Submitted:
11 September 2023
Posted:
13 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Identification and Characterization of PLD Family Members
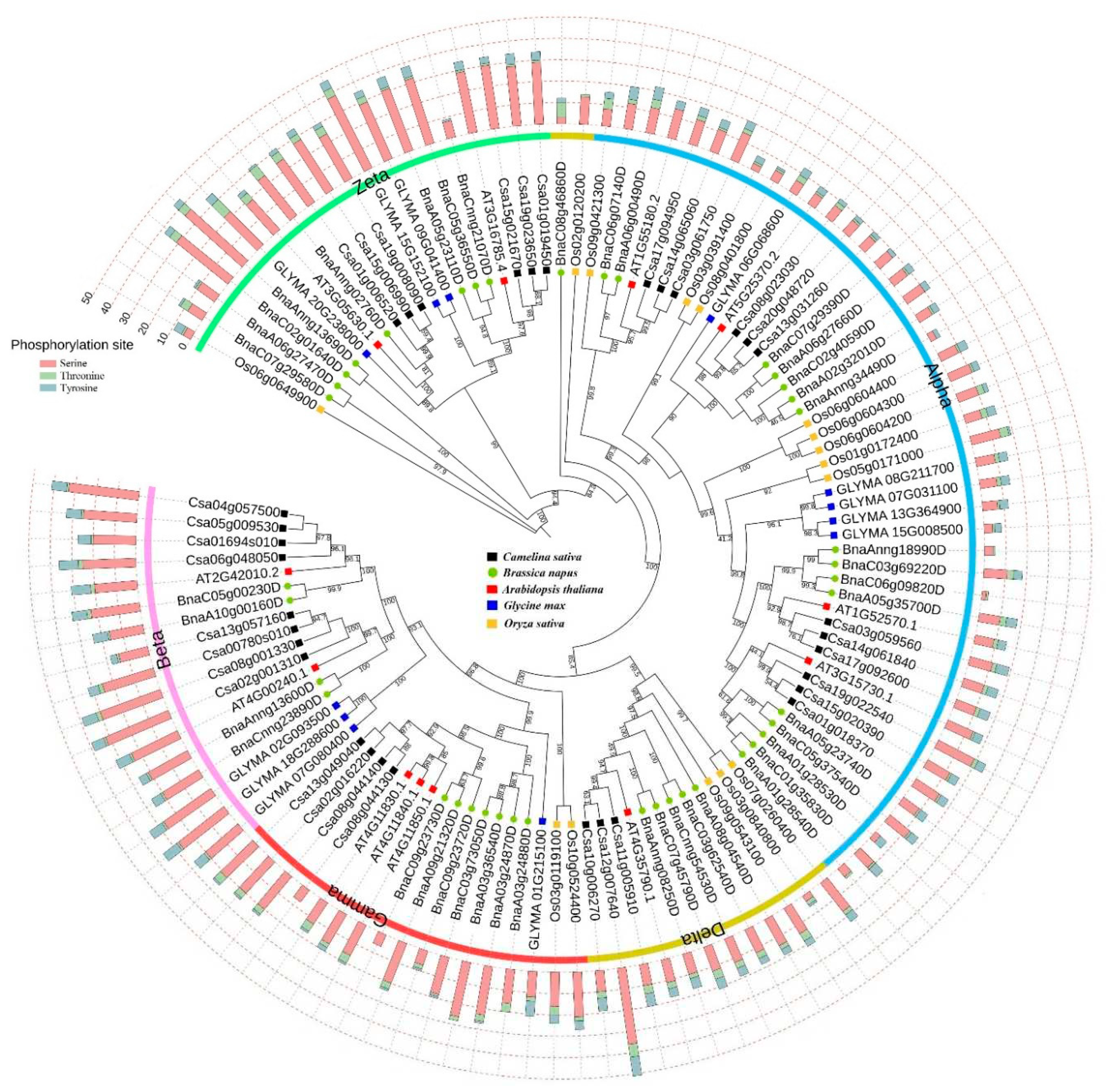
2.2. Phylogenetic Analysis of PLDs and Prediction of Phosphorylation Sites
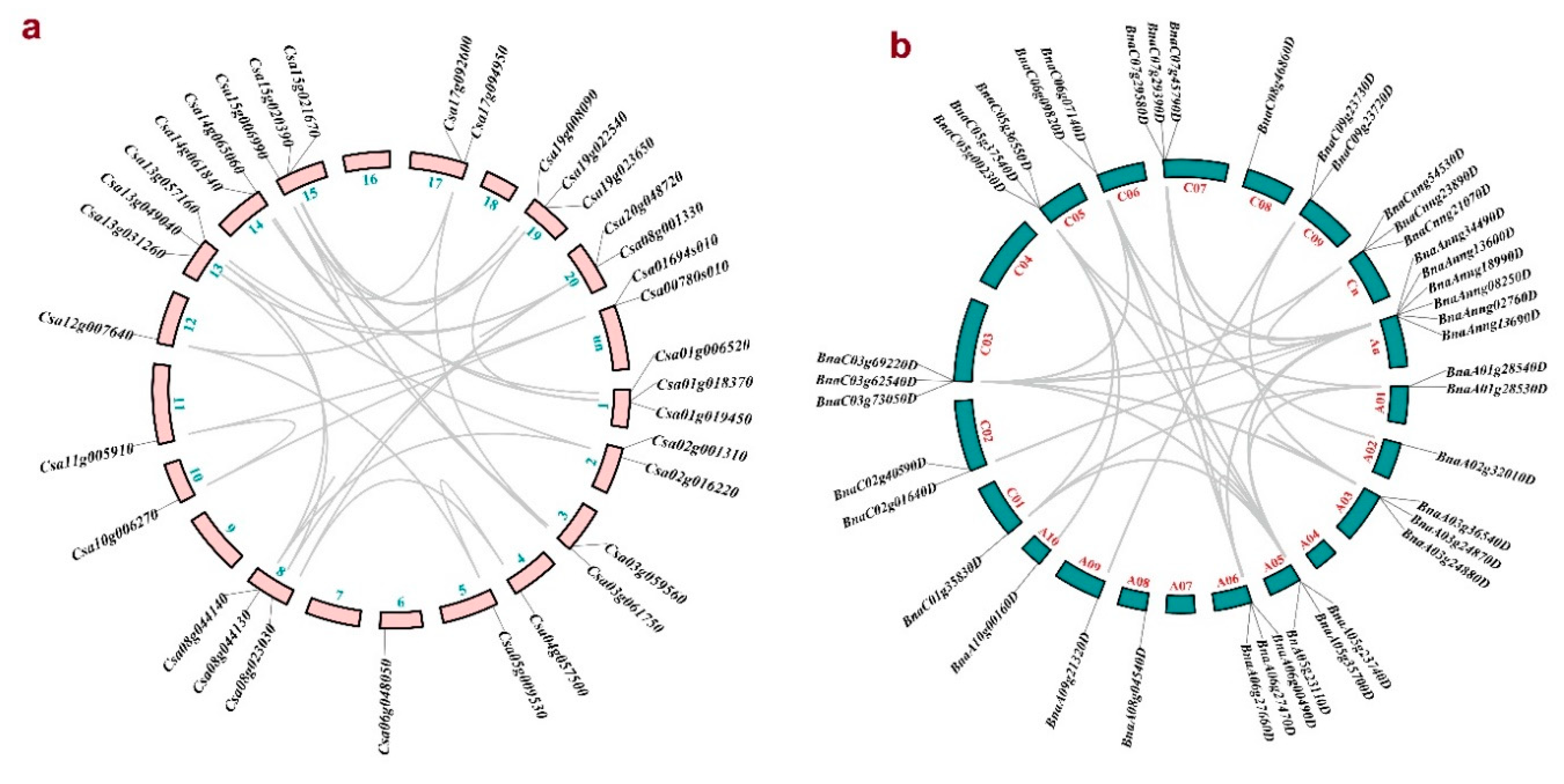
2.3. Duplication Analysis of CsPLD and BnPLD Genes
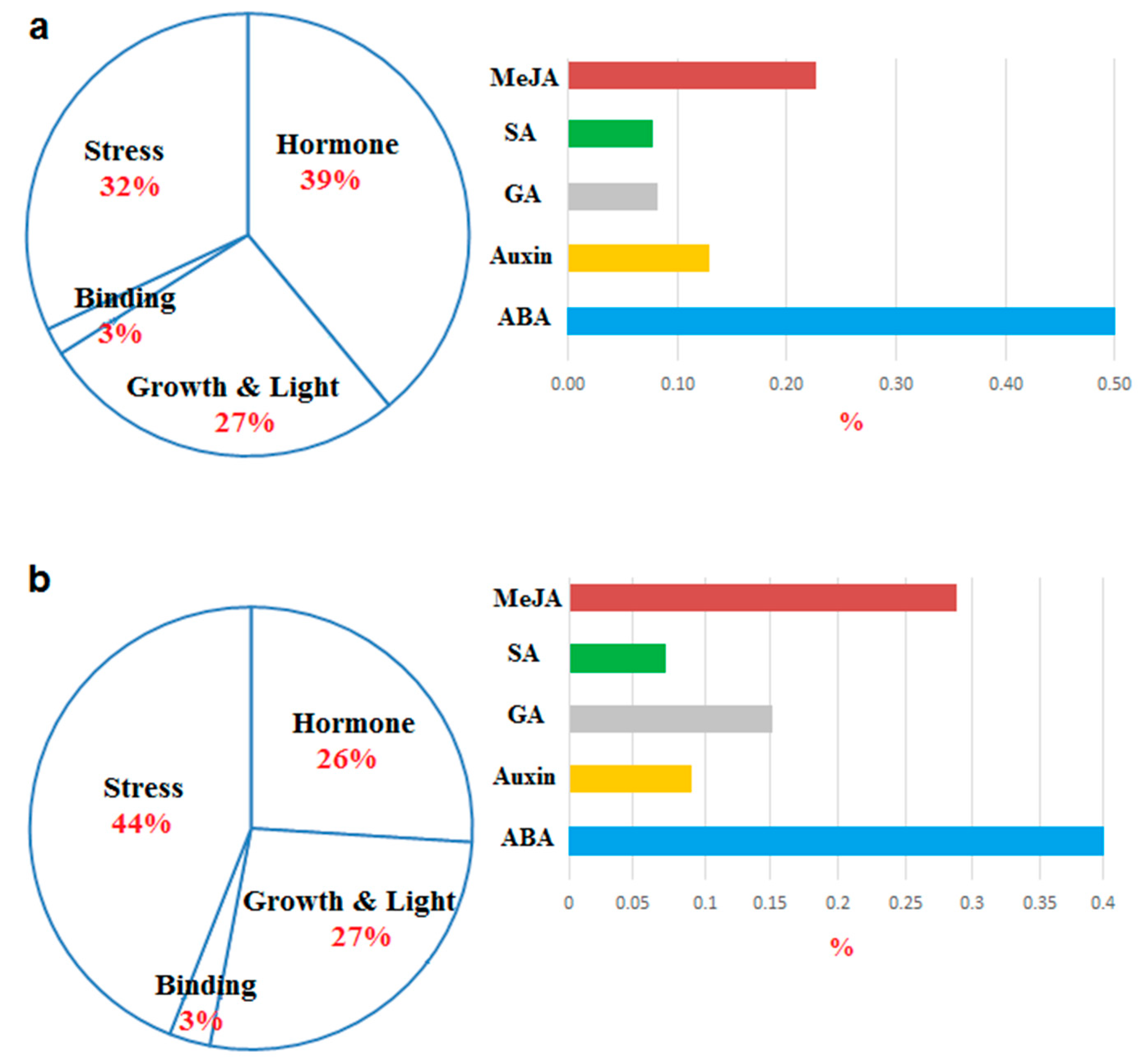
2.4. Conserved Motifs and Promoter Analysis
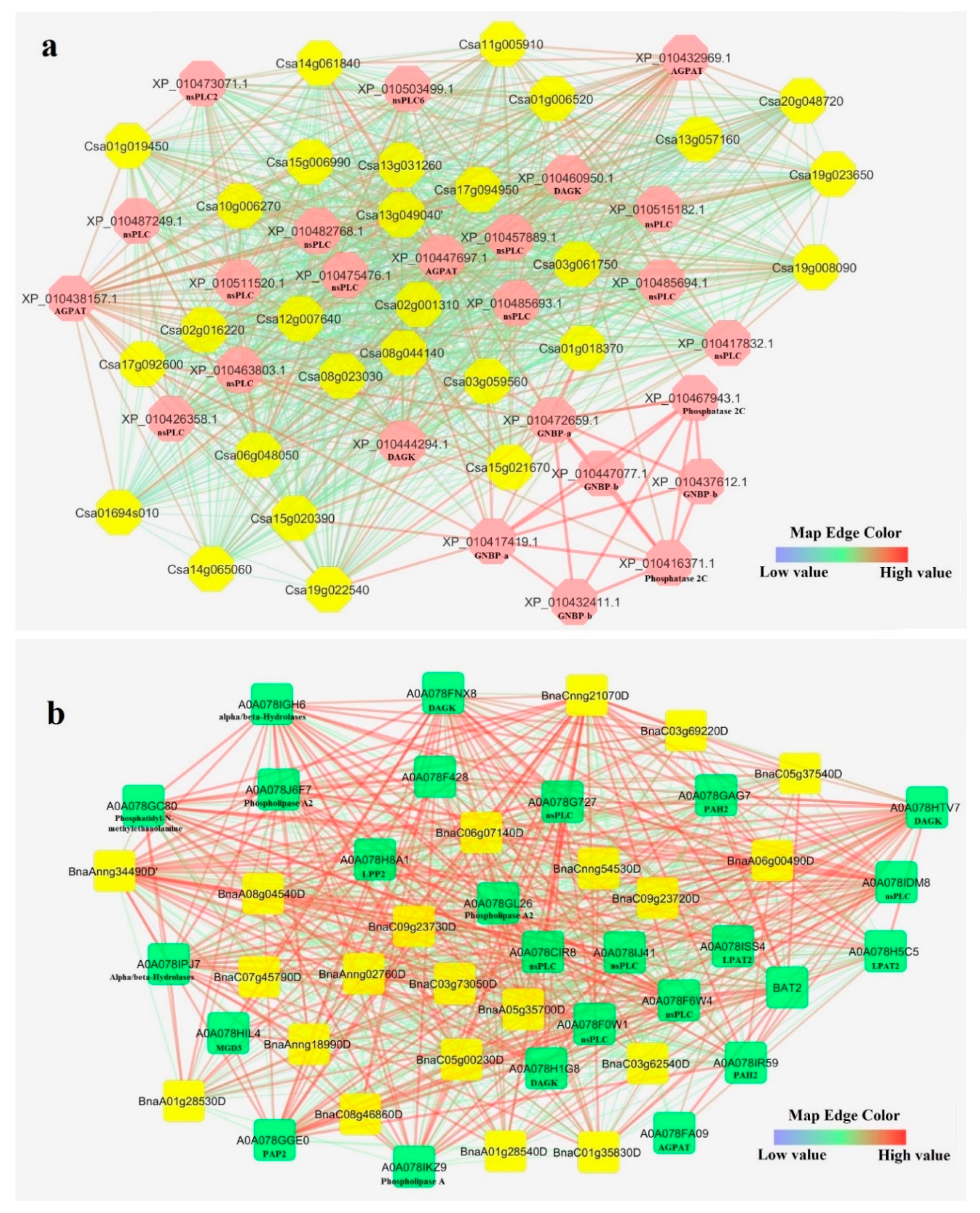
2.5. Interaction Networks and Structure Analysis of CsPLD and BnPLD
2.6. Expression Analysis of CsPLD and BnPLD in Different Tissues
2.7. Plant Materials and Treatment
2.8. Expression Analysis Using qPCR
3. Results
3.1. Physiochemical Properties of PLDs
3.2. Phylogenetic Analysis and Prediction of Post-Translational Modifications of PLD Gene Family
3.3. Conserved Motifs in PLDs
3.4. Duplication Events in PLD Gene Family
3.5. Promoter Analysis
3.6. Interaction Network of CsPLDs and BnPLDS
3.7. 3D Structure and Pocket Sites of Candidate PLD Subfamilies
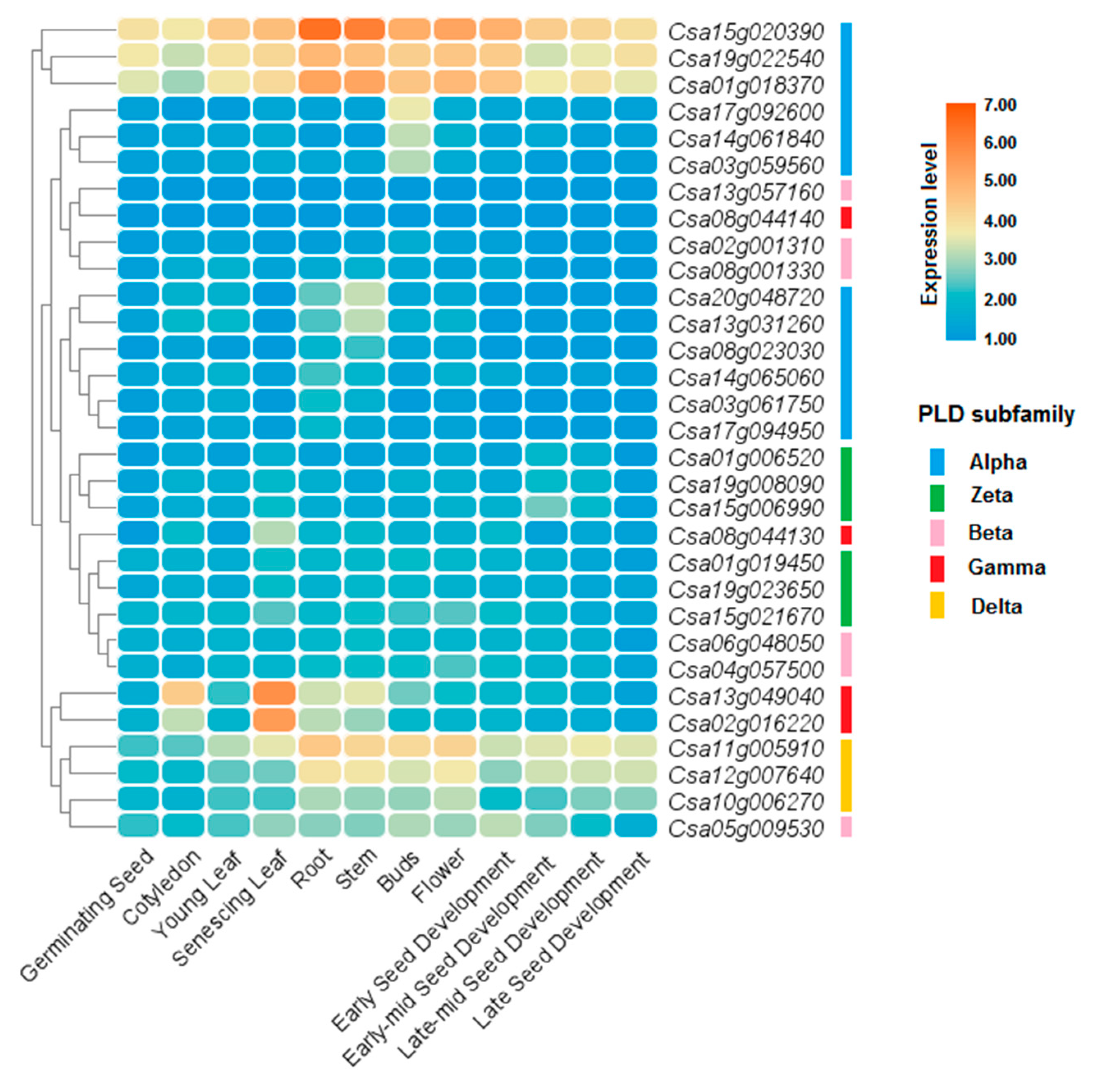
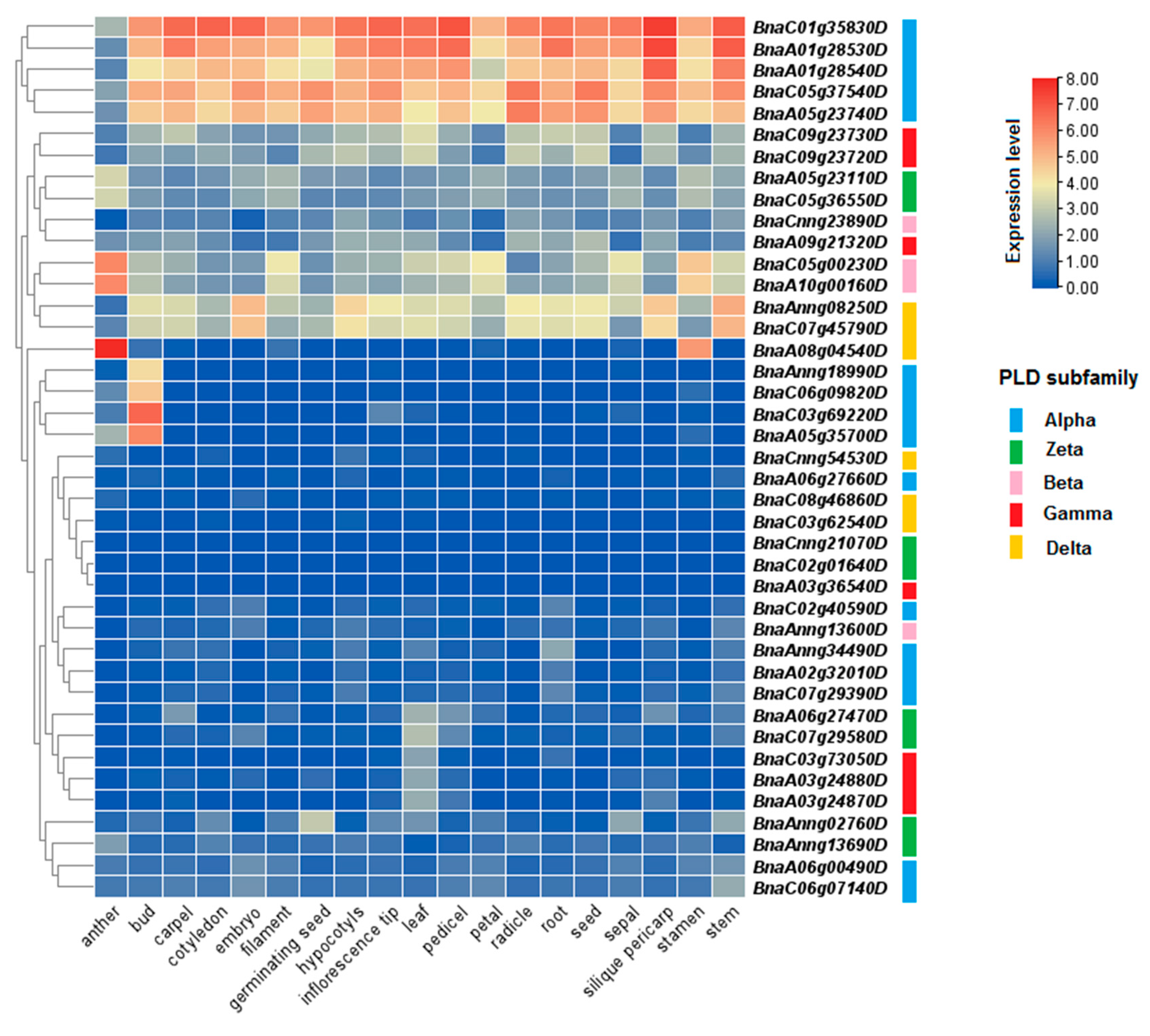
3.8. Expression Profile of PLDs in Different Tissues and Organs
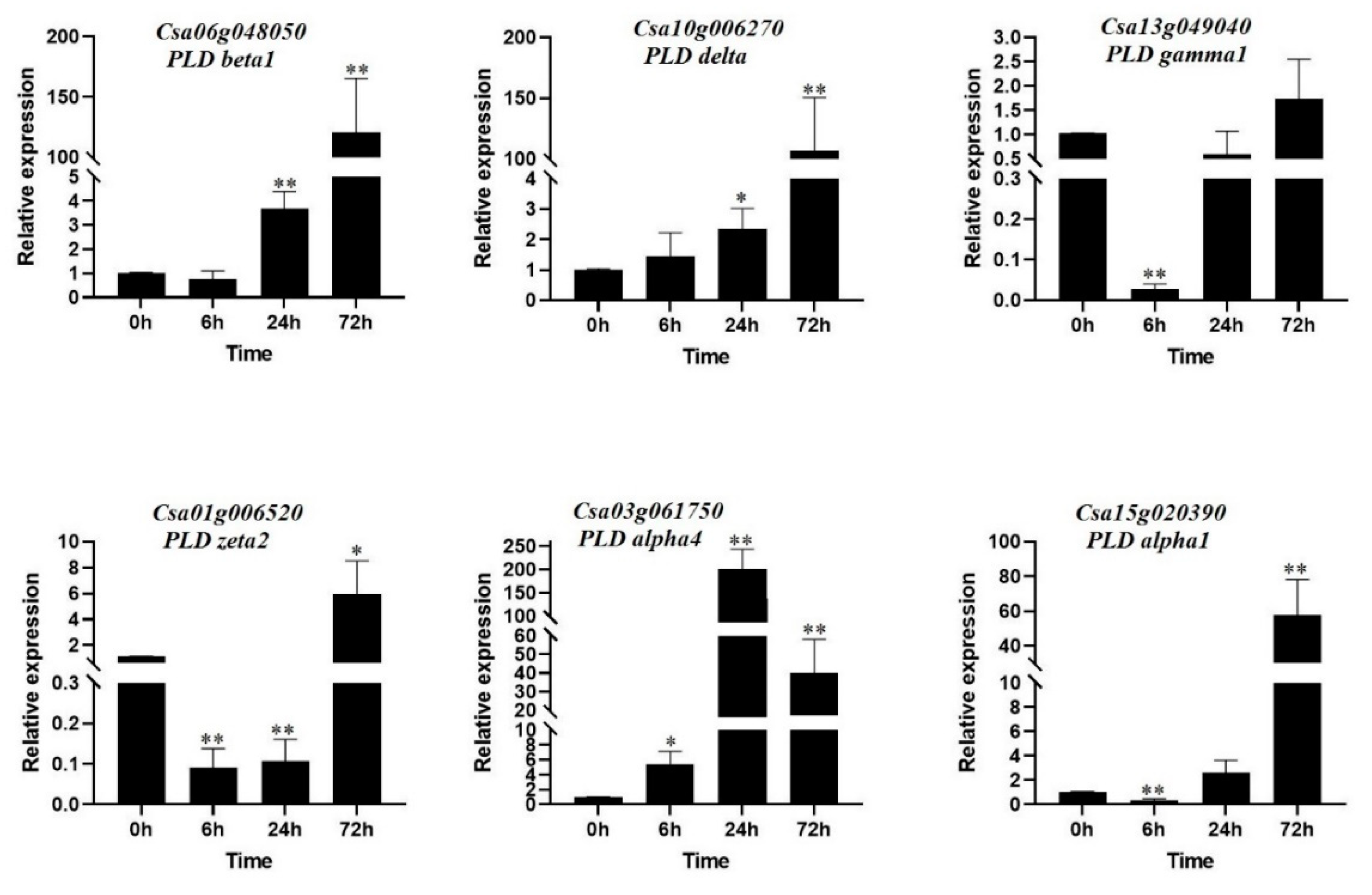
3.9. Expression Profile of CsPLD in Response to Salt Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Data Availability Statement
Conflicts of Interest
References
- Sadat, M.A.; Ullah, M.W.; Hossain, M.S.; Ahmed, B.; Bashar, K.K. Genome-wide in silico identification of phospholipase D (PLD) gene family from Corchorus capsularis and Corchorus olitorius: Reveals their responses to plant stress. J. Genet. Eng. Biotechnol. 2022, 20. [CrossRef]
- Takáč, T.; Novák, D.; Šamaj, J. Recent advances in the cellular and developmental biology of phospholipases in plants. Front. Plant Sci. 2019, 10, 432553. [CrossRef]
- Yuan, Y.; Yu, J.; Kong, L.; Zhang, W.; Hou, X.; Cui, G. Genome-wide investigation of the PLD gene family in alfalfa (Medicago sativa L.): Identification, analysis and expression. BMC Genomics 2022, 23. [CrossRef]
- Sagar, S.; Singh, A. Emerging role of phospholipase C mediated lipid signaling in abiotic stress tolerance and development in plants. Plant Cell Rep. 2021, 40, 2123–2133. [CrossRef]
- Hong, Y.; Zhao, J.; Guo, L.; Kim, S.C.; Deng, X.; Wang, G.; Zhang, G.; Li, M.; Wang, X. Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 2016, 62, 55–74. [CrossRef]
- Tang, K.; Dong, C.; Liu, J. Genome-wide analysis and expression profiling of the phospholipase D gene family in Gossypium arboreum. Sci. China. Life Sci. 2016, 59, 130–141. [CrossRef]
- Deepika, D.; Singh, A. Plant phospholipase D: Novel structure, regulatory mechanism, and multifaceted functions with biotechnological application. Crit. Rev. Biotechnol. 2022, 42, 106–124. [CrossRef]
- Iakimova, E.T.; Michaeli, R.; Woltering, E.J. Involvement of phospholipase D-related signal transduction in chemical-induced programmed cell death in tomato cell cultures. Protoplasma 2013, 250, 1169–1183. [CrossRef]
- Pleskot, R.; Li, J.; Žárský, V.; Potocký, M.; Staiger, C.J. Regulation of cytoskeletal dynamics by phospholipase D and phosphatidic acid. Trends Plant Sci. 2013, 18, 496–504. [CrossRef]
- Pejchar, P.; Sekereš, J.; Novotný, O.; Žárský, V.; Potocký, M. Functional analysis of phospholipase Dδ family in tobacco pollen tubes. Plant J. 2020, 103, 212–226. [CrossRef]
- Mancuso, S.; Marras, A.M.; Mugnai, S.; Schlicht, M.; Žársky, V.; Li, G.; Song, L.; Xue, H.W.; Baluška, F. Phospholipase Dζ2 Drives Vesicular Secretion of Auxin for Its Polar Cell-Cell Transport in the Transition Zone of the Root Apex. Plant Signal. Behav. 2007, 2, 240. [CrossRef]
- Distéfano, A.M.; Scuffi, D.; García-Mata, C.; Lamattina, L.; Laxalt, A.M. Phospholipase Dδ is involved in nitric oxide-induced stomatal closure. Planta 2012, 236, 1899–1907. [CrossRef]
- Li, G.; Lin, F.; Xue, H.W. Genome-wide analysis of the phospholipase D family in Oryza sativa and functional characterization of PLDβ1 in seed germination. Cell Res. 2007 1710 2007, 17, 881–894. [CrossRef]
- Wang, C.; Zien, C.A.; Afitlhile, M.; Welti, R.; Hildebrand, D.F.; Wang, X. Involvement of Phospholipase D in Wound-Induced Accumulation of Jasmonic Acid in Arabidopsis. Plant Cell 2000, 12, 2237. [CrossRef]
- Wang, X. Regulatory Functions of Phospholipase D and Phosphatidic Acid in Plant Growth, Development, and Stress Responses. Plant Physiol. 2005, 139, 566–573. [CrossRef]
- Wang, X. Plant phospholipases. Annu. Rev. Plant Biol. 2001, 52, 211–231.
- Liu, Q.; Zhang, C.; Yang, Y.; Hu, X. Genome-wide and molecular evolution analyses of the phospholipase D gene family in Poplar and Grape. BMC Plant Biol. 2010, 10. [CrossRef]
- Wang, X.; Xu, L.; Zheng, L. Cloning and expression of phosphatidylcholine-hydrolyzing phospholipase D from Ricinus communis L. J. Biol. Chem. 1994, 269, 20312–20317. [CrossRef]
- Du, D.; Cheng, T.; Pan, H.; Yang, W.; Wang, J.; Zhang, Q. Genome-wide identification, molecular evolution and expression analyses of the phospholipase D gene family in three Rosaceae species. Sci. Hortic. (Amsterdam). 2013, 153, 13–21. [CrossRef]
- Bolser, D.; Staines, D.M.; Pritchard, E.; Kersey, P. Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomics Data. Methods Mol. Biol. 2016, 1374, 115–140. [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, 2005; pp. 571–607. [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating Maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [CrossRef]
- Blom, N.; Sicheritz-Pontén, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [CrossRef]
- Musavizadeh, Z.; Najafi-Zarrini, H.; Kazemitabar, S.K.; Hashemi, S.H.; Faraji, S.; Barcaccia, G.; Heidari, P. Genome-Wide Analysis of Potassium Channel Genes in Rice: Expression of the OsAKT and OsKAT Genes under Salt Stress. Genes (Basel). 2021, 12, 784. [CrossRef]
- Puresmaeli, F.; Heidari, P.; Lawson, S. Insights into the Sulfate Transporter Gene Family and Its Expression Patterns in Durum Wheat Seedlings under Salinity. Genes (Basel). 2023, 14, 333. [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012, 40. [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [CrossRef]
- Li, L.; Zhang, C.; Zhang, M.; Yang, C.; Bao, Y.; Wang, D.; Chen, Y.; Chen, Q. Genome-Wide Analysis and Expression Profiling of the Phospholipase D Gene Family in Solanum tuberosum. Biology (Basel). 2021, 10. [CrossRef]
- Qin, C.; Wang, X. The Arabidopsis Phospholipase D Family. Characterization of aCalcium-Independent and Phosphatidylcholine-Selective PLDζ1 withDistinct Regulatory Domains. Plant Physiol. 2002, 128, 1057. [CrossRef]
- Chen, L.; Cao, B.; Han, N.; Tao, Y.; Zhou, S.F.; Li, W.C.; Fu, F.L. Phospholipase D family and its expression in response to abiotic stress in maize. Plant Growth Regul. 2017, 81, 197–207. [CrossRef]
- Faraji, S.; Ahmadizadeh, M.; Heidari, P. Genome-wide comparative analysis of Mg transporter gene family between Triticum turgidum and Camelina sativa. BioMetals 2021, 4. [CrossRef]
- Gao, X.; Liu, J.; Yu, L.; Yuan, B.; … X.W. Genome-wide identification and expression analysis of APX gene family in Taraxacum kok-saghyz. Acta Bot.-Boreali-Occident. Sin. 2019, 39, 1935–1942.
- Juretic, N.; Hoen, D.R.; Huynh, M.L.; Harrison, P.M.; Bureau, T.E. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005, 15, 1292–1297. [CrossRef]
- Faraji, S.; Filiz, E.; Kazemitabar, S.K.; Vannozzi, A.; Palumbo, F.; Barcaccia, G.; Heidari, P. The AP2/ERF Gene Family in Triticum durum: Genome-Wide Identification and Expression Analysis under Drought and Salinity Stresses. Genes (Basel). 2020, 11, 1464. [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49–e49. [CrossRef]
- Heidari, P.; Hasanzadeh, S.; Faraji, S.; Ercisli, S.; Mora-Poblete, F. Genome-Wide Characterization of the Sulfate Transporter Gene Family in Oilseed Crops: Camelina sativa and Brassica napus. Plants 2023, 12, 628. [CrossRef]
- Liu, B.; Yao, L.; Wang, W.; Gao, J.; Chen, F.; Wang, S.; Xu, Y.; Tang, L.; Jia, Y. Molecular cloning and characterization of phospholipase D from Jatropha curcas. Mol. Biol. Rep. 2010, 37, 939–946. [CrossRef]
- McDermott, M.I.; McDermott, M.; Wakelam, M.J.; Morris, A.J. Phospholipase d. cdnsciencepub.comM McDermott, MJO Wakelam, AJ MorrisBiochemistry Cell Biol. 2004•cdnsciencepub.com 2004, 82, 225–253.
- Ben Othman, A.; Ellouzi, H.; Planchais, S.; Vos, D. De; Faiyue, B. Phospholipases Df1 and Df2 have distinct roles in growth and antioxidant systems in Arabidopsis thaliana responding to salt stress. Planta 2017, 246, 721–735. [CrossRef]
- Guo L., Devaiah S.P., Narasimhan R., Pan X., Zhang Y., Zhang W., et al. Cytosolic Glyceraldehyde-3-Phosphate Dehydrogenases Interact with Phospholipase DΔ to Transduce Hydrogen Peroxide Signals in the Arabidopsis Response to. Plant Cell 2012, 24, 2200–2212. [CrossRef]
- Wang, P.; Shen, L.; Guo, J.; Jing, W.; Qu, Y.; Li, W.; … R.B.-T.P.; 2019, undefined Phosphatidic acid directly regulates PINOID-dependent phosphorylation and activation of the PIN-FORMED2 auxin efflux transporter in response to salt stress. Acad. Wang, L Shen, J Guo, W Jing, Y Qu, W Li, R Bi, W Xuan, Q Zhang, W ZhangThe Plant Cell, 2019•academic.oup.com. [CrossRef]
- Ahmadizadeh, M.; Rezaee, S.; Heidari, P. Genome-wide characterization and expression analysis of fatty acid desaturase gene family in Camelina sativa. Gene Reports 2020, 21, 100894. [CrossRef]
- Yaghobi, M.; Heidari, P. Genome-Wide Analysis of Aquaporin Gene Family in Triticum turgidum and Its Expression Profile in Response to Salt Stress. Genes (Basel). 2023, 14, 202. [CrossRef]
- Arab, M.; Najafi Zarrini, H.; Nematzadeh, G.; Heidari, P.; Hashemipetroudi, S.H.; Kuhlmann, M. Comprehensive Analysis of Calcium Sensor Families, CBL and CIPK, in Aeluropus littoralis and Their Expression Profile in Response to Salinity. Genes (Basel). 2023, 14, 753. [CrossRef]
- Darwish, E.; Testerink, C.; Khalil, M.; El-Shihy, O.; Munnik, T. Phospholipid Signaling Responses in Salt-Stressed Rice Leaves. Plant Cell Physiol. 2009, 50, 986–997. [CrossRef]
- Li, W.; Li, M.; Zhang, W.; Welti, R.; Wang, X. The plasma membrane-bound phospholipase Ddelta enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2004, 22, 427–433. [CrossRef]


| Plant | Protein length | Exon number | MW (KDa) | pI | GRAVY | Stability |
|---|---|---|---|---|---|---|
| Camelina sativa | 350 - 1596 | 3 - 27 | 40.02 - 181.62 | 5.57 - 9.09 | -0.655 , -0.316 | 33% |
| Brassica napus | 213 - 1465 | 2 - 22 | 24.66 - 164.78 | 5.10 - 10.30 | -0.682 , -0.247 | 61% |
| Glycine max | 711 - 1126 | 3 - 22 | 81.22 - 127.63 | 5.48 - 7.65 | -0.532 , -0.374 | 25% |
| Arabidopsis thaliana | 762 - 1108 | 4 - 20 | 86.99 - 124.73 | 5.53 - 8.36 | -0.555 , -0.343 | 75% |
| Oryza sativa | 355 - 1046 | 1 - 12 | 39.31 - 116.93 | 5.56 - 11.23 | -0.622 , -0.147 | 67% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





