Submitted:
05 September 2023
Posted:
06 September 2023
You are already at the latest version
Abstract
Keywords:
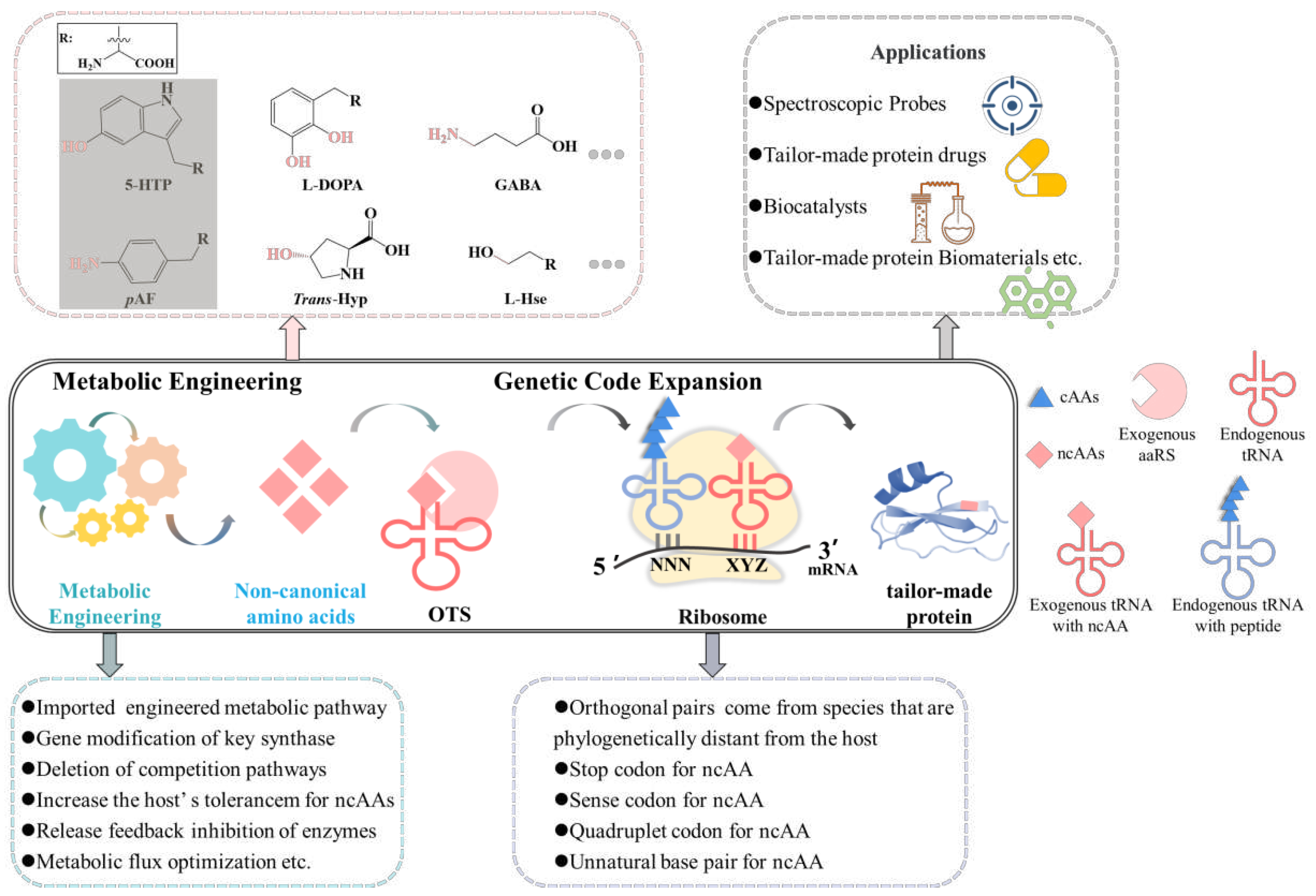
1. Introduction
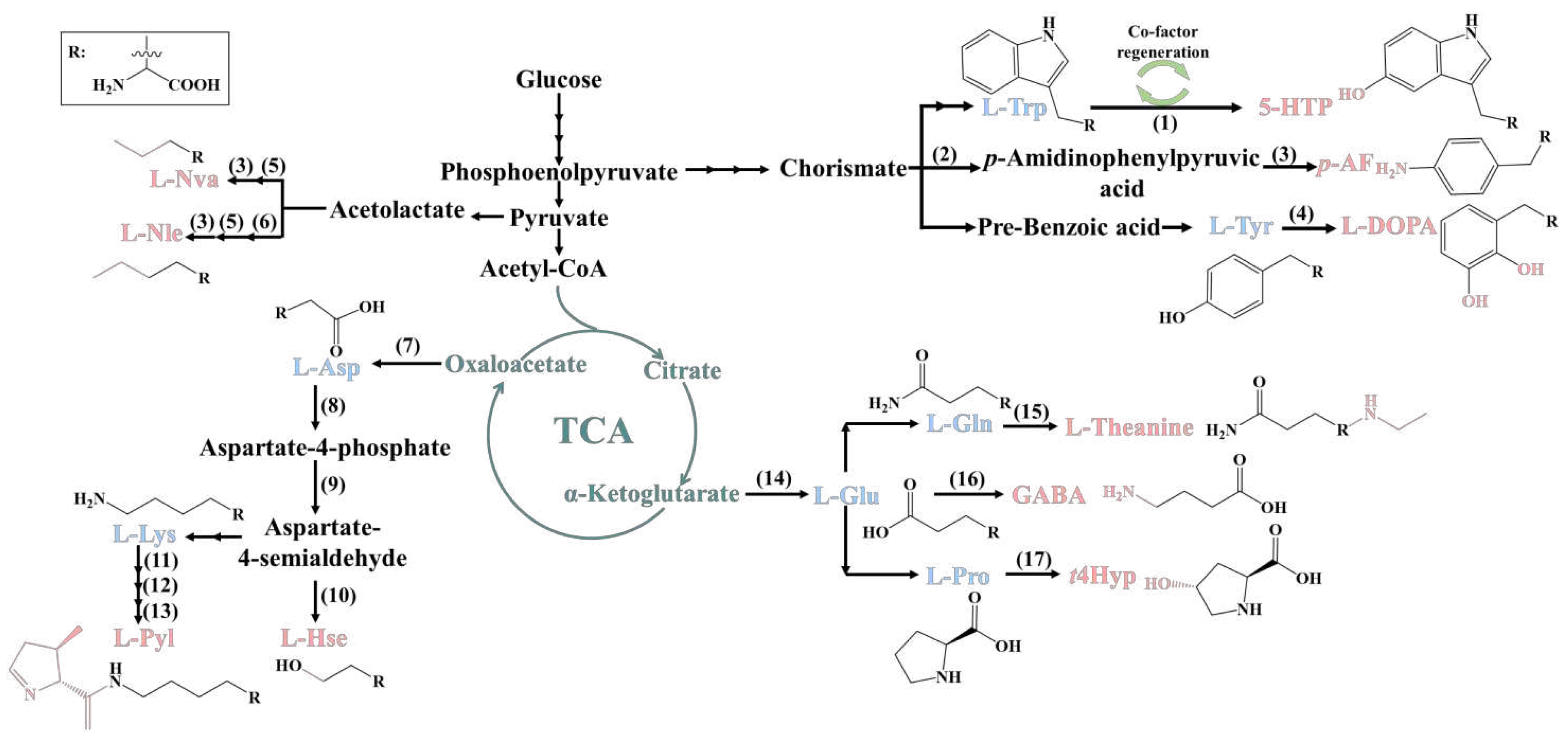
2. Biosynthesis of ncAAs
2.1. 5-Hydroxytryptophan
2.2. L-Homoserine
2.3. Trans-4-hydroxyproline
2.4. Other ncAAs
3. Methods for ncAAs incorporation into proteins
3.1. GCE methods for ncAAs incorporation into proteins
3.1.1. GCE based on stop codon suppression (SCS)
3.1.2. GCE based on synonymous codon compression
3.1.3. GCE based on other approaches
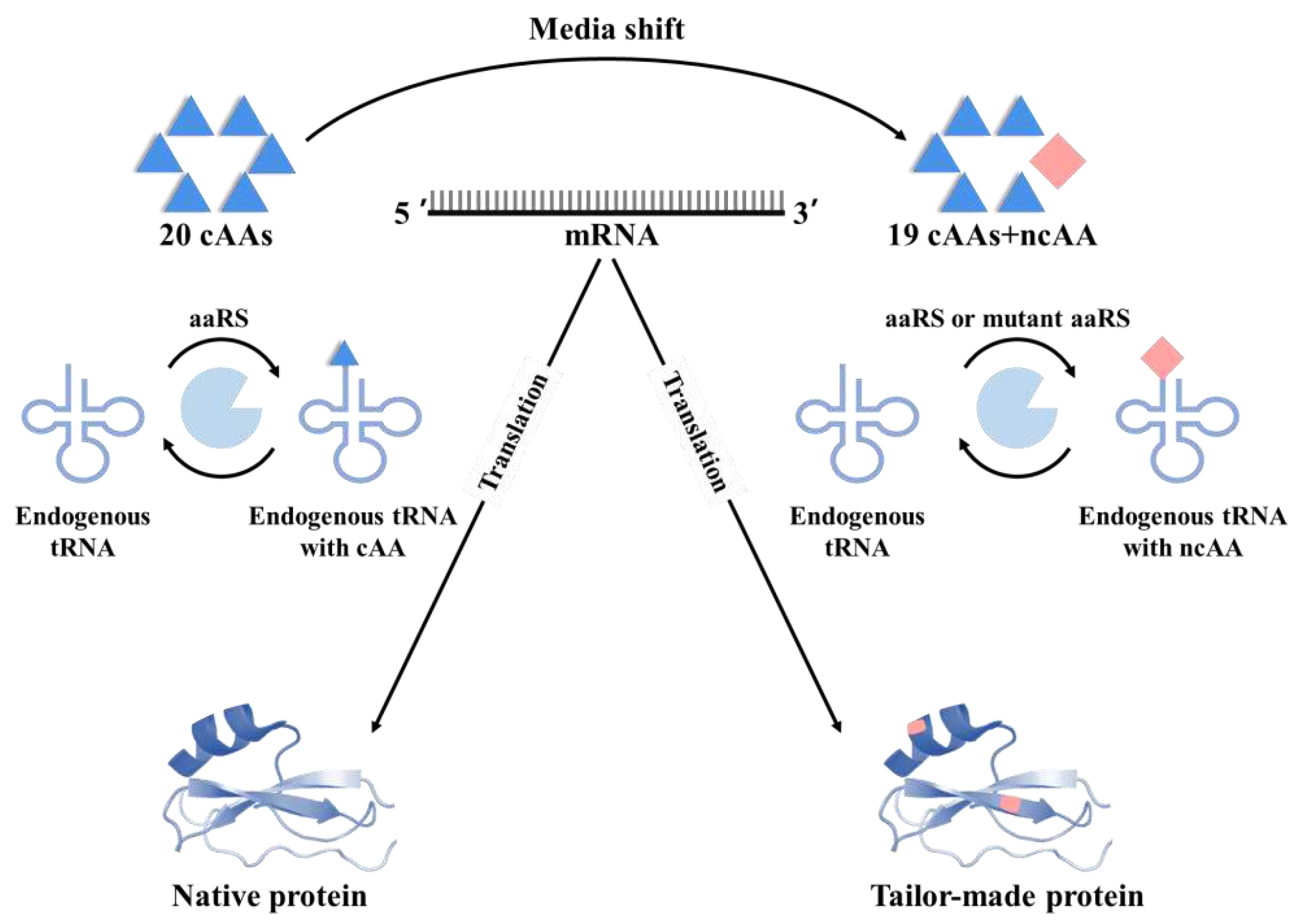
3.2. Selective pressure incorporation (SPI)
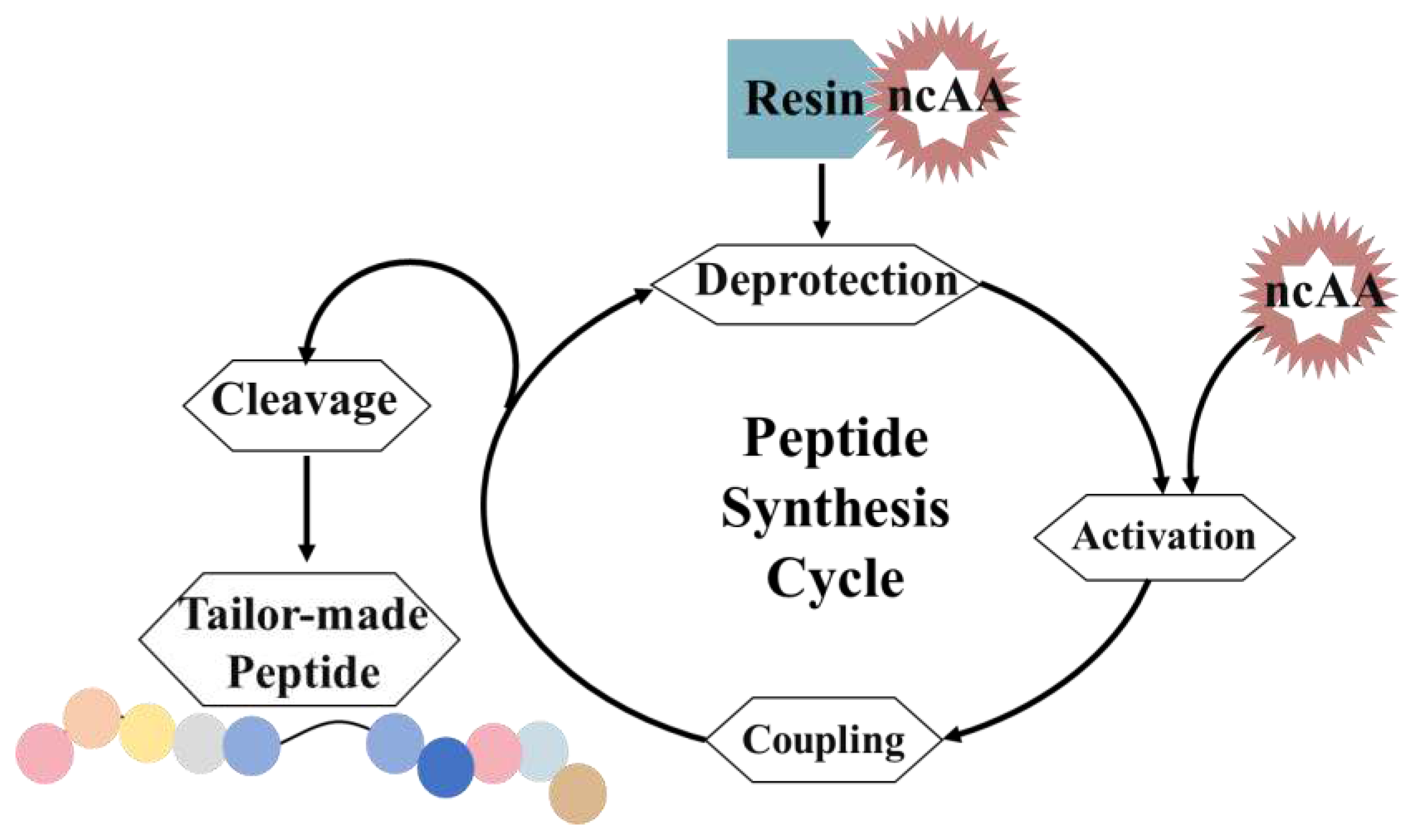
3.3. Solid-phase peptide synthesis (SPPS)
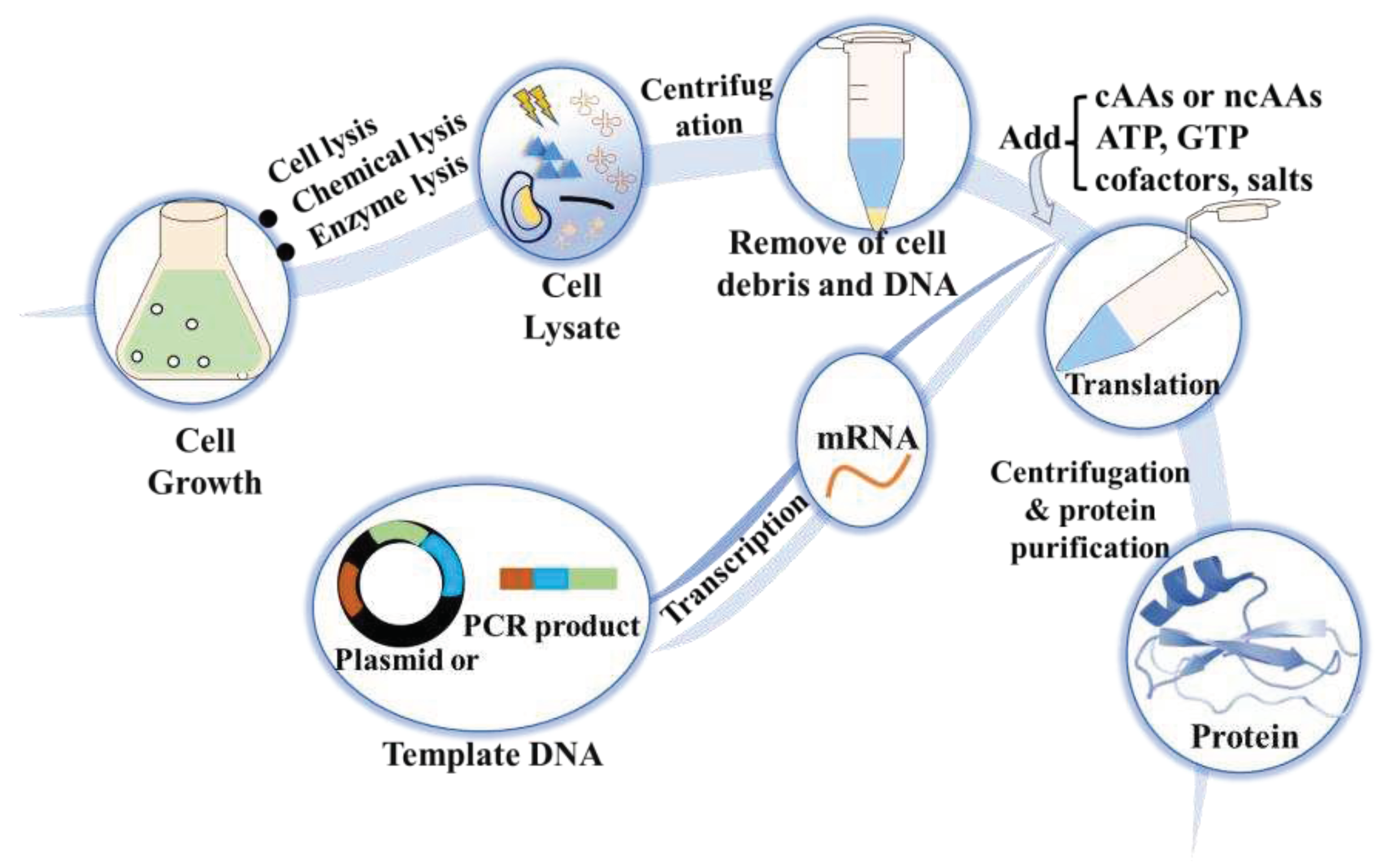
3.4. Cell-free protein synthesis (CFPS)
3.5. Other methods for the incorporation of ncAAs
4. The applications of tailor-made proteins
4.1. Tailor-made protein materials
4.2. Tailor-made protein probes
4.3. Tailor-made protein drugs
4.4. Applications of ncAAs in vaccine
5. Perspectives
Funding
References
- Wang, L. Genetically encoding new bioreactivity. N Biotechnol 2017, 38, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Des Soye, B.J.; Patel, J.R.; Isaacs, F.J.; Jewett, M.C. Repurposing the translation apparatus for synthetic biology. Curr Opin Chem Biol 2015, 28, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Macek, B.; Forchhammer, K.; Hardouin, J.; Weber-Ban, E.; Grangeasse, C.; Mijakovic, I. Protein post-translational modifications in bacteria. Nat Rev Microbiol 2019, 17, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Bu, N.; Lu, Y. Recent advances in cell-free unnatural protein synthesis. Sheng Wu Gong Cheng Xue Bao 2018, 34, 1371–1385. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tang, J.; Wang, L.; Tian, Z.; Cardenas, A.; Fang, X.; Chatterjee, A.; Xiao, H. Creation of Bacterial cells with 5-Hydroxytryptophan as a 21st Amino Acid Building Block. Chem 2020, 6, 2717–2727. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.-F.; Liu, Z.-Q.; Jin, L.-Q.; Tang, X.-L.; Shen, Z.-Y.; Yin, H.-H.; Zheng, Y.-G. Metabolic engineering of Escherichia coli for microbial production of L-methionine. Biotechnol Bioeng 2017, 114, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.-K.; Kim, J.H.; Yoon, J.H.; Park, H.-M.; Choi, S.J.; Song, G.H.; Lee, J.C.; Yang, Y.-L.; Shin, H.K.; Kim, J.N.; et al. O-Succinyl-L-homoserine-based C4-chemical production: succinic acid, homoserine lactone, γ-butyrolactone, γ-butyrolactone derivatives, and 1,4-butanediol. J Ind Microbiol Biotechnol 2014, 41, 1517–1524. [Google Scholar] [CrossRef]
- Kang, Z.; Zhang, J.; Zhou, J.; Qi, Q.; Du, G.; Chen, J. Recent advances in microbial production of δ-aminolevulinic acid and vitamin B12. Biotechnol Adv 2012, 30, 1533–1542. [Google Scholar] [CrossRef]
- Zhu, H.Q.; Tang, X.L.; Zheng, R.C.; Zheng, Y.G. Recent advancements in enzyme engineering via site-specific incorporation of unnatural amino acids. World J Microbiol Biotechnol 2021, 37, 213. [Google Scholar] [CrossRef] [PubMed]
- Smolskaya, S.; Andreev, Y.A. Site-Specific Incorporation of Unnatural Amino Acids into Escherichia coli Recombinant Protein: Methodology Development and Recent Achievement. Biomolecules 2019, 9, 255. [Google Scholar] [CrossRef]
- Narancic, T.; Almahboub, S.A.; O’Connor, K.E. Unnatural amino acids: production and biotechnological potential. world j microbiol biotechnol 2019, 35, 1–11. [Google Scholar] [CrossRef]
- Zou, H.; Li, L.; Zhang, T.; Shi, M.; Zhang, N.; Huang, J.; Xian, M. Biosynthesis and biotechnological application of non-canonical amino acids: complex and unclear. Biotechnol Adv 2018, 36, 1917–1927. [Google Scholar] [CrossRef] [PubMed]
- Karbalaei-Heidari, H.R.; Budisa, N. Combating antimicrobial resistance with new-to-nature lanthipeptides created by genetic code expansion. Front Microbiol 2020, 11, 590522. [Google Scholar] [CrossRef]
- Tsubogo, T.; Kano, Y.; Ikemoto, K.; Yamashita, Y.; Kobayashi, S. Synthesis of optically active, unnatural α-substituted glutamic acid derivatives by a chiral calcium-catalyzed 1,4-addition reaction. Tetrahedron Asymmetry 2010, 21, 1221–1225. [Google Scholar] [CrossRef]
- Li, B.; Zhang, J.; Xu, Y.; Yang, X.; Li, L. Improved synthesis of unnatural amino acids for peptide stapling. Tetrahedron Lett 2017, 58, 2374–2377. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, D.; Xu, P.; Dong, X.; Wang, X.; Dai, Z. The asymmetric alkylation reaction of glycine derivatives catalyzed by the novel chiral phase transfer catalysts. Tetrahedron Lett 2015, 56, 1067–1071. [Google Scholar] [CrossRef]
- Anderhuber, N.; Fladischer, P.; Gruber-Khadjawi, M.; Mairhofer, J.; Striedner, G.; Wiltschi, B. High-level biosynthesis of norleucine in E. coli for the economic labeling of proteins. J Biotechnol 2016, 235, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T.; Huisman, G.W.; Kazlauskas, R.J.; Lutz, S.; Moore, J.C.; Robins, K. Engineering the third wave of biocatalysis. Nature 2012, 485, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, W.; Shi, F.; Huang, L.; Lian, J.; Qu, L.; Cai, J.; Xu, Z. Metabolic pathway engineering for high-level production of 5-hydroxytryptophan in Escherichia coli. Metab Eng 2018, 48, 279–287. [Google Scholar] [CrossRef]
- Xu, D.; Fang, M.; Wang, H.; Huang, L.; Xu, Q.; Xu, Z. Enhanced production of 5-hydroxytryptophan through the regulation of L-tryptophan biosynthetic pathway. Appl Microbiol Biotechnol 2020, 104, 2481–2488. [Google Scholar] [CrossRef]
- Lin, Y.; Sun, X.; Yuan, Q.; Yan, Y. Engineering bacterial phenylalanine 4-hydroxylase for microbial synthesis of human neurotransmitter precursor 5-hydroxytryptophan. ACS Synth Biol 2014, 3, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Mora-Villalobos, J.A.; Zeng, A.P. Synthetic pathways and processes for effective production of 5-hydroxytryptophan and serotonin from glucose in Escherichia coli. J Biol Eng 2018, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mora-Villalobos, J.A.; Zeng, A.P. Protein and pathway engineering for the biosynthesis of 5-hydroxytryptophan in Escherichia coli. Eng Life Sci 2017, 17, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Mu, Q.; Zhang, S.; Mao, X.; Tao, Y.; Yu, B. Highly efficient production of L-homoserine in Escherichia coli by engineering a redox balance route. Metab Eng 2021, 67, 321–329. [Google Scholar] [CrossRef]
- Sun, B.Y.; Wang, F.Q.; Zhao, J.; Tao, X.Y.; Liu, M.; Wei, D.Z. Engineering Escherichia coli for L-homoserine production. J Basic Microbiol 2022. [CrossRef]
- Li, N.; Xu, S.; Du, G.; Chen, J.; Zhou, J. Efficient production of L-homoserine in Corynebacterium glutamicum ATCC 13032 by redistribution of metabolic flux. Biochem Eng J 2020, 161, 107665. [Google Scholar] [CrossRef]
- Li, N.; Zeng, W.; Zhou, J.; Xu, S. O-Acetyl-L-homoserine production enhanced by pathway strengthening and acetate supplementation in Corynebacterium glutamicum. Biotechnol Biofuels Bioprod 2022, 15, 27. [Google Scholar] [CrossRef]
- Cai, M.; Zhao, Z.; Li, X.; Xu, Y.; Xu, M.; Rao, Z. Development of a nonauxotrophic L-homoserine hyperproducer in Escherichia coli by systems metabolic engineering. Metab Eng 2022, 73, 270–279. [Google Scholar] [CrossRef]
- Huang, J.-F.; Zhang, B.; Shen, Z.-Y.; Liu, Z.-Q.; Zheng, Y.-G. Metabolic engineering of E. coli for the production of O-succinyl-L-homoserine with high yield. 3 Biotech 2018, 8, 310. [Google Scholar] [CrossRef]
- Li, H.; Wang, B.; Zhu, L.; Cheng, S.; Li, Y.; Zhang, L.; Ding, Z.Y.; Gu, Z.H.; Shi, G.Y. Metabolic engineering of Escherichia coli W3110 for L-homoserine production. Process Biochem 2016, 51, 1973–1983. [Google Scholar] [CrossRef]
- Yi, Y.; Sheng, H.; Li, Z.; Ye, Q. Biosynthesis of trans-4-hydroxyproline by recombinant strains of Corynebacterium glutamicum and Escherichia coli. BMC Biotechnol 2014, 14, 44. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Shang, X.; Wang, B.; Hu, Q.; Liu, S.; Wen, T. Reconstruction of tricarboxylic acid cycle in Corynebacterium glutamicum with a genome-scale metabolic network model for trans-4-hydroxyproline production. Biotechnol Bioeng 2019, 116, 99–109. [Google Scholar] [CrossRef]
- Long, M.; Xu, M.; Ma, Z.; Pan, X.; You, J.; Hu, M.; Shao, Y.; Yang, T.; Zhang, X.; Rao, Z. Significantly enhancing production of trans-4-hydroxy-l-proline by integrated system engineering in Escherichia coli. Sci Adv 2020, 6, eaba2383. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-C.; Liu, J.; Zhao, J.; Ni, X.-M.; Zheng, P.; Guo, X.; Sun, C.-M.; Sun, J.-B.; Ma, Y.-H. Efficient production of trans-4-hydroxy-L-proline from glucose using a new trans-proline 4-hydroxylase in Escherichia coli. J Biosci 2018, 126, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Ghavami, S.; Pandi, A. CRISPR interference and its applications. Prog Mol Biol Transl Sci 2021, 180, 123–140. [Google Scholar] [CrossRef]
- James, C.M.; Ferguson, T.K.; Leykam, J.F.; Krzycki, J.A. The Amber Codon in the Gene Encoding the Monomethylamine Methyltransferase Isolated from Methanosarcina barkeri Is Translated as a Sense Codon*. J Biol Chem 2001, 276, 34252–34258. [Google Scholar] [CrossRef]
- Gaston, M.A.; Zhang, L.; Green-Church, K.B.; Krzycki, J.A. The complete biosynthesis of the genetically encoded amino acid pyrrolysine from lysine. Nature 2011, 471, 647–650. [Google Scholar] [CrossRef]
- Longstaff, D.G.; Larue, R.C.; Faust, J.E.; Mahapatra, A.; Zhang, L.; Green-Church, K.B.; Krzycki, J.A. A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine. Proc Natl Acad Sci USA 2007, 104, 1021–1026. [Google Scholar] [CrossRef]
- Srinivasan, G.; James, C.M.; Krzycki, J.A. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science 2002, 296, 1459–1462. [Google Scholar] [CrossRef]
- Ho, J.M.L.; Miller, C.A.; Smith, K.A.; Mattia, J.R.; Bennett, M.R. Improved pyrrolysine biosynthesis through phage assisted non-continuous directed evolution of the complete pathway. Nat Commun 2021, 12, 3914. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, H.; Liu, L.; Ko, K.; Kim, I. Screening of gamma-aminobutyric acid-producing lactic acid bacteria and the characteristic of glutamate decarboxylase from Levilactobacillus brevis F109-MD3 isolated from kimchi. J Appl Microbiol 2022, 132, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, X.; Shu, G.; Ma, W. Screening of gamma-aminobutyric acid-producing lactic acid bacteria and its application in Monascus-fermented rice production. Acta Sci Pol Technol Aliment 2020, 19, 387–394. [Google Scholar] [CrossRef]
- Kim, N.Y.; Kim, S.K.; Ra, C.H. Evaluation of gamma-aminobutyric acid (GABA) production by Lactobacillus plantarum using two-step fermentation. Bioprocess Biosyst Eng 2021, 44, 2099–2108. [Google Scholar] [CrossRef] [PubMed]
- Diez-Gutiérrez, L.; Vicente, L.S.; Sáenz, J.; Esquivel, A.; Barron, L.J.R.; Chávarri, M. Biosynthesis of gamma-aminobutyric acid by Lactiplantibacillus plantarum K16 as an alternative to revalue agri-food by-products. Sci Rep 2022, 12, 18904. [Google Scholar] [CrossRef] [PubMed]
- Praveschotinunt, P.; Dorval Courchesne, N.-M.; den Hartog, I.; Lu, C.; Kim, J.J.; Nguyen, P.Q.; Joshi, N.S. Tracking of Engineered Bacteria In Vivo Using Nonstandard Amino Acid Incorporation. ACS Synth Biol 2018, 7, 1640–1650. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, D.L.; Tsuji, P.A.; Carlson, B.A.; Gladyshev, V.N. Selenium and selenocysteine: roles in cancer, health, and development. Trends Biochem Sci 2014, 39, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.L.; Simonović, M. Synthesis and decoding of selenocysteine and human health. Croat Med J 2012, 53, 535–550. [Google Scholar] [CrossRef]
- Long, M.R.; Ong, W.K.; Reed, J.L. Computational methods in metabolic engineering for strain design. Curr Opin Biotechnol 2015, 34, 135–141. [Google Scholar] [CrossRef]
- Masuo, S.; Zhou, S.; Kaneko, T.; Takaya, N. Bacterial fermentation platform for producing artificial aromatic amines. Sci Rep 2016, 6, 25764. [Google Scholar] [CrossRef]
- Zomorrodi, A.R.; Hemez, C.; Arranz-Gibert, P.; Wu, T.; Isaacs, F.J.; Segrè, D. Computational design and engineering of an Escherichia coli strain producing the nonstandard amino acid para-aminophenylalanine. iScience 2022, 25, 104562. [Google Scholar] [CrossRef]
- Hecht, S.M.; Alford, B.L.; Kuroda, Y.; Kitano, S. "Chemical aminoacylation" of tRNA's. J Biol Chem 1978, 253, 4517–4520. [Google Scholar] [CrossRef] [PubMed]
- Gamper, H.; Hou, Y.M. A Label-Free Assay for Aminoacylation of tRNA. Genes 2020, 11. [Google Scholar] [CrossRef]
- Noren, C.J.; Anthony-Cahill, S.J.; Griffith, M.C.; Schultz, P.G. A general method for site-specific incorporation of unnatural amino acids into proteins. Science 1989, 244, 182–188. [Google Scholar] [CrossRef]
- Pastore, A.J.; Ficaretta, E.; Chatterjee, A.; Davidson, V.L. Substitution of the sole tryptophan of the cupredoxin, amicyanin, with 5-hydroxytryptophan alters fluorescence properties and energy transfer to the type 1 copper site. J Inorg Biochem 2022, 234, 111895. [Google Scholar] [CrossRef]
- Lajoie, M.J.; Rovner, A.J.; Goodman, D.B.; Aerni, H.-R.; Haimovich, A.D.; Kuznetsov, G.; Mercer, J.A.; Wang, H.H.; Carr, P.A.; Mosberg, J.A.; et al. Genomically recoded organisms expand biological functions. Science 2013, 342, 357–360. [Google Scholar] [CrossRef]
- Yi, H.; Zhang, J.; Ke, F.; Guo, X.; Yang, J.; Xie, P.; Liu, L.; Wang, Q.; Gao, X. Comparative Analyses of the Transcriptome and Proteome of Escherichia coli C321.△A and Further Improving Its Noncanonical Amino Acids Containing Protein Expression Ability by Integration of T7 RNA Polymerase. Front Microbiol 2021, 12, 744284. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.A.; Mozhdehi, D.; Dzuricky, M.J.; Isaacs, F.J.; Brustad, E.M.; Chilkoti, A. Active targeting of cancer cells by nanobody decorated polypeptide micelle with bio-orthogonally conjugated drug. Nano Lett 2019, 19, 247–254. [Google Scholar] [CrossRef]
- Wang, K.; Fredens, J.; Brunner, S.F.; Kim, S.H.; Chia, T.; Chin, J.W. Defining synonymous codon compression schemes by genome recoding. Nature 2016, 539, 59–64. [Google Scholar] [CrossRef]
- Fredens, J.; Wang, K.; de la Torre, D.; Funke, L.F.H.; Robertson, W.E.; Christova, Y.; Chia, T.; Schmied, W.H.; Dunkelmann, D.L.; Beránek, V.; et al. Total synthesis of Escherichia coli with a recoded genome. Nature 2019, 569, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Robertson, W.E.; Funke, L.F.H.; de la Torre, D.; Fredens, J.; Elliott, T.S.; Spinck, M.; Christova, Y.; Cervettini, D.; Boge, F.L.; Liu, K.C.; et al. Sense codon reassignment enables viral resistance and encoded polymer synthesis. Science 2021, 372, 1057–1062. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Brule, C.E.; Grayhack, E.J. Synonymous codons: choose wisely for expression. Trends Genet 2017, 33, 283–297. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Ray, S.K.; Banerjee, R. Synonymous codons influencing gene expression in organisms. Rese Rep Biochem 2016, 2016, 57–65. [Google Scholar] [CrossRef]
- Wang, N.; Shang, X.; Cerny, R.; Niu, W.; Guo, J. Systematic Evolution and Study of UAGN Decoding tRNAs in a Genomically Recoded Bacteria. Sci Rep 2016, 6, 21898. [Google Scholar] [CrossRef] [PubMed]
- Hohsaka, T.; Ashizuka, Y.; Murakami, H.; Sisido, M. Five-base codons for incorporation of nonnatural amino acids into proteins. Nucleic Acids Res 2001, 29, 3646–3651. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Davis, L.; Baxter, K.; Tynan, A.; Goutou, A.; Greiss, S. Using a quadruplet codon to expand the genetic code of an animal. Nucleic Acids Res 2022, 50, 4801–4812. [Google Scholar] [CrossRef]
- de la Torre, D.; Chin, J.W. Reprogramming the genetic code. Nat Rev Genet 2021, 22, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Gamper, H.; Masuda, I.; Hou, Y.-M. Genome Expansion by tRNA +1 Frameshifting at Quadruplet Codons. J Mol Biol 2022, 434, 167440. [Google Scholar] [CrossRef]
- Guo, J.; Niu, W. Genetic Code Expansion Through Quadruplet Codon Decoding. J Mol Biol 2022, 434, 167346. [Google Scholar] [CrossRef]
- Bain, J.D.; Switzer, C.; Chamberlin, R.; Benner, S.A. Ribosome-mediated incorporation of a non-standard amino acid into a peptide through expansion of the genetic code. Nature 1992, 356, 537–539. [Google Scholar] [CrossRef]
- Malyshev, D.A.; Dhami, K.; Lavergne, T.; Chen, T.; Dai, N.; Foster, J.M.; Corrêa, I.R.; Romesberg, F.E. A semi-synthetic organism with an expanded genetic alphabet. Nature 2014, 509, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Benner, S.A.; Karalkar, N.B.; Hoshika, S.; Laos, R.; Shaw, R.W.; Matsuura, M.; Fajardo, D.; Moussatche, P. Alternative Watson-Crick Synthetic Genetic Systems. Cold Spring Harb Perspect Biol 2016, 8, a023770. [Google Scholar] [CrossRef]
- Kimoto, M.; Hirao, I. Genetic alphabet expansion technology by creating unnatural base pairs. Chem Soc Rev 2020, 49, 7602–7626. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, M.; Hirao, I. Genetic Code Engineering by Natural and Unnatural Base Pair Systems for the Site-Specific Incorporation of Non-Standard Amino Acids Into Proteins. Front Mol Biosci 2022, 9, 851646. [Google Scholar] [CrossRef] [PubMed]
- Mehl, R.A.; Anderson, J.C.; Santoro, S.W.; Wang, L.; Martin, A.B.; King, D.S.; Horn, D.M.; Schultz, P.G. Generation of a Bacterium with a 21 Amino Acid Genetic Code. J Am Chem Soc 2003, 125, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Budisa, N. Prolegomena to Future Experimental Efforts on Genetic Code Engineering by Expanding Its Amino Acid Repertoire. Angew Chem Int Ed 2004, 43, 6426–6463. [Google Scholar] [CrossRef] [PubMed]
- Hoesl, M.G.; Budisa, N. In vivo incorporation of multiple noncanonical amino acids into proteins. Angew Chem Int Ed Engl 2011, 50, 2896–2902. [Google Scholar] [CrossRef]
- Baumann, T.; Schmitt, F.J.; Pelzer, A.; Spiering, V.J.; Freiherr von Sass, G.J.; Friedrich, T.; Budisa, N. Engineering 'Golden' Fluorescence by Selective Pressure Incorporation of Non-canonical Amino Acids and Protein Analysis by Mass Spectrometry and Fluorescence. J Vis Exp 2018. [CrossRef]
- Johnson, J.A.; Lu, Y.Y.; Van Deventer, J.A.; Tirrell, D.A. Residue-specific incorporation of non-canonical amino acids into proteins: recent developments and applications. Curr Opin Chem Biol 2010, 14, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Viel, J.H.; Chen, J.; Kuipers, O.P. Synthesis and Characterization of Heterodimers and Fluorescent Nisin Species by Incorporation of Methionine Analogues and Subsequent Click Chemistry. ACS Synth Biol 2020, 9, 2525–2536. [Google Scholar] [CrossRef]
- Baumann, T.; Nickling, J.H.; Bartholomae, M.; Buivydas, A.; Kuipers, O.P.; Budisa, N. Prospects of In vivo Incorporation of Non-canonical Amino Acids for the Chemical Diversification of Antimicrobial Peptides. Front Microbiol 2017, 8, 124. [Google Scholar] [CrossRef]
- Nickling, J.H.; Baumann, T.; Schmitt, F.J.; Bartholomae, M.; Kuipers, O.P.; Friedrich, T.; Budisa, N. Antimicrobial Peptides Produced by Selective Pressure Incorporation of Non-canonical Amino Acids. J Vis Exp 2018. [CrossRef]
- Münzker, L.; Oddo, A.; Hansen, P.R. Chemical synthesis of antimicrobial peptides. Methods Mol Biol 2017, 1548, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. Comprehensive Organic Name Reactions; Wiley: 2010.
- Tsuchiya, K.; Numata, K. Chemoenzymatic synthesis of polypeptides containing the unnatural amino acid 2-aminoisobutyric acid. Chem Commun 2017, 53, 7318–7321. [Google Scholar] [CrossRef]
- Jakas, A.; Vlahoviček-Kahlina, K.; Ljolić-Bilić, V.; Horvat, L.; Kosalec, I. Design and synthesis of novel antimicrobial peptide scaffolds. Bioorg Chem 2020, 103, 104178. [Google Scholar] [CrossRef]
- Mueller, L.K.; Baumruck, A.C.; Zhdanova, H.; Tietze, A.A. Challenges and Perspectives in Chemical Synthesis of Highly Hydrophobic Peptides. Front Bioeng Biotechnol 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Behrendt, R.; White, P.; Offer, J. Advances in Fmoc solid-phase peptide synthesis. J Pept Sci 2016, 22, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Yue, K.; Trung, T.N.; Zhu, Y.; Kaldenhoff, R.; Kai, L. Co-Translational Insertion of Aquaporins into Liposome for Functional Analysis via an E. coli Based Cell-Free Protein Synthesis System. Cells 2019, 8, 1325. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.Z.; Gregorio, N.E.; Jewett, M.C.; Watts, K.R.; Oza, J.P. Escherichia coli-Based Cell-Free Protein Synthesis: Protocols for a robust, flexible, and accessible platform technology. J Vis Exp 2019. [CrossRef]
- Zemella, A.; Thoring, L.; Hoffmeister, C.; Kubick, S. Cell-Free Protein Synthesis: Pros and Cons of Prokaryotic and Eukaryotic Systems. Chembiochem 2015, 16, 2420–2431. [Google Scholar] [CrossRef]
- Purkayastha, A.; Iyappan, K.; Kang, T.J. Multiple Gene Expression in Cell-Free Protein Synthesis Systems for Reconstructing Bacteriophages and Metabolic Pathways. Microorganisms 2022, 10, 2477. [Google Scholar] [CrossRef]
- Anastasina, M.; Terenin, I.; Butcher, S.J.; Kainov, D.E. A technique to increase protein yield in a rabbit reticulocyte lysate translation system. Biotechniques 2014, 56, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Wright, T.H.; Bower, B.J.; Chalker, J.M.; Bernardes, G.J.L.; Wiewiora, R.; Ng, W.-L.; Raj, R.; Faulkner, S.; Vallée, M.R.J.; Phanumartwiwath, A.; et al. Posttranslational mutagenesis: A chemical strategy for exploring protein side-chain diversity. Science 2016, 354, aag1465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zang, C.; An, G.; Shang, M.; Cui, Z.; Chen, G.; Xi, Z.; Zhou, C. Cysteine-specific protein multi-functionalization and disulfide bridging using 3-bromo-5-methylene pyrrolones. Nat Commun 2020, 11, 1015. [Google Scholar] [CrossRef] [PubMed]
- Chalker, J.M.; Bernardes, G.J.L.; Lin, Y.A.; Davis, B.G. Chemical Modification of Proteins at Cysteine: Opportunities in Chemistry and Biology. Chemistry 2009, 4, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Silverman, H.G.; Roberto, F.F. Understanding marine mussel adhesion. Mar Biotechnol 2007, 9, 661–681. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Li, Y.; Zhang, Y.; Liao, Y.; Liu, W. High-strength photoresponsive hydrogels enable surface-mediated gene delivery and light-induced reversible cell adhesion/detachment. Langmuir 2014, 30, 11823–11832. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Kan, Y.; Rapp, M.; Danner, E.; Wei, W.; Das, S.; Miller, D.R.; Chen, Y.; Waite, J.H.; Israelachvili, J.N. Adaptive hydrophobic and hydrophilic interactions of mussel foot proteins with organic thin films. Proc Natl Acad Sci 2013, 110, 15680–15685. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cao, Y. The molecular mechanisms underlying mussel adhesion. Nanoscale Adv 2019, 1, 4246–4257. [Google Scholar] [CrossRef] [PubMed]
- Hauf, M.; Richter, F.; Schneider, T.; Faidt, T.; Martins, B.M.; Baumann, T.; Durkin, P.; Dobbek, H.; Jacobs, K.; Möglich, A.; et al. Photoactivatable mussel-based underwater adhesive proteins by an expanded genetic code. ChemBioChem 2017, 18, 1819–1823. [Google Scholar] [CrossRef]
- Xiang, Z.; Ren, H.; Hu, Y.S.; Coin, I.; Wei, J.; Cang, H.; Wang, L. Adding an unnatural covalent bond to proteins through proximity-enhanced bioreactivity. Nat Methods 2013, 10, 885–888. [Google Scholar] [CrossRef]
- Zappala, F.; Tsourkas, A. Site-specific photocrosslinking to immunoglobulin G using photoreactive antibody-binding domains. In Bioconjugation, Springer: 2019; pp. 275–286.
- Elia, N. Using unnatural amino acids to selectively label proteins for cellular imaging: a cell biologist viewpoint. Febs J 2021, 288, 1107–1117. [Google Scholar] [CrossRef]
- Pagar, A.D.; Jeon, H.; Khobragade, T.P.; Sarak, S.; Giri, P.; Lim, S.; Yoo, T.H.; Ko, B.J.; Yun, H. Non-Canonical Amino Acid-Based Engineering of (R)-Amine Transaminase. Front Chem 2022, 10, 839636. [Google Scholar] [CrossRef]
- Miyazaki, R.; Akiyama, Y.; Mori, H. A photo-cross-linking approach to monitor protein dynamics in living cells. Biochim Biophys Acta Gen Subj 2020, 1864, 129317. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.M.; Sungwienwong, I.; Petersson, E.J. The development of intrinsically fluorescent unnatural amino acids for in vivo incorporation into proteins. Biophys 2019, 116, 473a. [Google Scholar] [CrossRef]
- Curnew, L.J.F.; McNicholas, K.; Green, B.; Barry, J.; Wallace, H.L.; Wang, L.; Davidson, C.; Pezacki, J.P.; Russell, R.S. Visualizing HCV core protein via fluorescent unnatural amino acid incorporation. Proceedings 2020, 50, 129. [Google Scholar]
- Wang, Y.; Chen, X.; Cai, W.; Tan, L.; Yu, Y.; Han, B.; Li, Y.; Xie, Y.; Su, Y.; Luo, X.; et al. Expanding the structural diversity of protein building blocks with noncanonical amino acids biosynthesized from aromatic thiols. Angew Chem Int Ed Engl 2021, 60, 10040–10048. [Google Scholar] [CrossRef]
- Benedini, L. Advanced Protein Drugs and Formulations. Curr Protein Pept Sci 2022, 23, 2–5. [Google Scholar] [CrossRef]
- Li, Q.; Chen, Q.; Klauser, P.C.; Li, M.; Zheng, F.; Wang, N.; Li, X.; Zhang, Q.; Fu, X.; Wang, Q.; et al. Developing covalent protein drugs via proximity-enabled reactive therapeutics. Cell 2020, 182, 85–97. [Google Scholar] [CrossRef]
- Liu, J.; Cao, L.; Klauser, P.C.; Cheng, R.; Berdan, V.Y.; Sun, W.; Wang, N.; Ghelichkhani, F.; Yu, B.; Rozovsky, S.; et al. A Genetically Encoded Fluorosulfonyloxybenzoyl-L-lysine for Expansive Covalent Bonding of Proteins via SuFEx Chemistry. J Am Chem Soc 2021, 143, 10341–10351. [Google Scholar] [CrossRef]
- Lindberg, J.; Nilvebrant, J.; Nygren, P.; Lehmann, F. Progress and Future Directions with Peptide-Drug Conjugates for Targeted Cancer Therapy. Molecules 2021, 26, 6042. [Google Scholar] [CrossRef]
- Fu, Z.; Li, S.; Han, S.; Shi, C.; Zhang, Y. Antibody drug conjugate: the "biological missile" for targeted cancer therapy. Signal Transduct Target Ther 2022, 7, 93. [Google Scholar] [CrossRef]
- Jin, Y.; Schladetsch, M.A.; Huang, X.; Balunas, M.J.; Wiemer, A.J. Stepping forward in antibody-drug conjugate development. Pharmacol Ther 2022, 229, 107917. [Google Scholar] [CrossRef]
- Hallam, T.J.; Wold, E.; Wahl, A.; Smider, V.V. Antibody conjugates with unnatural amino acids. Mol Pharm 2015, 12, 1848–1862. [Google Scholar] [CrossRef]
- Grünewald, J.; Tsao, M.-L.; Perera, R.; Dong, L.; Niessen, F.; Wen, B.G.; Kubitz, D.M.; Smider, V.V.; Ruf, W.; Nasoff, M.; et al. Immunochemical termination of self-tolerance. Proc Natl Acad Sci USA 2008, 105, 11276–11280. [Google Scholar] [CrossRef] [PubMed]
- Grünewald, J.; Hunt, G.S.; Dong, L.; Niessen, F.; Wen, B.G.; Tsao, M.L.; Perera, R.; Kang, M.; Laffitte, B.A.; Azarian, S.; et al. Mechanistic studies of the immunochemical termination of self-tolerance with unnatural amino acids. Proc Natl Acad Sci U S A 2009, 106, 4337–4342. [Google Scholar] [CrossRef]
- Wang, N.; Li, Y.; Niu, W.; Sun, M.; Cerny, R.; Li, Q.; Guo, J. Construction of a live-attenuated HIV-1 vaccine through genetic code expansion. Angew Chem Int Ed Engl 2014, 53, 4867–4871. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Wang, N.; Kang, G.; Niu, W.; Li, Q.; Guo, J. Controlling Multicycle Replication of Live-Attenuated HIV-1 Using an Unnatural Genetic Switch. ACS Synth Biol 2017, 6, 721–731. [Google Scholar] [CrossRef]
- Si, L.; Xu, H.; Zhou, X.; Zhang, Z.; Tian, Z.; Wang, Y.; Wu, Y.; Zhang, B.; Niu, Z.; Zhang, C.; et al. Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science 2016, 354, 1170–1173. [Google Scholar] [CrossRef]
- Chang, Z.; Liu, D.; Yang, Z.; Wu, J.; Zhuang, W.; Niu, H.; Ying, H. Efficient Xylitol Production from Cornstalk Hydrolysate Using Engineered Escherichia coli Whole Cells. J Agric Food Chem 2018, 66, 13209–13216. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H.C. The origin of the genetic code. J. Mol. Biol 1968, 38, 367–379. [Google Scholar] [CrossRef]
- Kim, S.; Sung, B.H.; Kim, S.C.; Lee, H.S. Genetic incorporation of l-dihydroxyphenylalanine (DOPA) biosynthesized by a tyrosine phenol-lyase. Chem Commun 2018, 54, 3002–3005. [Google Scholar] [CrossRef]
- Marchand, J.A.; Neugebauer, M.E.; Ing, M.C.; Lin, C.I.; Pelton, J.G.; Chang, M.C.Y. Discovery of a pathway for terminal-alkyne amino acid biosynthesis. Nature 2019, 567, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.S.; Brunner, S.F.; Huguenin-Dezot, N.; Liang, A.D.; Schmied, W.H.; Rogerson, D.T.; Chin, J.W. Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing. Nat Methods 2017, 14, 729–736. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




