-2312(99)00127-7. [CrossRef]
- K. Chumachenko, A. Iosifidis, M. Gabbouj, Feedforward neural networks initialization based on discriminant learning, Neural Networks 146, pp. 220-229, 2022. https://doi.org/10.1016/j.neunet.2021.11.020. [CrossRef]
- M.D. Shahjahan, M. Kazuyuki, Neural network training algorithm with possitive correlation, IEEE Trans. Inf & Syst. 88, pp. 2399-2409, 2005. https://doi.org/10.1093/ietisy/e88-d.10.2399. [CrossRef]
- N.K. Treadgold, T.D. Gedeon, Simulated annealing and weight decay in adaptive learning: the SARPROP algorithm, IEEE Trans. on Neural Networks 9, pp. 662-668, 1998. https://doi.org/10.1109/72.701179. [CrossRef]
- C.S. Leung, K.W. Wong, P.F. Sum, L.W. Chan, A pruning method for the recursive least squared algorithm, Neural networks 14, pp. 147-174, 2001. https://doi.org/10.1016/S0893-6080(00)00093-9. [CrossRef]
- M. O’Neill, C. Ryan, Grammatical evolution, IEEE Trans. Evol. Comput. 5,pp. 349–358, 2001. https://doi.org/10.1109/4235.942529. [CrossRef]
- J. W. Backus. The Syntax and Semantics of the Proposed International Algebraic Language of the Zurich ACM-GAMM Conference. Proceedings of the International Conference on Information Processing, UNESCO, 1959, pp.125-132.
- C. Ryan, J. Collins, M. O’Neill, Grammatical evolution: Evolving programs for an arbitrary language. In: Banzhaf, W., Poli, R., Schoenauer, M., Fogarty, T.C. (eds) Genetic Programming. EuroGP 1998. Lecture Notes in Computer Science, vol 1391. Springer, Berlin, Heidelberg, 1998.
- M. O’Neill, M., C. Ryan, Evolving Multi-line Compilable C Programs. In: Poli, R., Nordin, P., Langdon, W.B., Fogarty, T.C. (eds) Genetic Programming. EuroGP 1999. Lecture Notes in Computer Science, vol 1598. Springer, Berlin, Heidelberg, 1999.
- C. Ryan, M. O’Neill, J.J. Collins, Grammatical evolution: Solving trigonometric identities, proceedings of Mendel. Vol. 98. 1998.
- A.O. Puente, R. S. Alfonso, M. A. Moreno, Automatic composition of music by means of grammatical evolution, In: APL ’02: Proceedings of the 2002 conference on APL: array processing languages: lore, problems, and applications July 2002 Pages 148–155. https://doi.org/10.1145/602231.602249. [CrossRef]
- Lídio Mauro Limade Campo, R. Célio Limã Oliveira,Mauro Roisenberg, Optimization of neural networks through grammatical evolution and a genetic algorithm, Expert Systems with Applications 56, pp. 368-384, 2016. https://doi.org/10.1016/j.eswa.2016.03.012. [CrossRef]
- K. Soltanian, A. Ebnenasir, M. Afsharchi, Modular Grammatical Evolution for the Generation of Artificial Neural Networks, Evolutionary Computation 30, pp 291–327, 2022. https://doi.org/10.1162/evco_a_00302. [CrossRef]
- I. Dempsey, M.O’ Neill, A. Brabazon, Constant creation in grammatical evolution, International Journal of Innovative Computing and Applications 1 , pp 23–38, 2007. https://doi.org/10.1007/978-3-540-24855-2_56. [CrossRef]
- E. Galván-López, J.M. Swafford, M. O’Neill, A. Brabazon, Evolving a Ms. PacMan Controller Using Grammatical Evolution. In: , et al. Applications of Evolutionary Computation. EvoApplications 2010. Lecture Notes in Computer Science, vol 6024. Springer, Berlin, Heidelberg, 2010.
- N. Shaker, M. Nicolau, G. N. Yannakakis, J. Togelius, M. O’Neill, Evolving levels for Super Mario Bros using grammatical evolution, 2012 IEEE Conference on Computational Intelligence and Games (CIG), 2012, pp. 304-31. https://doi.org/10.1109/CIG.2012.6374170. [CrossRef]
- D. Martínez-Rodríguez, J. M. Colmenar, J. I. Hidalgo, R.J. Villanueva Micó, S. Salcedo-Sanz, Particle swarm grammatical evolution for energy demand estimation, Energy Science and Engineering 8, pp. 1068-1079, 2020. https://doi.org/10.1002/ese3.568. [CrossRef]
- N. R. Sabar, M. Ayob, G. Kendall, R. Qu, Grammatical Evolution Hyper-Heuristic for Combinatorial Optimization Problems, IEEE Transactions on Evolutionary Computation 17, pp. 840-861, 2013. https://doi.org/10.1109/TEVC.2013.2281527. [CrossRef]
- C. Ryan, M. Kshirsagar, G. Vaidya, G. et al. Design of a cryptographically secure pseudo random number generator with grammatical evolution. Sci Rep 12, 8602, 2022. https://doi.org/10.1038/s41598-022-11613-x. [CrossRef]
- P.J. Pereira, P. Cortez, R. Mendes, Multi-objective Grammatical Evolution of Decision Trees for Mobile Marketing user conversion prediction, Expert Systems with Applications 168, 114287, 2021. https://doi.org/10.1016/j.eswa.2020.114287. [CrossRef]
- F. Castejón, E.J. Carmona, Automatic design of analog electronic circuits using grammatical evolution, Applied Soft Computing 62, pp. 1003-1018, 2018. https://doi.org/10.1016/j.asoc.2017.09.036. [CrossRef]
- P. Kaelo, M.M. Ali, Integrated crossover rules in real coded genetic algorithms, European Journal of Operational Research 176, pp. 60-76, 2007. https://doi.org/10.1016/j.ejor.2005.07.025. [CrossRef]
- M.J.D Powell, A Tolerant Algorithm for Linearly Constrained Optimization Calculations, Mathematical Programming 45, pp. 547-566, 1989. https://doi.org/10.1007/BF01589118. [CrossRef]
- M. Kelly, R. Longjohn, K. Nottingham, The UCI Machine Learning Repository. 2023. Available online: https://archive.ics.uci.edu (accessed on 20 September 2023).
- J. Alcalá-Fdez, A. Fernandez, J. Luengo, J. Derrac,S. García, L. Sánchez, F. Herrera. KEEL Data-Mining Software Tool: Data Set Repository, Integration of Algorithms and Experimental Analysis Framework. Journal of Multiple-Valued Logic and Soft Computing 17, pp. 255-287, 2011.
- Weiss, Sholom M. and Kulikowski, Casimir A., Computer Systems That Learn: Classification and Prediction Methods from Statistics, Neural Nets, Machine Learning, and Expert Systems, Morgan Kaufmann Publishers Inc, 1991.
- J.R. Quinlan, Simplifying Decision Trees. International Journal of Man-Machine Studies 27, pp. 221-234, 1987. https://doi.org/10.1016/S0020-7373(87)80053-6. [CrossRef]
- T. Shultz, D. Mareschal, W. Schmidt, Modeling Cognitive Development on Balance Scale Phenomena, Machine Learning 16, pp. 59-88, 1994. https://doi.org/10.1007/BF00993174. [CrossRef]
- Z.H. Zhou,Y. Jiang, NeC4.5: neural ensemble based C4.5, in IEEE Transactions on Knowledge and Data Engineering 16, pp. 770-773, 2004. https://doi.org/10.1109/TKDE.2004.11. [CrossRef]
- R. Setiono , W.K. Leow, FERNN: An Algorithm for Fast Extraction of Rules from Neural Networks, Applied Intelligence 12, pp. 15-25, 2000. https://doi.org/10.1023/A:1008307919726. [CrossRef]
- G. Demiroz, H.A. Govenir, N. Ilter, Learning Differential Diagnosis of Eryhemato-Squamous Diseases using Voting Feature Intervals, Artificial Intelligence in Medicine. 13, pp. 147–165, 1998. https://doi.org/10.1016/S0933-3657(98)00028-1. [CrossRef]
- I. Kononenko, E. Šimec, M. Robnik-Šikonja, Overcoming the Myopia of Inductive Learning Algorithms with RELIEFF, Applied Intelligence 7, pp. 39–55, 1997. https://doi.org/10.1023/A:1008280620621. [CrossRef]
- B. Hayes-Roth, B., F. Hayes-Roth. Concept learning and the recognition and classification of exemplars. Journal of Verbal Learning and Verbal Behavior 16, pp. 321-338, 1977. https://doi.org/10.1016/S0022-5371(77)80054-6. [CrossRef]
- R.M. French, N. Chater, Using noise to compute error surfaces in connectionist networks: a novel means of reducing catastrophic forgetting, Neural Comput. 14, pp. 1755-1769, 2002. https://doi.org/10.1162/08997660260028700. [CrossRef]
- J.G. Dy , C.E. Brodley, Feature Selection for Unsupervised Learning, The Journal of Machine Learning Research 5, pp 845--889, 2004.
- S. J. Perantonis, V. Virvilis, Input Feature Extraction for Multilayered Perceptrons Using Supervised Principal Component Analysis, Neural Processing Letters 10, pp 243–252, 1999. https://doi.org/10.1023/A:1018792728057. [CrossRef]
- J. Garcke, M. Griebel, Classification with sparse grids using simplicial basis functions, Intell. Data Anal. 6, pp. 483-502, 2002. https://doi.org/10.3233/IDA-2002-6602. [CrossRef]
- M. Elter, R. Schulz-Wendtland, T. Wittenberg, The prediction of breast cancer biopsy outcomes using two CAD approaches that both emphasize an intelligible decision process, Med Phys. 34, pp. 4164-72, 2007. https://doi.org/10.1118/1.2786864. [CrossRef]
- M.A. Little, P.E. McSharry, E.J. Hunter, J. Spielman, L.O. Ramig, Suitability of dysphonia measurements for telemonitoring of Parkinson’s disease. IEEE Trans Biomed Eng. 56, pp. 1015-1022, 2009. https://doi.org/10.1038/npre.2008.2298.1. [CrossRef]
- J.W. Smith, J.E. Everhart, W.C. Dickson, W.C. Knowler, R.S. Johannes, Using the ADAP learning algorithm to forecast the onset of diabetes mellitus, In: Proceedings of the Symposium on Computer Applications and Medical Care IEEE Computer Society Press, pp.261-265, 1988.
- D.D. Lucas, R. Klein, J. Tannahill, D. Ivanova, S. Brandon, D. Domyancic, Y. Zhang, Failure analysis of parameter-induced simulation crashes in climate models, Geoscientific Model Development 6, pp. 1157-1171, 2013. https://doi.org/10.5194/gmd-6-1157-2013. [CrossRef]
- N. Giannakeas, M.G. Tsipouras, A.T. Tzallas, K. Kyriakidi, Z.E. Tsianou, P. Manousou, A. Hall, E.C. Karvounis, V. Tsianos, E. Tsianos, A clustering based method for collagen proportional area extraction in liver biopsy images (2015) Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, 2015-November, art. no. 7319047, pp. 3097-3100. https://doi.org/10.1109/EMBC.2015.7319047. [CrossRef]
- T. Hastie, R. Tibshirani, Non-parametric logistic and proportional odds regression, JRSS-C (Applied Statistics) 36, pp. 260–276, 1987. https://doi.org/10.2307/2347785. [CrossRef]
- M. Dash, H. Liu, P. Scheuermann, K. L. Tan, Fast hierarchical clustering and its validation, Data & Knowledge Engineering 44, pp 109–138, 2003. https://doi.org/10.1016/S0169-023X(02)00138-6. [CrossRef]
- W.H. Wolberg, O.L. Mangasarian, Multisurface method of pattern separation for medical diagnosis applied to breast cytology, Proc Natl Acad Sci U S A. 87, pp. 9193–9196, 1990. https://doi.org/10.1073/pnas.87.23.9193. [CrossRef]
- M. Raymer, T.E. Doom, L.A. Kuhn, W.F. Punch, Knowledge discovery in medical and biological datasets using a hybrid Bayes classifier/evolutionary algorithm. IEEE transactions on systems, man, and cybernetics. Part B, Cybernetics : a publication of the IEEE Systems, Man, and Cybernetics Society, 33 , pp. 802-813, 2003. https://doi.org/10.1109/TSMCB.2003.816922. [CrossRef]
- P. Zhong, M. Fukushima, Regularized nonsmooth Newton method for multi-class support vector machines, Optimization Methods and Software 22, pp. 225-236, 2007. https://doi.org/10.1080/10556780600834745. [CrossRef]
- R.G. Andrzejak, K. Lehnertz, F. Mormann, C. Rieke, P. David, and C. E. Elger, Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state, Phys. Rev. E 64, pp. 1-8, 2001. https://doi.org/10.1103/PhysRevE.64.061907. [CrossRef]
- M. Koivisto, K. Sood, Exact Bayesian Structure Discovery in Bayesian Networks, The Journal of Machine Learning Research 5, pp. 549--573, 2004.
- R.G. Andrzejak, K. Lehnertz, F. Mormann, C. Rieke, P. David, and C. E. Elger, Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state, Phys. Rev. E 64, pp. 1-8, 2001. https://doi.org/10.1103/PhysRevE.64.061907. [CrossRef]
- W. J Nash, T.L. Sellers, S.R. Talbot, A.J. Cawthor, W.B. Ford, The Population Biology of Abalone (_Haliotis_ species) in Tasmania. I. Blacklip Abalone (_H. rubra_) from the North Coast and Islands of Bass Strait, Sea Fisheries Division, Technical Report No. 48 (ISSN 1034-3288), 1994.
- T.F. Brooks, D.S. Pope, A.M. Marcolini, Airfoil self-noise and prediction. Technical report, NASA RP-1218, July 1989.
- J.S. Simonoff, Smooting Methods in Statistics, Springer - Verlag, 1996.
- I.Cheng Yeh, Modeling of strength of high performance concrete using artificial neural networks, Cement and Concrete Research. 28, pp. 1797-1808, 1998. https://doi.org/10.1016/S0008-8846(98)00165-3. [CrossRef]
- D. Harrison and D.L. Rubinfeld, Hedonic prices and the demand for clean ai, J. Environ. Economics & Management 5, pp. 81-102, 1978.
- J.S. Simonoff, Smooting Methods in Statistics, Springer - Verlag, 1996.
- R.D. King, S. Muggleton, R. Lewis, M.J.E. Sternberg, Proc. Nat. Acad. Sci. USA 89, pp. 11322--11326, 1992.
- K. O. Stanley, R. Miikkulainen, Evolving Neural Networks through Augmenting Topologies, Evolutionary Computation 10, pp. 99-127, 2002. [CrossRef]
- E. Cantu-Paz, D.E. Goldberg, Efficient parallel genetic algorithms: theory and practice, Computer methods in applied mechanics and engineering 186, pp. 221-238, 2000. https://doi.org/10.1016/S0045-7825(99)00385-0. [CrossRef]
- T. Harada, E. Alba, Parallel genetic algorithms: a useful survey, ACM Computing Surveys (CSUR) 53, pp. 1-39, 2022. https://doi.org/10.1145/3400031. [CrossRef]
- W. Gropp, E. Lusk, N. Doss, A. Skjellum, A high-performance, portable implementation of the MPI message passing interface standard, Parallel Computing 22, pp. 789-828, 1996. https://doi.org/10.1016/0167-8191(96)00024-5. [CrossRef]
- R. Chandra, L. Dagum, D. Kohr, D. Maydan,J. McDonald and R. Menon, Parallel Programming in OpenMP, Morgan Kaufmann Publishers Inc., 2001.


| Expression | Chromosome | Operation |
|---|---|---|
| 9,8,6,4,15,9,16,23,8 | 9 mod 2=1 | |
| <expr>,<expr> | 8,6,4,15,9,16,23,8 | 8 mod 2=0 |
| (<xlist>,<lcommand>,<rcommand>),<expr> | 6,4,15,9,16,23,8 | 6 mod 8=6 |
| (x7,<lcommand>,<rcommand>),<expr> | 4,15,9,16,23,8 | 4 % 3=1 |
| (x7,EXPAND,<rcommand>),<expr> | 15,9,16,23,8 | 15%3=0 |
| (x7,EXPAND,NOTHING),<expr> | 9,16,23,8 | 9 %2 =1 |
| (x7,EXPAND,NOTHING),(<xlist>,<lcommand>,<rcommand>) | 16,23,8 | 16%8=0 |
| (x7,EXPAND,NOTHING),(x1,<lcommand>,<rcommand>) | 23,8 | 23%3=2 |
| (x7,EXPAND,NOTHING),(x1,DIVIDE,<rcommand>) | 8 | 8%3=2 |
| (x7,EXPAND,NOTHING),(x1,DIVIDE,EXPAND) |
| PARAMETER | VALUE |
|---|---|
| H | 10 |
| 200 | |
| 50 | |
| 200 | |
| 0.10 | |
| 0.01 |
| DATASET | GENETIC | ADAM | RPROP | NEAT | PROPOSED |
|---|---|---|---|---|---|
| Appendicitis | 18.10% | 16.50% | 16.30% | 17.20% | 17.00% |
| Australian | 32.21% | 35.65% | 36.12% | 31.98% | 24.55% |
| Balance | 8.97% | 7.87% | 8.81% | 23.14% | 16.71% |
| Cleveland | 51.60% | 67.55% | 61.41% | 53.44% | 47.91% |
| Dermatology | 30.58% | 26.14% | 15.12% | 32.43% | 8.93% |
| Hayes Roth | 56.18% | 59.70% | 37.46% | 50.15% | 32.21% |
| Heart | 28.34% | 38.53% | 30.51% | 39.27% | 17.40% |
| HouseVotes | 6.62% | 7.48% | 6.04% | 10.89% | 3.48% |
| Ionosphere | 15.14% | 16.64% | 13.65% | 19.67% | 7.14% |
| Liverdisorder | 31.11% | 41.53% | 40.26% | 30.67% | 28.90% |
| Lymography | 23.26% | 29.26% | 24.67% | 33.70% | 17.86% |
| Mammographic | 19.88% | 46.25% | 18.46% | 22.85% | 17.32% |
| Parkinsons | 18.05% | 24.06% | 22.28% | 18.56% | 14.35% |
| Pima | 32.19% | 34.85% | 34.27% | 34.51% | 25.58% |
| Popfailures | 5.94% | 5.18% | 4.81% | 7.05% | 4.58% |
| Regions2 | 29.39% | 29.85% | 27.53% | 33.23% | 28.32% |
| Saheart | 34.86% | 34.04% | 34.90% | 34.51% | 27.43% |
| Segment | 57.72% | 49.75% | 52.14% | 66.72% | 20.68% |
| Wdbc | 8.56% | 35.35% | 21.57% | 12.88% | 5.23% |
| Wine | 19.20% | 29.40% | 30.73% | 25.43% | 5.35% |
| Z_F_S | 10.73% | 47.81% | 29.28% | 38.41% | 6.56% |
| ZO_NF_S | 8.41% | 47.43% | 6.43% | 43.75% | 3.60% |
| ZONF_S | 2.60% | 11.99% | 27.27% | 5.44% | 2.21% |
| ZOO | 16.67% | 14.13% | 15.47% | 20.27% | 6.10% |
| AVERAGE | 23.60% | 31.54% | 25.65% | 29.42% | 16.23% |
| DATASET | GENETIC | ADAM | RPROP | NEAT | PROPOSED |
|---|---|---|---|---|---|
| ABALONE | 7.17 | 4.30 | 4.55 | 9.88 | 4.48 |
| AIRFOIL | 0.003 | 0.005 | 0.002 | 0.067 | 0.002 |
| BASEBALL | 103.60 | 77.90 | 92.05 | 100.39 | 51.39 |
| BK | 0.027 | 0.03 | 1.599 | 0.15 | 0.02 |
| BL | 5.74 | 0.28 | 4.38 | 0.05 | 0.002 |
| CONCRETE | 0.0099 | 0.078 | 0.0086 | 0.081 | 0.004 |
| DEE | 1.013 | 0.63 | 0.608 | 1.512 | 0.23 |
| DIABETES | 19.86 | 3.03 | 1.11 | 4.25 | 0.41 |
| HOUSING | 43.26 | 80.20 | 74.38 | 56.49 | 24.55 |
| FA | 1.95 | 0.11 | 0.14 | 0.19 | 0.01 |
| MB | 3.39 | 0.06 | 0.055 | 0.061 | 0.048 |
| MORTGAGE | 2.41 | 9.24 | 9.19 | 14.11 | 0.65 |
| PY | 5.41 | 0.09 | 0.039 | 0.075 | 0.025 |
| QUAKE | 0.040 | 0.06 | 0.041 | 0.298 | 0.038 |
| TREASURY | 2.929 | 11.16 | 10.88 | 15.52 | 0.84 |
| WANKARA | 0.012 | 0.02 | 0.0003 | 0.005 | 0.0002 |
| AVERAGE | 12.30 | 11.70 | 12.44 | 12.70 | 5.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





