Submitted:
02 October 2023
Posted:
03 October 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
- Building an ML model that will predict diabetes using socio-demographic characteristics rather than clinical attributes. Because all people, especially from lower-income countries do not know the clinical features.
- Revealing significant risk factors that indicate diabetes.
- Proposing a best fit clinically usable framework to predict diabetes at an early stage.
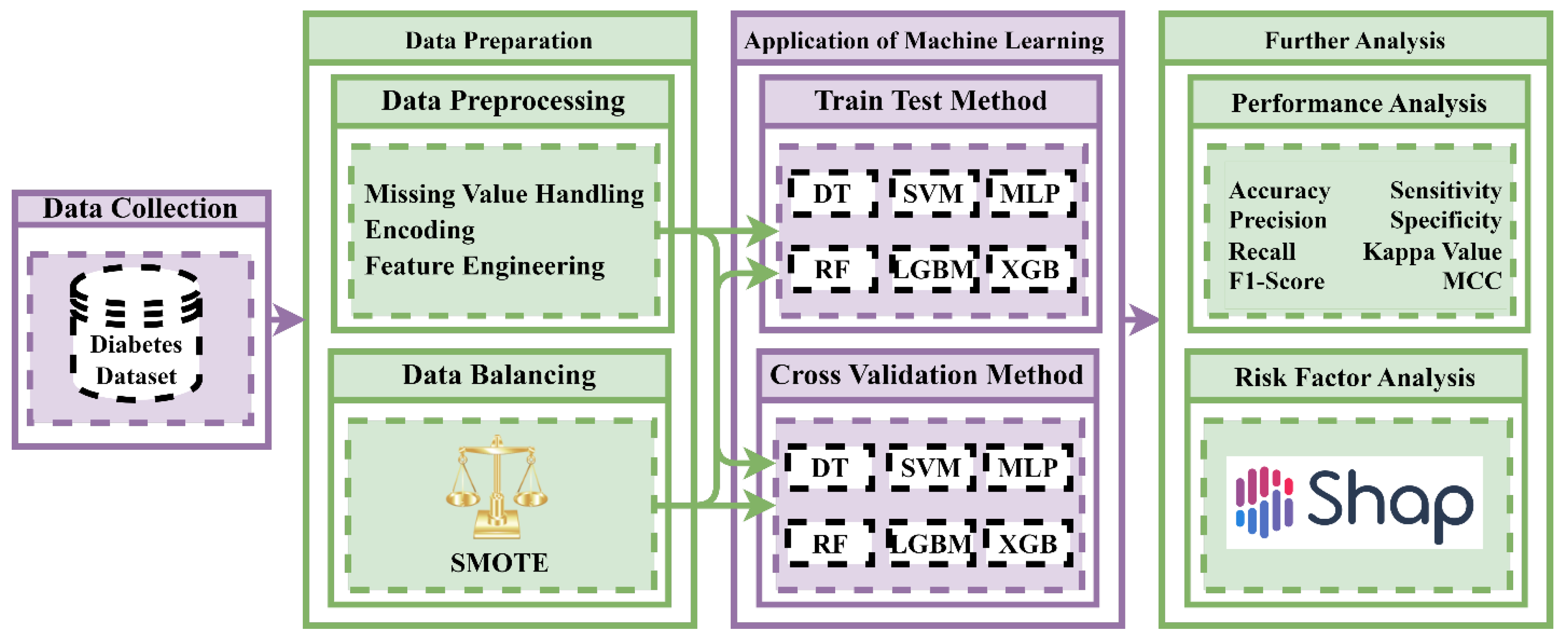
2. Materials and Methods
2.1. Dataset Description
| Attributes | Data Type | Interpretation |
| age | numeric | Age of the patient |
| gender | nominal | Whether the patient male/female |
| polyuria | nominal | Is not whether the patient had frequent urination |
| polydipsia | nominal | Determine whether or not the patient had excessive thirst/drinking |
| sudden_weight_loss | nominal | Whether or not the patient experienced a period of sudden reduced weight |
| weakness | nominal | Whether the patient experienced a moment of weakness |
| polyphagia | nominal | Whether or not the patient experienced extreme hunger |
| genital_thrush | nominal | Whether or not the patient had a yeast infection |
| visual_blurring | nominal | Whether the patient experienced unclear seeing. |
| itching | nominal | If the patient had an experienced of itch |
| irritability | nominal | If or not the patient had an experienced of irritability |
| delayed_healing | nominal | Whether the patient observed a delay in recovery after being injured |
| partial_paresis | nominal | If the patient experienced a period of muscle wasting or a group of failing muscles |
| muscle_stiffness | nominal | Whether or not the patient experienced of muscle stiffness |
| alopecia | nominal | Patient had hair loss or not |
| obesity | nominal | Considering his body mass index, determine yet if the patient is obese or not. |
| class | nominal | Presence of Diabetes (Positive/Negative) |
2.2. Data Preprocessing
2.3. Data Balancing Techniques
2.4. Performance Evaluation Metrics
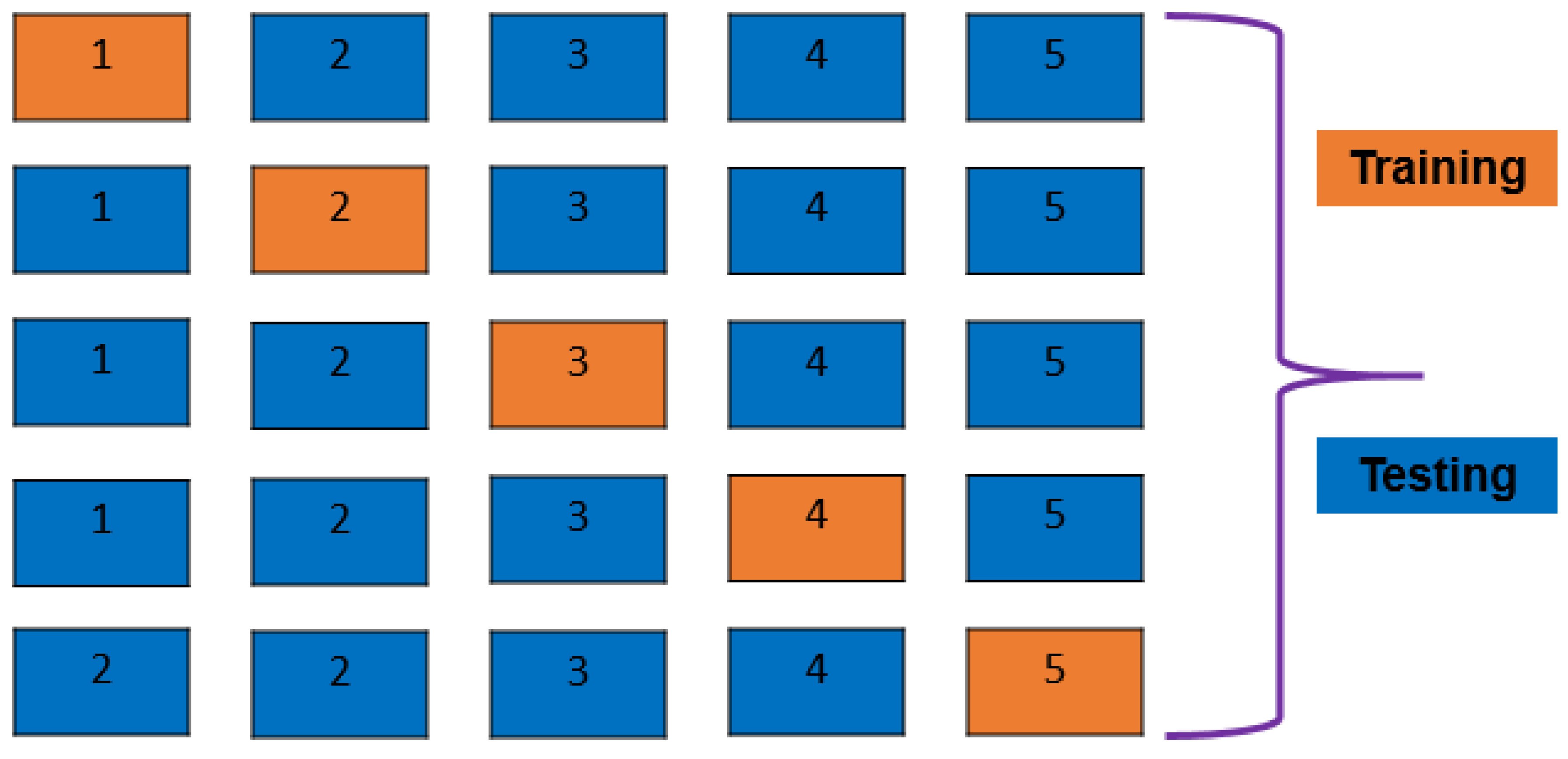
2.5. K-Fold Cross-validation and Train Test Split
2.6. Machine Learning Algorithms
2.6.1. Decision Tree Classifier
2.6.2. Random Forest Classifier
2.6.3. Support Vector Machine
2.6.4. XGBoost Classifier
2.6.5. LightGBM Classifier
2.6.6. Multi-Layer Perceptron
2.7. Feature Importance and Model Explanation
3. Result Analysis & Discussion
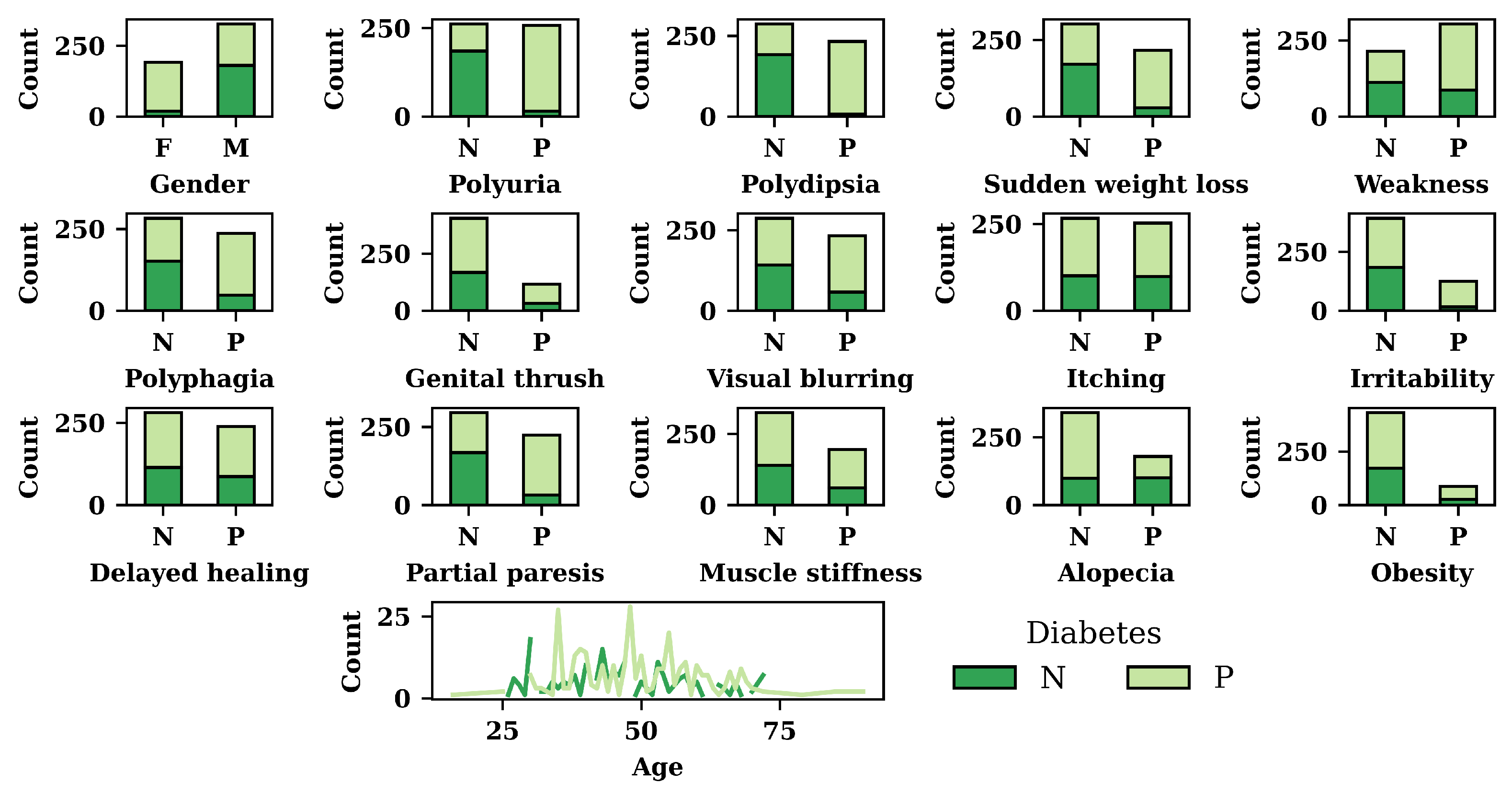
3.1. Exploratory Data Analysis (EDA) result
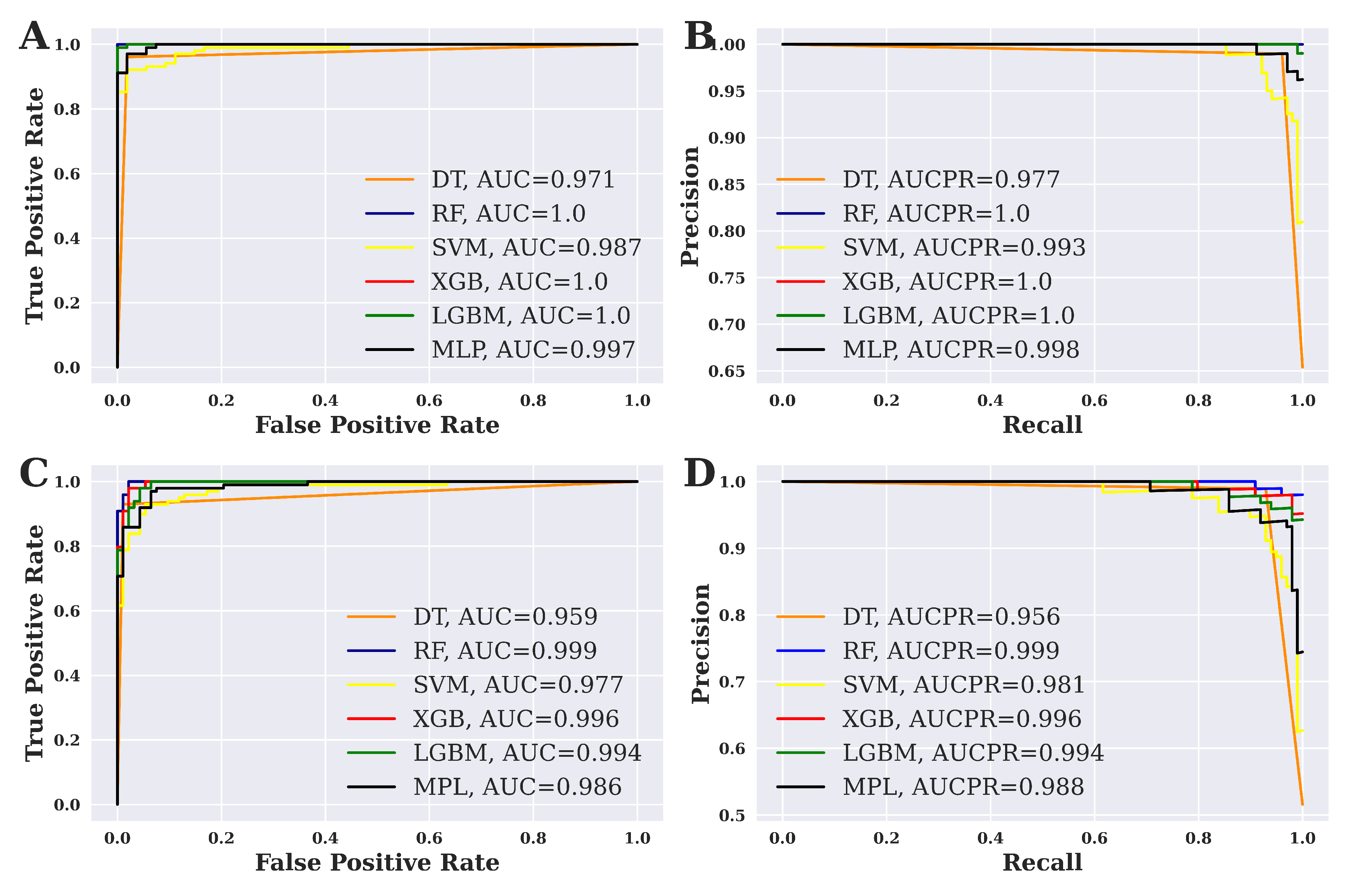
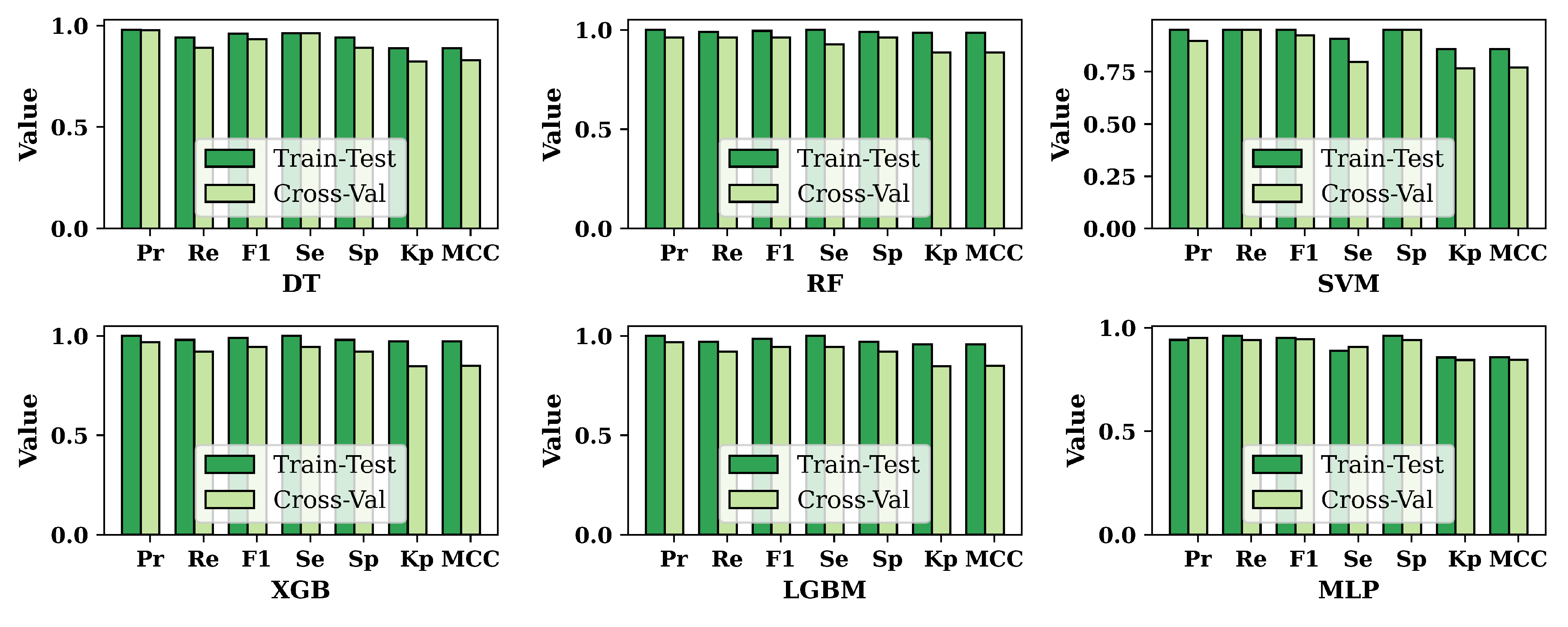
3.2. Performance Evaluation of ML Models
| Algorithm | Accuracy | Precision | Recall | F1-Score | Sensitivity | Specificity | Kappa Statistics | MCC |
| SVM | 92.19% | 0.9117 | 0.9394 | 0.9254 | 0.9032 | 0.9394 | 0.8434 | 0.8438 |
| MLP | 93.23% | 0.9388 | 0.9293 | 0.934 | 0.9355 | 0.9293 | 0.8645 | 0.8645 |
| LGBM | 94.27% | 0.9782 | 0.9091 | 0.9424 | 0.9785 | 0.9091 | 0.8855 | 0.8879 |
| XGB | 96.35% | 0.9791 | 0.9495 | 0.9641 | 0.9785 | 0.9495 | 0.9271 | 0.9275 |
| DT | 97.39% | 0.9896 | 0.9596 | 0.9743 | 0.9892 | 0.9596 | 0.9479 | 0.9484 |
| RF | 98.44% | 0.98 | 0.9899 | 0.9849 | 0.9785 | 0.9899 | 0.9687 | 0.9687 |
| Algorithm | Accuracy | Precision | Recall | F1-Score | Sensitivity | Specificity | Kappa Statistics | MCC |
| MLP | 93.59% | 0.9423 | 0.9608 | 0.9514 | 0.8889 | 0.9608 | 0.8571 | 0.8575 |
| SVM | 93.59% | 0.951 | 0.951 | 0.951 | 0.9074 | 0.951 | 0.8584 | 0.8584 |
| DT | 94.87% | 0.9796 | 0.9412 | 0.96 | 0.9629 | 0.9412 | 0.8886 | 0.89 |
| LGBM | 98.08% | 1 | 0.9706 | 0.9851 | 1 | 0.9706 | 0.958 | 0.9589 |
| XGB | 98.72% | 1 | 0.9804 | 0.9901 | 1 | 0.9804 | 0.9719 | 0.9723 |
| RF | 99.37% | 1 | 0.9902 | 0.9951 | 1 | 0.9902 | 0.9859 | 0.986 |
| Algorithm | Accuracy | Precision | Recall | F1-Score | Sensitivity | Specificity | Kappa statistics | MCC |
| DT | 91.61% | 0.9784 | 0.8921 | 0.9333 | 0.9629 | 0.8921 | 0.8228 | 0.8291 |
| RF | 94.87% | 0.9608 | 0.9608 | 0.9608 | 0.9259 | 0.9608 | 0.8867 | 0.8867 |
| SVM | 89.74% | 0.8981 | 0.9509 | 0.9238 | 0.7963 | 0.9509 | 0.7673 | 0.7703 |
| XGB | 92.95% | 0.9691 | 0.9216 | 0.9447 | 0.9444 | 0.9216 | 0.8475 | 0.8496 |
| LGBM | 92.95% | 0.9691 | 0.9216 | 0.9447 | 0.9444 | 0.9216 | 0.8475 | 0.8496 |
| MLP | 92.95% | 0.9505 | 0.9412 | 0.9458 | 0.9074 | 0.9412 | 0.8449 | 0.845 |
| Algorithm | Accuracy | Precision | Recall | F1-Score | Sensitivity | Specificity | Kappa Statistics | MCC |
| DT | 91.67% | 0.9278 | 0.9091 | 0.9184 | 0.9247 | 0.9091 | 0.8333 | 0.8334 |
| RF | 93.23% | 0.9574 | 0.9091 | 0.9326 | 0.957 | 0.9091 | 0.8647 | 0.8658 |
| SVM | 87.50% | 0.8947 | 0.8586 | 0.8763 | 0.8925 | 0.8586 | 0.7501 | 0.7507 |
| XGB | 92.71% | 0.9474 | 0.9091 | 0.9278 | 0.9462 | 0.9091 | 0.8542 | 0.8549 |
| LGBM | 93.23% | 0.9574 | 0.9091 | 0.9326 | 0.957 | 0.9091 | 0.8647 | 0.8658 |
| MLP | 91.14% | 0.9271 | 0.8989 | 0.9128 | 0.9247 | 0.8989 | 0.8229 | 0.8233 |
3.3. Overall Performance Evaluation for ML Methods

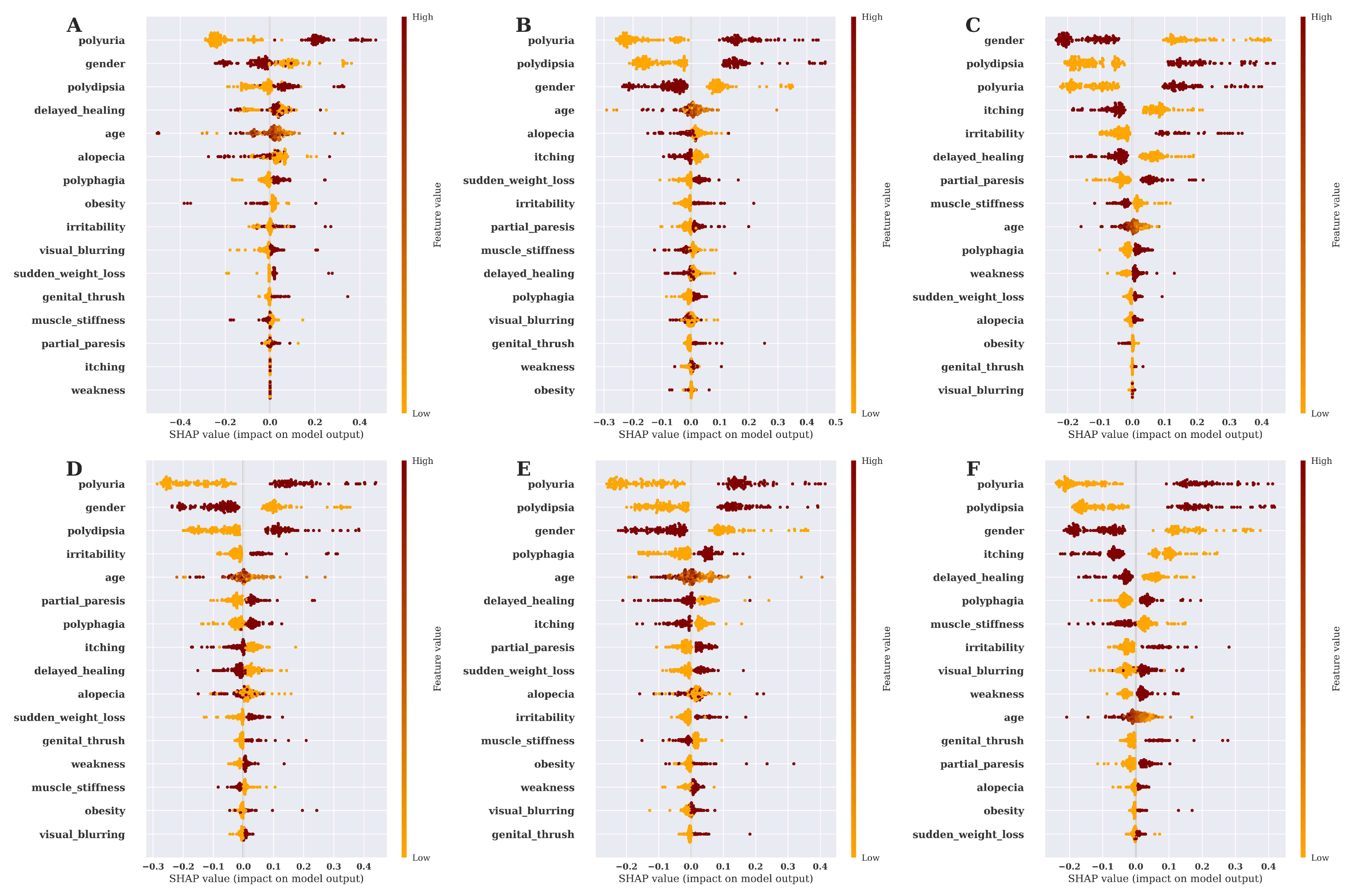
3.4. Risk Factor Analysis and Model Explanation Based on SHAP Value
3.5. Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Roglic, Gojka. "WHO Global report on diabetes: A summary." International Journal of Noncommunicable Diseases 1.1 (2016): 3. [CrossRef]
- Balfe, Myles, et al. "What’s distressing about having type 1 diabetes? A qualitative study of young adults’ perspectives." BMC endocrine disorders 13 (2013): 1-14. [CrossRef]
- Khanam, Jobeda Jamal, and Simon Y. Foo. "A comparison of machine learning algorithms for diabetes prediction." ICT Express 7.4 (2021): 432-439. [CrossRef]
- Islam, M. M., et al. "Likelihood prediction of diabetes at early stage using data mining techniques." Computer Vision and Machine Intelligence in Medical Image Analysis. Springer, Singapore, 2020. 113-125. [CrossRef]
- Krishnamoorthi, Raja, et al. "A novel diabetes healthcare disease prediction framework using machine learning techniques." Journal of Healthcare Engineering 2022 (2022). [CrossRef]
- Islam, Md Shafiqul, et al. "Advanced techniques for predicting the future progression of type 2 diabetes." IEEE Access 8 (2020): 120537-120547. [CrossRef]
- Hasan, Md Kamrul, et al. "Diabetes prediction using ensembling of different machine learning classifiers." IEEE Access 8 (2020): 76516-76531. [CrossRef]
- Fazakis, Nikos, et al. "Machine learning tools for long-term type 2 diabetes risk prediction." IEEE Access 9 (2021): 103737-103757. [CrossRef]
- Ahmed, Usama, et al. "Prediction of diabetes empowered with fused machine learning." IEEE Access 10 (2022): 8529-8538. [CrossRef]
- Maniruzzaman, Md, et al. "Classification and prediction of diabetes disease using machine learning paradigm." Health information science and systems 8.1 (2020): 1-14. [CrossRef]
- Barakat, Nahla, Andrew P. Bradley, and Mohamed Nabil H. Barakat. "Intelligible support vector machines for diagnosis of diabetes mellitus." IEEE transactions on information technology in biomedicine 14.4 (2010): 1114-1120. [CrossRef]
- Dataset: Available online: https://www.kaggle.com/datasets/andrewmvd/early-diabetes-classification (accessed on 17 November 2022).
- Sanni, Rachana R., and H. S. Guruprasad. "Analysis of performance metrics of heart failured patients using Python and machine learning algorithms." Global transitions proceedings 2.2 (2021): 233-237. [CrossRef]
- Silva, Fabrício R., et al. "Sensitivity and specificity of machine learning classifiers for glaucoma diagnosis using Spectral Domain OCT and standard automated perimetry." Arquivos brasileiros de oftalmologia 76 (2013): 170-174. [CrossRef]
- Chicco, Davide, Niklas Tötsch, and Giuseppe Jurman. "The Matthews correlation coefficient (MCC) is more reliable than balanced accuracy, bookmaker informedness, and markedness in two-class confusion matrix evaluation." BioData mining 14.1 (2021): 1-22. [CrossRef]
- Chicco, Davide, and Giuseppe Jurman. "The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation." BMC genomics 21.1 (2020): 1-13. [CrossRef]
- Erickson, Bradley J., and Felipe Kitamura. "Magician’s corner: 9. Performance metrics for machine learning models." Radiology: Artificial Intelligence 3.3 (2021): e200126. [CrossRef]
- Mohamed, Amr E. "Comparative study of four supervised machine learning techniques for classification." International Journal of Applied 7.2 (2017): 1-15.
- Priyam, Anuja, et al. "Comparative analysis of decision tree classification algorithms." International Journal of current engineering and technology 3.2 (2013): 334-337.
- Azar, Ahmad Taher, et al. "A random forest classifier for lymph diseases." Computer methods and programs in biomedicine 113.2 (2014): 465-473. [CrossRef]
- Song, Yan-Yan, and L. U. Ying. "Decision tree methods: applications for classification and prediction." Shanghai archives of psychiatry 27.2 (2015): 130. [CrossRef]
- Liaw, Andy, and Matthew Wiener. "Classification and regression by randomForest." R news 2.3 (2002): 18-22.
- Zhang, Yongli. "Support vector machine classification algorithm and its application." Information Computing and Applications: Third International Conference, ICICA 2012, Chengde, China, September 14-16, 2012. Proceedings, Part II 3. Springer Berlin Heidelberg, 2012. [CrossRef]
- Ramraj, Santhanam, et al. "Experimenting XGBoost algorithm for prediction and classification of different datasets." International Journal of Control Theory and Applications 9.40 (2016): 651-662.
- XGBoost Documentation : Available online: https://xgboost.readthedocs.io/en/stable/ (accessed on 24 December 2022).
- Rufo, Derara Duba, et al. "Diagnosis of diabetes mellitus using gradient boosting machine (LightGBM)." Diagnostics 11.9 (2021): 1714.
- Abdurrahman, Muhammad Hafizh, Budhi Irawan, and Casi Setianingsih. "A review of light gradient boosting machine method for hate speech classification on twitter." 2020 2nd International Conference on Electrical, Control and Instrumentation Engineering (ICECIE). IEEE, 2020. [CrossRef]
- Desai, Meha, and Manan Shah. "An anatomization on breast cancer detection and diagnosis employing multi-layer perceptron neural network (MLP) and Convolutional neural network (CNN)." Clinical eHealth 4 (2021): 1-11. [CrossRef]
- Maulidevi, Nur Ulfa, and Kridanto Surendro. "SMOTE-LOF for noise identification in imbalanced data classification." Journal of King Saud University-Computer and Information Sciences 34.6 (2022): 3413-3423. [CrossRef]
- Marcílio, Wilson E., and Danilo M. Eler. "From explanations to feature selection: assessing SHAP values as feature selection mechanism." 2020 33rd SIBGRAPI conference on Graphics, Patterns and Images (SIBGRAPI). Ieee, 2020. [CrossRef]
- Bowen, Dillon, and Lyle Ungar. "Generalized SHAP: Generating multiple types of explanations in machine learning." arXiv preprint arXiv:2006.07155 (2020). [CrossRef]
- Chawla, Nitesh V., et al. "SMOTE: synthetic minority over-sampling technique." Journal of artificial intelligence research 16 (2002): 321-357.
- Zou, Quan, et al. "Predicting diabetes mellitus with machine learning techniques." Frontiers in genetics 9 (2018): 515. [CrossRef]
- Tan, Jimin, et al. "A critical look at the current train/test split in machine learning." arXiv preprint arXiv:2106.04525 (2021). [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





