Submitted:
01 September 2023
Posted:
05 September 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
References
- Tian, X.; Zhang, S.; Zhou, L.; Seyhan, A.A.; Hernandez Borrero, L.; Zhang, Y.; El-Deiry, W.S. Targeting the Integrated Stress Response in Cancer Therapy. Front Pharmacol 2021, 12, 747837. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Zhang, Y.; Ron, D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999, 397, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.H.; Petryshyn, R.; London, I.M. Characterization of double-stranded-RNA-activated kinase that phosphorylates alpha subunit of eukaryotic initiation factor 2 (eIF-2 alpha) in reticulocyte lysates. Proc Natl Acad Sci U S A 1980, 77, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; London, I.M. Regulation of protein synthesis by heme-regulated eIF-2 alpha kinase. Trends Biochem Sci 1995, 20, 105–108. [Google Scholar] [CrossRef]
- Vattem, K.M.; Wek, R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 2004, 101, 11269–11274. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Zhang, Y.; Zeng, H.; Novoa, I.; Lu, P.D.; Calfon, M.; Sadri, N.; Yun, C.; Popko, B.; Paules, R.; et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 2003, 11, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Cordova, R.A.; Misra, J.; Amin, P.H.; Klunk, A.J.; Damayanti, N.P.; Carlson, K.R.; Elmendorf, A.J.; Kim, H.G.; Mirek, E.T.; Elzey, B.D.; et al. GCN2 eIF2 kinase promotes prostate cancer by maintaining amino acid homeostasis. Elife 2022, 11. [Google Scholar] [CrossRef]
- Tameire, F.; Verginadis, I.; Leli, N.M.; Polte, C.; Conn, C.S.; Ojha, R.; Salas Salinas, C.; Chinga, F.; Monroy, A.M.; Fu, W.; et al. ATF4 couples MYC-dependent translational activity to bioenergetic demands during tumour progression. Nat Cell Biol 2019, 21, 889–899. [Google Scholar] [CrossRef]
- Gold, L.T.; Masson, G.R. GCN2: roles in tumour development and progression. Biochem Soc Trans 2022, 50, 737–745. [Google Scholar] [CrossRef]
- Jin, Y.; Saatcioglu, F. Targeting the Unfolded Protein Response in Hormone-Regulated Cancers. Trends Cancer 2020, 6, 160–171. [Google Scholar] [CrossRef]
- Jiang, H.Y.; Wek, R.C. Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J Biol Chem 2005, 280, 14189–14202. [Google Scholar] [CrossRef]
- Ye, J.; Kumanova, M.; Hart, L.S.; Sloane, K.; Zhang, H.; De Panis, D.N.; Bobrovnikova-Marjon, E.; Diehl, J.A.; Ron, D.; Koumenis, C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J 2010, 29, 2082–2096. [Google Scholar] [CrossRef] [PubMed]
- Broer, A.; Rahimi, F.; Broer, S. Deletion of Amino Acid Transporter ASCT2 (SLC1A5) Reveals an Essential Role for Transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to Sustain Glutaminolysis in Cancer Cells. J Biol Chem 2016, 291, 13194–13205. [Google Scholar] [CrossRef]
- Nakamura, A.; Nambu, T.; Ebara, S.; Hasegawa, Y.; Toyoshima, K.; Tsuchiya, Y.; Tomita, D.; Fujimoto, J.; Kurasawa, O.; Takahara, C.; et al. Inhibition of GCN2 sensitizes ASNS-low cancer cells to asparaginase by disrupting the amino acid response. Proc Natl Acad Sci U S A 2018, 115, E7776–E7785. [Google Scholar] [CrossRef] [PubMed]
- Okano, N.; Kawai, K.; Yamauchi, Y.; Kobayashi, T.; Naruge, D.; Nagashima, F.; Endou, H.; Furuse, J. First-in-human phaseⅠstudy of JPH203 in patients with advanced solid tumors. Journal of Clinical Oncology 2018, 36, 419–419. [Google Scholar] [CrossRef]
- Huang, Z.; Wu, Y.; Zhou, X.; Xu, J.; Zhu, W.; Shu, Y.; Liu, P. Efficacy of therapy with bortezomib in solid tumors: a review based on 32 clinical trials. Future Oncol 2014, 10, 1795–1807. [Google Scholar] [CrossRef]
- Lane, H.A.; Breuleux, M. Optimal targeting of the mTORC1 kinase in human cancer. Curr Opin Cell Biol 2009, 21, 219–229. [Google Scholar] [CrossRef]
- Ables, G.P.; Hens, J.R.; Nichenametla, S.N. Methionine restriction beyond life-span extension. Ann N Y Acad Sci 2016, 1363, 68–79. [Google Scholar] [CrossRef]
- Holbeck, S.L.; Camalier, R.; Crowell, J.A.; Govindharajulu, J.P.; Hollingshead, M.; Anderson, L.W.; Polley, E.; Rubinstein, L.; Srivastava, A.; Wilsker, D.; et al. The National Cancer Institute ALMANAC: A Comprehensive Screening Resource for the Detection of Anticancer Drug Pairs with Enhanced Therapeutic Activity. Cancer Res 2017, 77, 3564–3576. [Google Scholar] [CrossRef]
- Kato, Y.; Kunimasa, K.; Takahashi, M.; Harada, A.; Nagasawa, I.; Osawa, M.; Sugimoto, Y.; Tomida, A. GZD824 Inhibits GCN2 and Sensitizes Cancer Cells to Amino Acid Starvation Stress. Mol Pharmacol 2020, 98, 669–676. [Google Scholar] [CrossRef]
- Dorsch, D.; Hölzemann, G.; Calderini, M.; Wegener, A.; Pöschke, O. Triazolo[4,5-d]pyrimidine derivatives for the treatment of diseases such as cancer. 2014.
- Broer, A.; Gauthier-Coles, G.; Rahimi, F.; van Geldermalsen, M.; Dorsch, D.; Wegener, A.; Holst, J.; Broer, S. Ablation of the ASCT2 (SLC1A5) gene encoding a neutral amino acid transporter reveals transporter plasticity and redundancy in cancer cells. J Biol Chem 2019, 294, 4012–4026. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Wang, W.; Aldahdooh, J.; Malyutina, A.; Shadbahr, T.; Tanoli, Z.; Pessia, A.; Tang, J. SynergyFinder Plus: Toward Better Interpretation and Annotation of Drug Combination Screening Datasets. Genomics Proteomics Bioinformatics 2022. [Google Scholar] [CrossRef] [PubMed]
- Marcotte, R.; Brown, K.R.; Suarez, F.; Sayad, A.; Karamboulas, K.; Krzyzanowski, P.M.; Sircoulomb, F.; Medrano, M.; Fedyshyn, Y.; Koh, J.L.Y.; et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov 2012, 2, 172–189. [Google Scholar] [CrossRef]
- Wengrod, J.; Wang, D.; Weiss, S.; Zhong, H.; Osman, I.; Gardner, L.B. Phosphorylation of eIF2α triggered by mTORC1 inhibition and PP6C activation is required for autophagy and is aberrant in PP6C-mutated melanoma. Sci Signal 2015, 8, ra27. [Google Scholar] [CrossRef]
- Broer, A.; Fairweather, S.; Broer, S. Disruption of Amino Acid Homeostasis by Novel ASCT2 Inhibitors Involves Multiple Targets. Front Pharmacol 2018, 9, 785. [Google Scholar] [CrossRef]
- Cormerais, Y.; Giuliano, S.; LeFloch, R.; Front, B.; Durivault, J.; Tambutte, E.; Massard, P.A.; de la Ballina, L.R.; Endou, H.; Wempe, M.F.; et al. Genetic Disruption of the Multifunctional CD98/LAT1 Complex Demonstrates the Key Role of Essential Amino Acid Transport in the Control of mTORC1 and Tumor Growth. Cancer Res 2016, 76, 4481–4492. [Google Scholar] [CrossRef]
- Meyer, H.; Bug, M.; Bremer, S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 2012, 14, 117–123. [Google Scholar] [CrossRef]
- Leung-Pineda, V.; Pan, Y.; Chen, H.; Kilberg, M.S. Induction of p21 and p27 expression by amino acid deprivation of HepG2 human hepatoma cells involves mRNA stabilization. Biochem J 2004, 379, 79–88. [Google Scholar] [CrossRef]
- Kwiatkowski, N.; Zhang, T.; Rahl, P.B.; Abraham, B.J.; Reddy, J.; Ficarro, S.B.; Dastur, A.; Amzallag, A.; Ramaswamy, S.; Tesar, B.; et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014, 511, 616–620. [Google Scholar] [CrossRef]
- Meulenbeld, H.J.; Mathijssen, R.H.; Verweij, J.; de Wit, R.; de Jonge, M.J. Danusertib, an aurora kinase inhibitor. Expert Opin Investig Drugs 2012, 21, 383–393. [Google Scholar] [CrossRef]
- Stretton, C.; Lipina, C.; Hyde, R.; Cwiklinski, E.; Hoffmann, T.M.; Taylor, P.M.; Hundal, H.S. CDK7 is a component of the integrated stress response regulating SNAT2 (SLC38A2)/System A adaptation in response to cellular amino acid deprivation. Biochim Biophys Acta Mol Cell Res 2019, 1866, 978–991. [Google Scholar] [CrossRef] [PubMed]
- Finicle, B.T.; Jayashankar, V.; Edinger, A.L. Nutrient scavenging in cancer. Nat Rev Cancer 2018, 18, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Gouirand, V.; Guillaumond, F.; Vasseur, S. Influence of the Tumor Microenvironment on Cancer Cells Metabolic Reprogramming. Front Oncol 2018, 8, 117. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, C.; Poppleton, H.; Kocak, M.; Hogg, T.L.; Fuller, C.; Hamner, B.; Oh, E.Y.; Gaber, M.W.; Finklestein, D.; Allen, M.; et al. A perivascular niche for brain tumor stem cells. Cancer Cell 2007, 11, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Nofal, M.; Wang, T.; Yang, L.; Jankowski, C.S.R.; Hsin-Jung Li, S.; Han, S.; Parsons, L.; Frese, A.N.; Gitai, Z.; Anthony, T.G.; et al. GCN2 adapts protein synthesis to scavenging-dependent growth. Cell Syst 2022, 13, 158–172.e159. [Google Scholar] [CrossRef]
- Broer, S.; Broer, A. Amino acid homeostasis and signalling in mammalian cells and organisms. Biochem J 2017, 474, 1935–1963. [Google Scholar] [CrossRef]
- Nigg, E.A. Cyclin-dependent kinase 7: at the cross-roads of transcription, DNA repair and cell cycle control? Curr Opin Cell Biol 1996, 8, 312–317. [Google Scholar] [CrossRef]
- Fisher, R.P. Cdk7: a kinase at the core of transcription and in the crosshairs of cancer drug discovery. Transcription 2019, 10, 47–56. [Google Scholar] [CrossRef]
- Gaccioli, F.; Huang, C.C.; Wang, C.; Bevilacqua, E.; Franchi-Gazzola, R.; Gazzola, G.C.; Bussolati, O.; Snider, M.D.; Hatzoglou, M. Amino acid starvation induces the SNAT2 neutral amino acid transporter by a mechanism that involves eukaryotic initiation factor 2alpha phosphorylation and cap-independent translation. J Biol Chem 2006, 281, 17929–17940. [Google Scholar] [CrossRef]
- Kartha, N.; Gianopulos, J.E.; Schrank, Z.; Cavender, S.M.; Dobersch, S.; Kynnap, B.D.; Wallace-Povirk, A.; Wladyka, C.L.; Santana, J.F.; Kim, J.C.; et al. Sirtuin 6 is required for the integrated stress response and resistance to inhibition of transcriptional cyclin-dependent kinases. Sci Transl Med 2023, 15, eabn9674. [Google Scholar] [CrossRef]
- Piecyk, M.; Triki, M.; Laval, P.A.; Duret, C.; Fauvre, J.; Cussonneau, L.; Machon, C.; Guitton, J.; Rama, N.; Gibert, B.; et al. The stress sensor GCN2 differentially controls ribosome biogenesis in colon cancer according to the nutritional context. Mol Oncol 2023. [Google Scholar] [CrossRef]
- King, R.W.; Deshaies, R.J.; Peters, J.M.; Kirschner, M.W. How proteolysis drives the cell cycle. Science 1996, 274, 1652–1659. [Google Scholar] [CrossRef] [PubMed]
- Cormerais, Y.; Vucetic, M.; Parks, S.K.; Pouyssegur, J. Amino Acid Transporters Are a Vital Focal Point in the Control of mTORC1 Signaling and Cancer. Int J Mol Sci 2020, 22. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, M.M.; Pan, Y.X.; Gjymishka, A.; Zhong, C.; Kaufman, R.J.; Kilberg, M.S. MEK signaling is required for phosphorylation of eIF2alpha following amino acid limitation of HepG2 human hepatoma cells. J Biol Chem 2008, 283, 10848–10857. [Google Scholar] [CrossRef] [PubMed]
- Franchi-Gazzola, R.; Visigalli, R.; Bussolati, O.; Dall’Asta, V.; Gazzola, G.C. Adaptive increase of amino acid transport system A requires ERK1/2 activation. J Biol Chem 1999, 274, 28922–28928. [Google Scholar] [CrossRef] [PubMed]
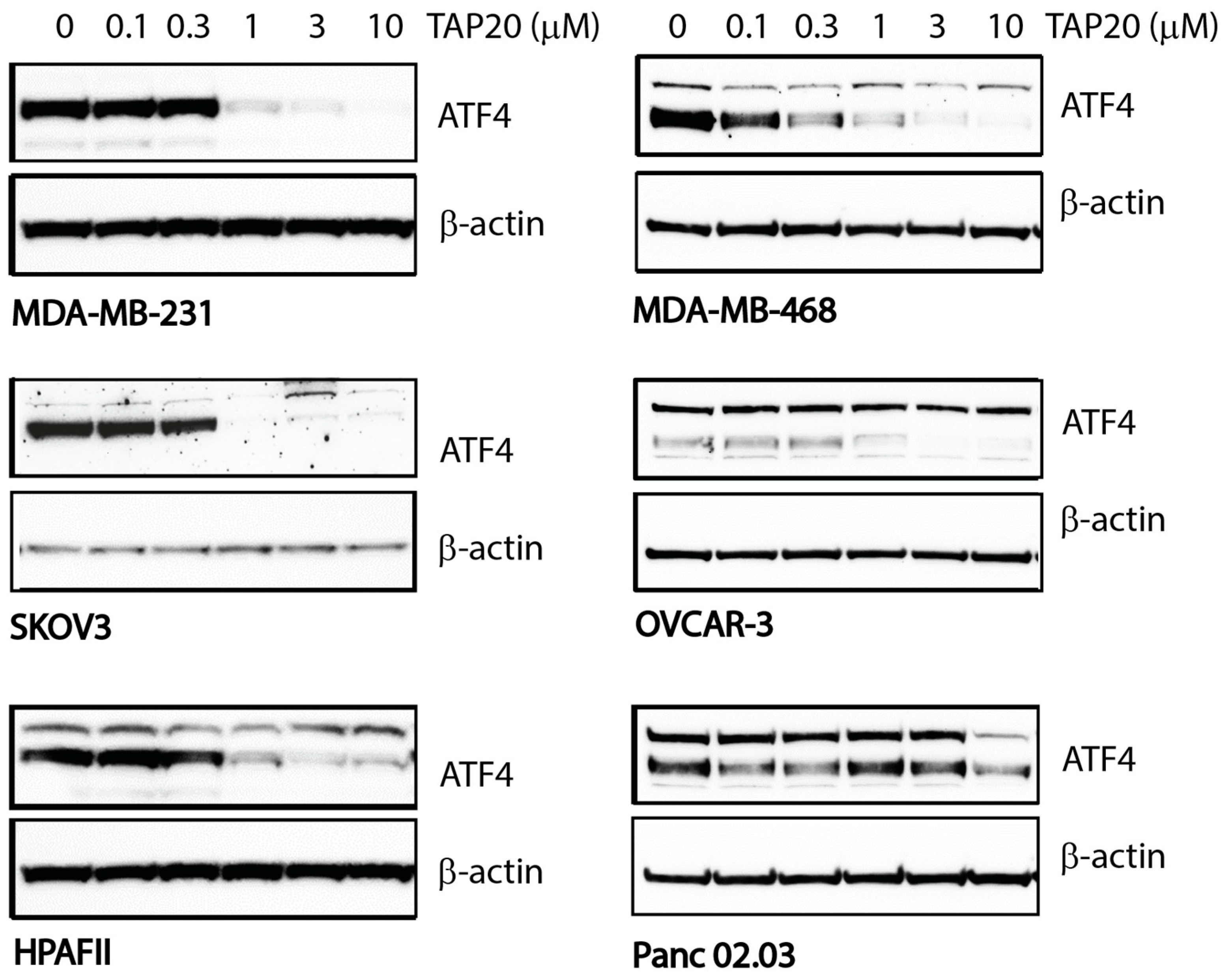
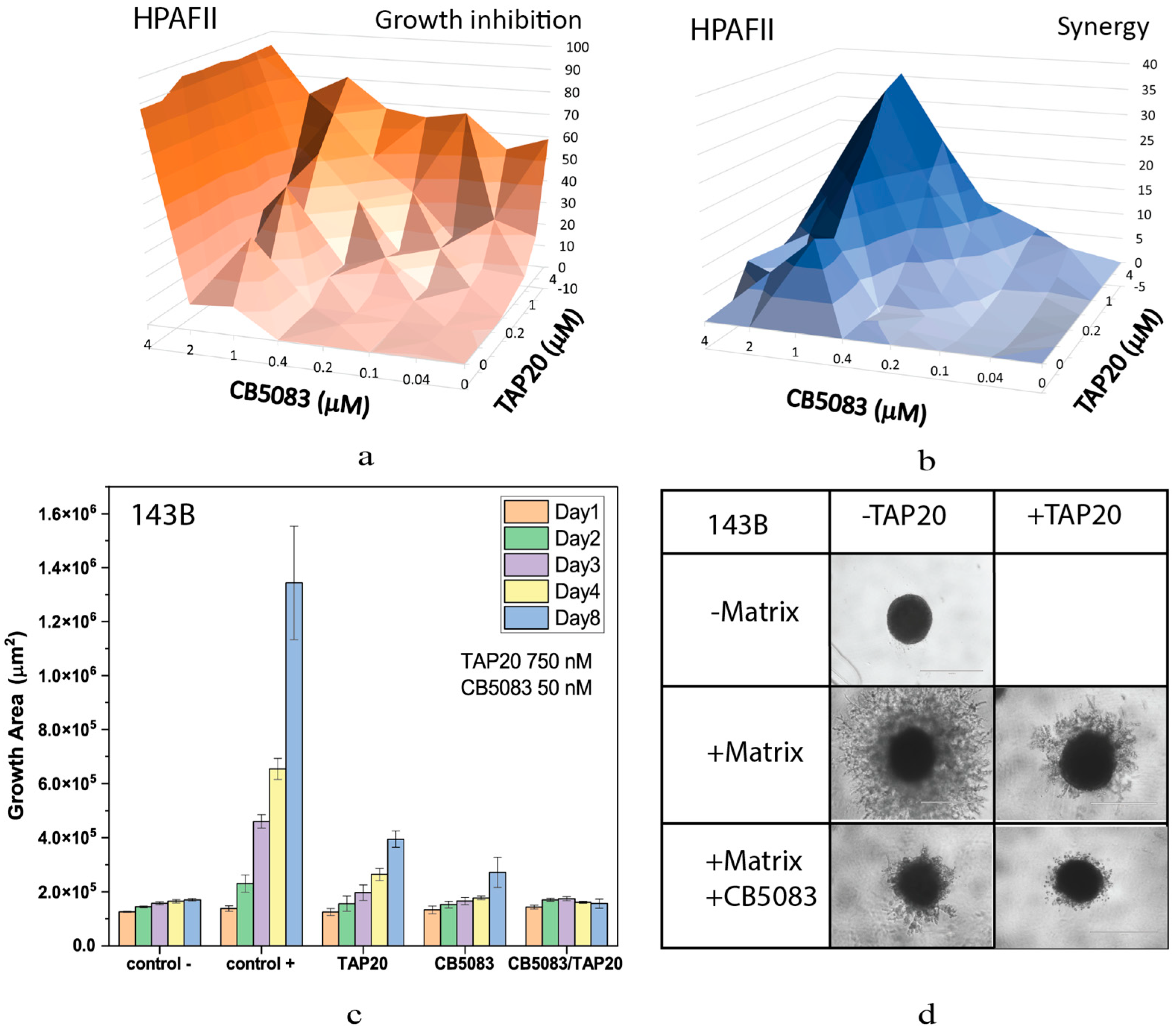
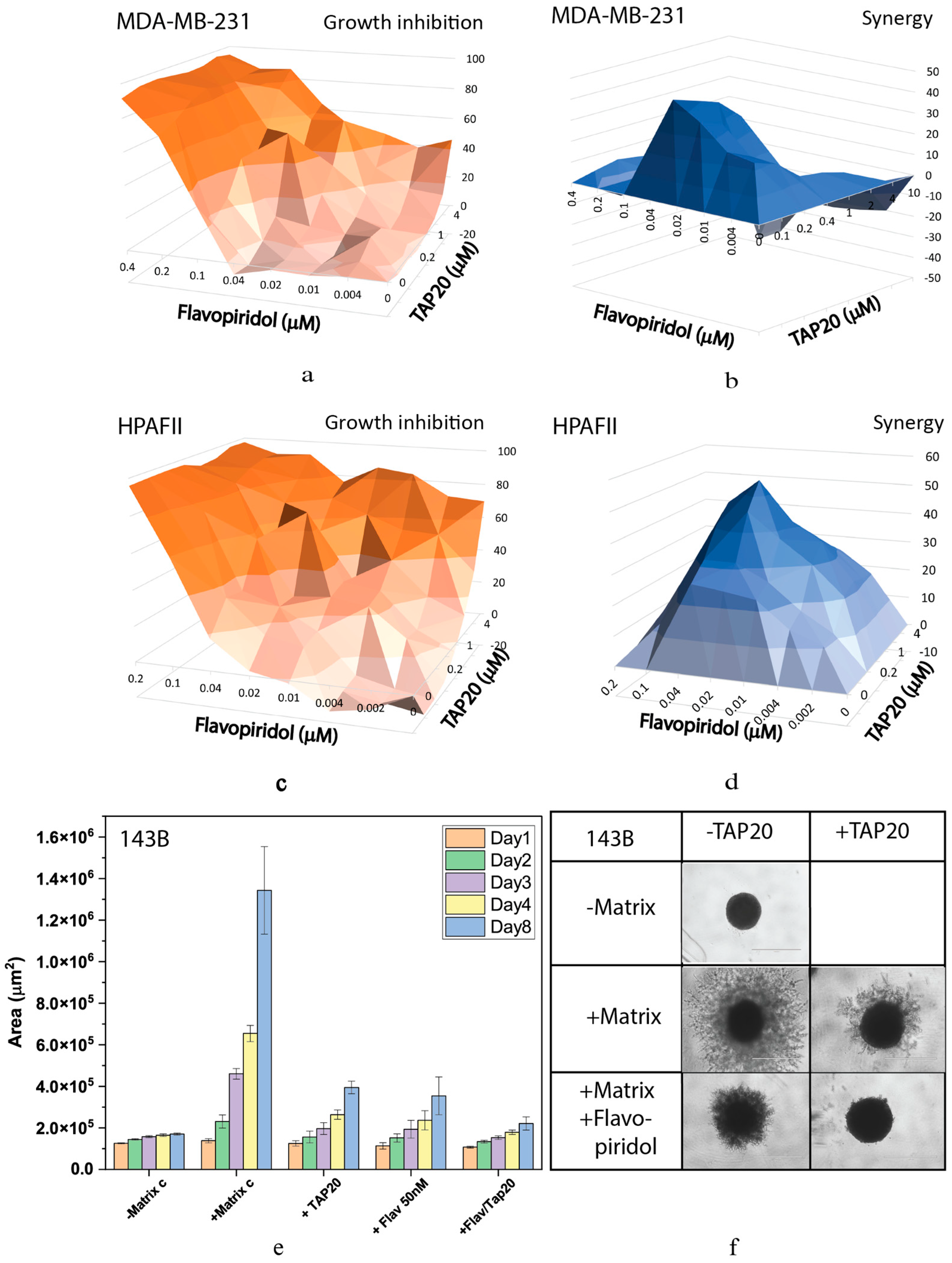
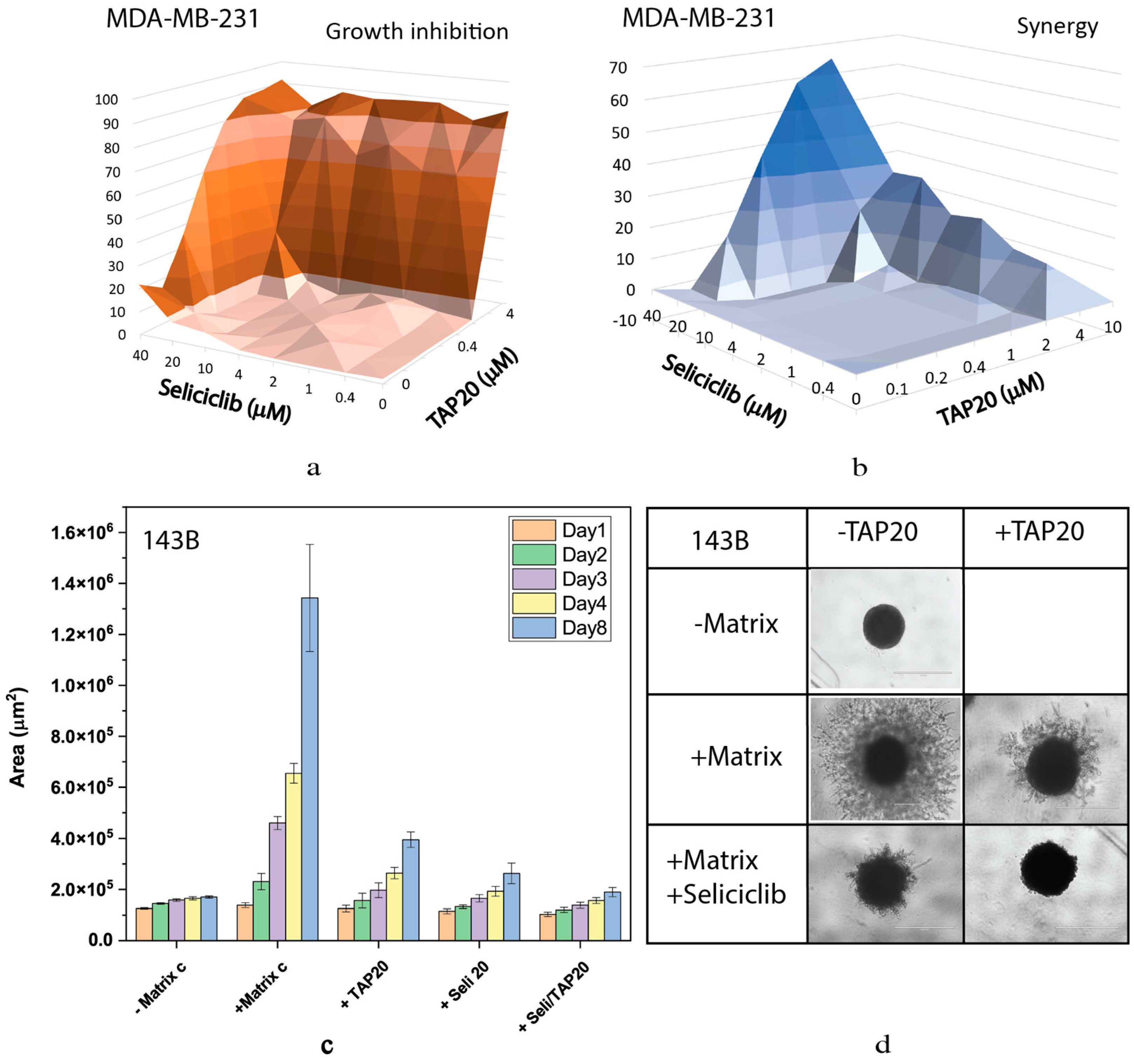
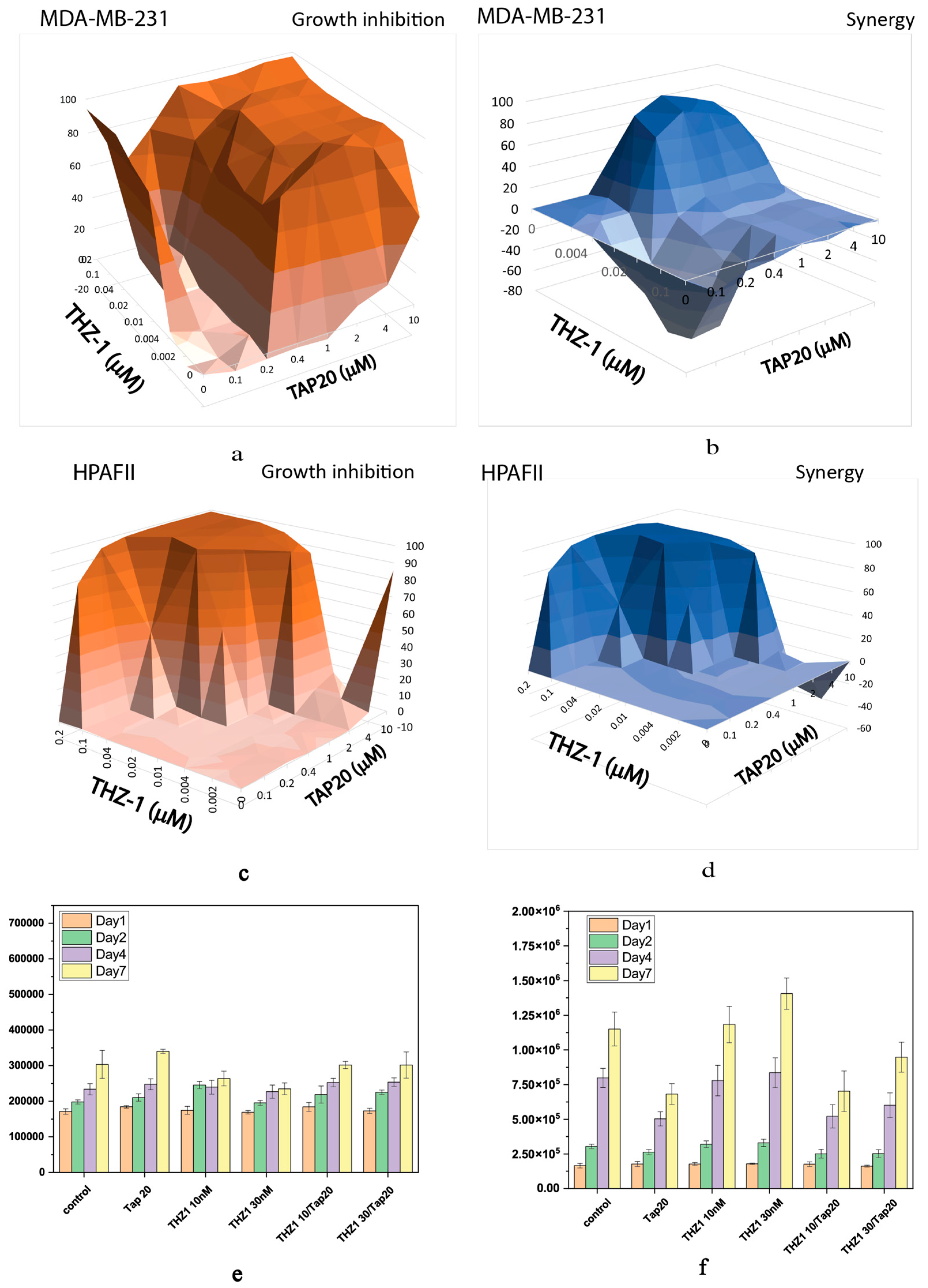
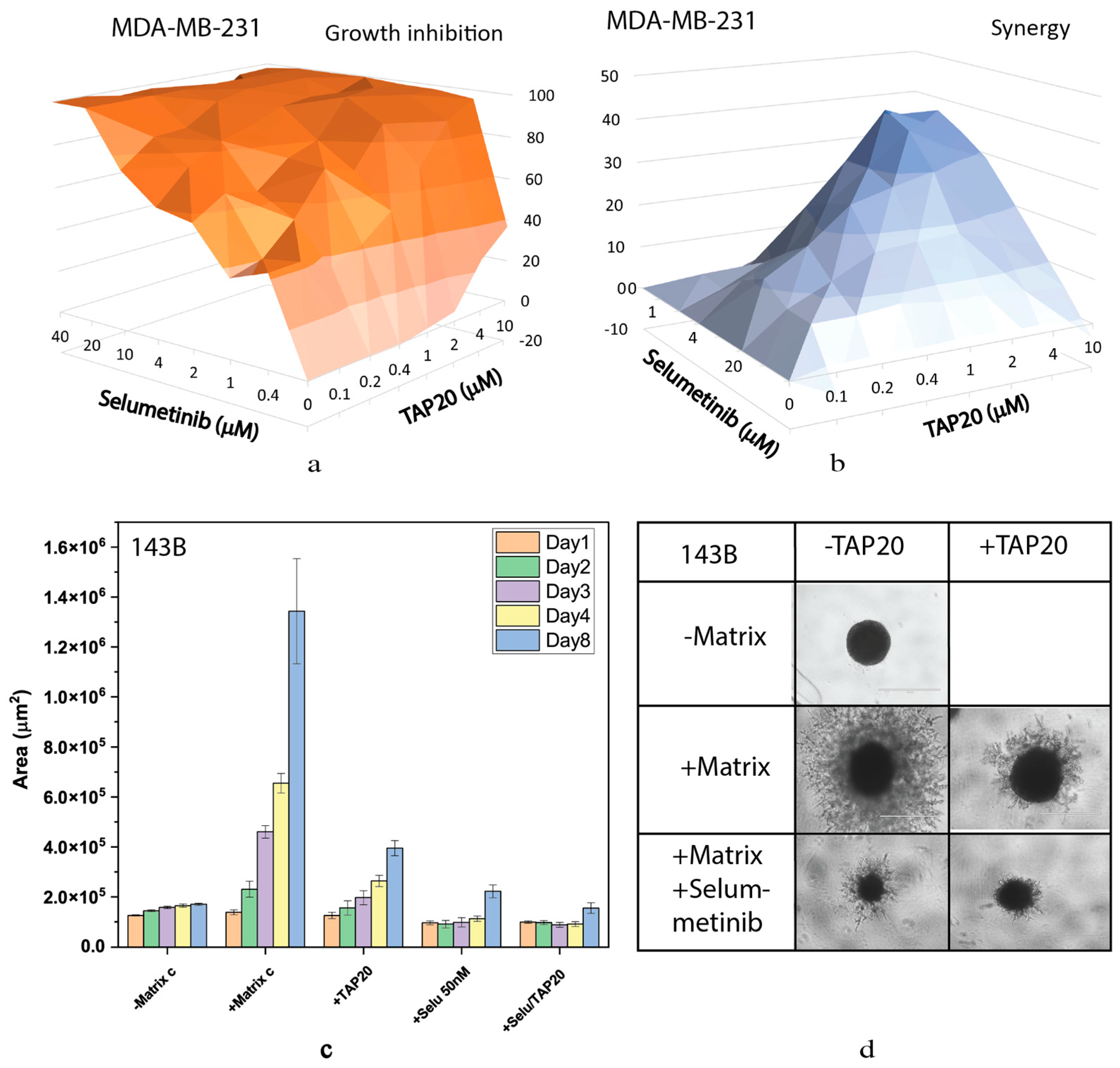
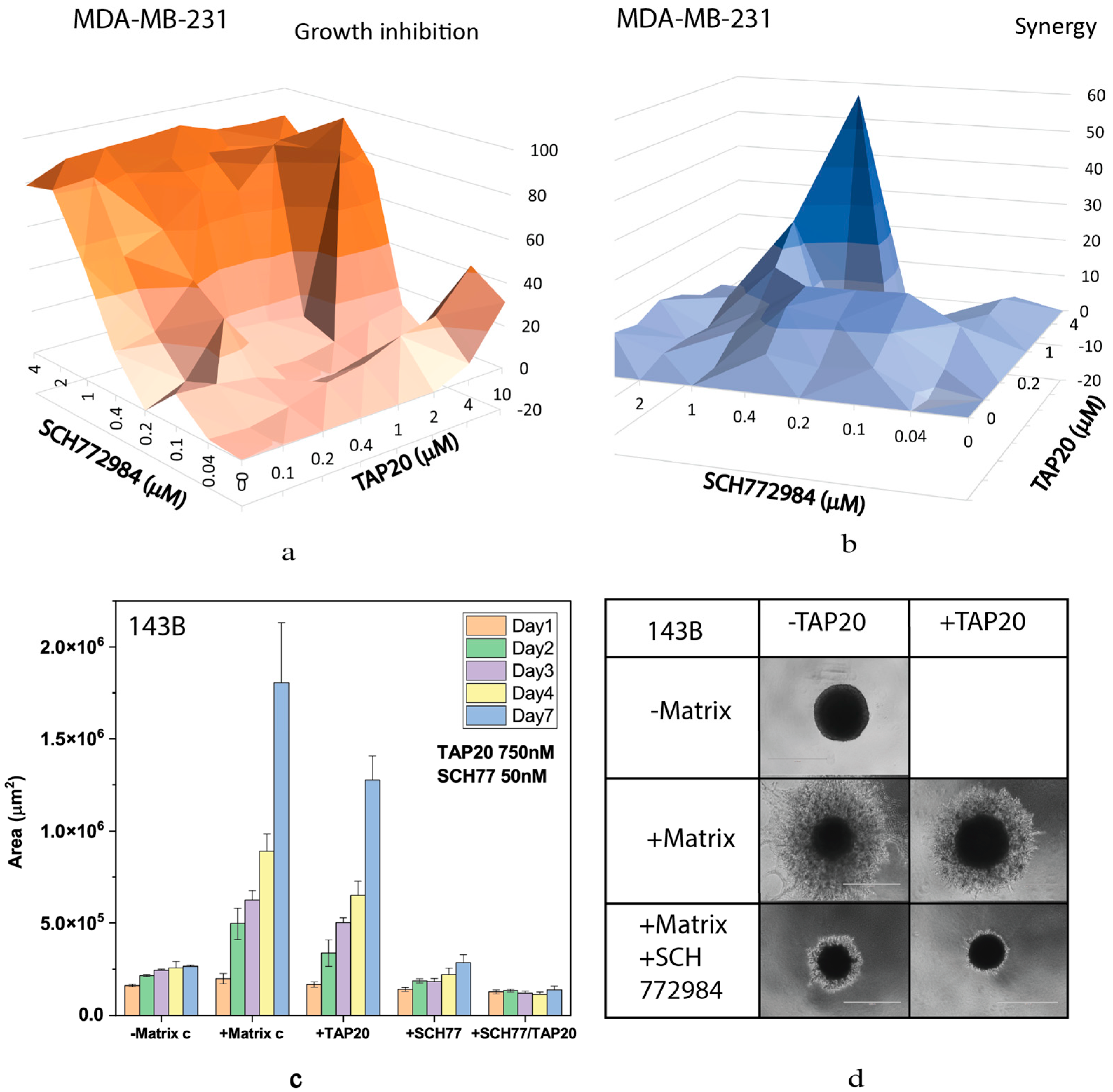
| Compound | Target | IC50 |
|---|---|---|
| Agerafenib | BRAFV600E | 14 nM |
| AMG PERK44 | PERK | 6 nM |
| AZD3965 | MCT1 | 1.6 nM |
| Bay876 | GLUT1 | 2 nM |
| Bortezomib | 20S proteasome | 0.6 nM |
| Bumetanide | NKCC1 | 680 nM |
| CB-839 | Glutaminase 1 | 25 nM |
| CB-5083 | p97AAA ATPase | 11 nM |
| Cetuximab | EGFR | 0.2 nM |
| Danusertib | Aurora-kinases | 13–79 nM |
| GSK2837808A | LDH-A | 2.6 nM |
| GZD824 | Bcr-Abl | 0.32 nM |
| Flavopiridol | CDK (non-specific) | 6–300 nM |
| 5-Fluorouracil | Thymidylate synthase | Irr1 |
| Halofuginone | Prolyl-tRNA synthetase | 18 nM |
| NCT-503 | PHGDH | 2500 nM |
| PLX8394 | BRAF/BRAFV600E | 14 nM/5 nM |
| Rapamycin | FKBP12 | 0.1 nM |
| SCH772984 | Erk1/2 | 4 nM/1 nM |
| Seliciclib | CDK (non-specific) | 200–800 nM |
| Selumetinib | MEK1/2 | 14 nM |
| Thapsigargin | SERCA | 0.4 nM |
| THZ-1 | CDK7 | Irr1 |
| V-9302 | SNAT2/LAT1 | n.d. |
| YH16899 | KRS-67LR interaction | 8600 nM |
| Compound | MDA-MB-231 | MDA-MB-468 | HPAFII | Panc02.03 | SKOV3 | OVCAR3 | ||
|---|---|---|---|---|---|---|---|---|
| Agerafenib | 4.3 | 4.3 | 1.6 | 3.7 | 4.7 | 6.5 | ||
| AMG PERK44 | >10 | >10 | 9 | >10 | >10 | >10 | ||
| AZD3965 | >10 | >10 | >10 | >10 | >10 | >10 | ||
| Bay876 | 10 | 10 | 0.25 | 0.15 | 1.4 | 0.16 | ||
| Bortezomib | 0.002 | 0.003 | 0.003 | 0.004 | 0.004 | 0.002 | ||
| Bumetanide | n.i. | n.d. | n.i. | n.d. | n.i. | n.d. | ||
| CB-839 | 10 | n.d. | >10 | n.d. | >10 | n.d. | ||
| CB-5083 | 0.76 | 0.36 | 0.19 | 0.12 | 1.2 | 0.36 | ||
| Cetuximab | >10 | n.d. | >10 | n.d. | >10 | n.d. | ||
| Danusertib | <0.1 | <0.1 | 0.8 | 0.36 | 2.2 | <0.1 | ||
| GSK2837808A | >10 | n.d. | >10 | n.d. | >10 | n.d. | ||
| GZD824 | 1 | 1.3 | <0.1 | 0.31 | 1.6 | 1.8 | ||
| Flavopiridol | 0.1 | 0.04 | <0.1 | 4 | 0.22 | 1.7 | ||
| 5-Fluorouracil | >10 | >10 | >10 | 8.9 | >10 | 1.7 | ||
| Halofuginone | 0.032 | 0.054 | 0.038 | 0.057 | 0.019 | 0.058 | ||
| NCT-503 | >10 | >10 | >10 | >10 | >10 | >10 | ||
| PLX8394 | 10 | 10 | 4.5 | 10 | 6 | 3.6 | ||
| Rapamycin | >10 | n.d. | >10 | n.d. | >10 | n.d. | ||
| SCH772984 | 1 | 1 | 0.45 | 0.4 | 8.4 | 5 | ||
| Seliciclib | >10 | >10 | 9.5 | 6.3 | >10 | 7.4 | ||
| Selumetinib | 10 | 6.3 | <0.1 | 0.88 | 10 | 1.4 | ||
| Thapsigargin | <0.1 | 0.005 | <0.1 | 0.004 | 0.52 | <0.003 | ||
| V-9302 | 1.1 | 1.1 | 1.1 | 2 | 2.6 | 1.1 | ||
| YH16899 | >10 | n.d. | >10 | n.d. | >10 | n.d |
| Compound | MDA-MB-231 | MDA-MB-468 | HPAFII | Panc02.03 | SKOV3 | OVCAR3 | ||
|---|---|---|---|---|---|---|---|---|
| Agerafenib | 3 | 3 | 1 | 0.3 | 3 | 3 | ||
| AMG PERK44 | 10 | 10 | 10 | 10 | 10 | 10 | ||
| AZD3965 | 10 | 10 | 10 | 10 | 10 | 3 | ||
| Bay876 | 10 | 3 | 0.1 | 0.03 | 1 | 0.1 | ||
| Bortezomib | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | ||
| Bumetanide | 10 | 10 | 10 | 10 | 10 | 10 | ||
| CB-839 | 10 | 10 | 10 | 10 | 10 | 10 | ||
| CB-5083 | 1 | 0.3 | 0.1 | 0.1 | 1 | 0.3 | ||
| Cetuximab | 0.033 | 0.033 | 0.033 | 0.033 | 0.033 | 0.033 | ||
| Danusertib | 0.01 | 0.03 | 0.3 | 0.1 | 1 | 0.01 | ||
| GSK2837808A | 10 | 10 | 10 | 10 | 10 | 10 | ||
| GZD824 | 0.3 | 0.3 | 0.01 | 1 | 0.1 | 0.1 | ||
| Flavopiridol | 0.1 | 0.03 | 0.03 | 3 | 0.1 | 1 | ||
| 5-Fluorouracil | 10 | 3 | 3 | 3 | 10 | 1 | ||
| Halofuginone | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | ||
| NCT-503 | 10 | 10 | 10 | 10 | 10 | 10 | ||
| PLX8394 | 10 | 3 | 3 | 3 | 3 | 1 | ||
| Rapamycin | 10 | 10 | 10 | 10 | 10 | 10 | ||
| SCH772984 | 1 | 0.3 | 0.3 | 0.1 | 3 | 0.1 | ||
| Seliciclib | 10 | 3 | 3 | 3 | 10 | 3 | ||
| Selumetinib | 10 | 3 | 0.03 | 0.3 | 10 | 0.3 | ||
| Thapsigargin | 0.01 | 0.001 | 0.003 | 0.001 | 0.1 | 0.001 | ||
| V-9302 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| YH16899 | 10 | 10 | 10 | 10 | 10 | 10 |
| Compound | MDA-MB-231 | MDA-MB-468 | HPAFII | Panc02.03 | SKOV3 | OVCAR3 |
|---|---|---|---|---|---|---|
| Agerafenib | 1.21 | 0.96 | 0.97 | 1.07 | 1.49 | 1.57 |
| AMG PERK44 | 0.88 | 0.94 | 0.66 | 0.97 | 1.07 | 1.3 |
| AZD3965 | 0.77 | 0.79 | 0.76 | 1.05 | 0.96 | 0.84 |
| Bay876 | 0.97 | 0.63 | 0.73 | 0.73 | 0.87 | 0.72 |
| Bortezomib | 0.79 | 0.82 | 0.62 | 0.83 | 0.89 | 0.91 |
| Bumetanide | 0.83 | 0.81 | 0.86 | 0.99 | 0.97 | 0.93 |
| CB-839 | 0.86 | 0.9 | 0.89 | 1.03 | 0.97 | 0.97 |
| CB-5083 | 0.85 | 0.89 | 0.51 | 0.96 | 1.17 | 0.81 |
| Cetuximab | 1.11 | 0.77 | 1.06 | 0.97 | 0.99 | 0.91 |
| Danusertib | 0.85 | 0.65 | 0.23 | 0.67 | 0.65 | 1.53 |
| GSK2837808A | 0.85 | 0.87 | 0.78 | 1.03 | 0.99 | 0.99 |
| GZD824 | 0.82 | 0.86 | 0.87 | 0.91 | 1.5 | 1.1 |
| Flavopiridol | 0.24 | 0.96 | 0.4 | 1.06 | 0.8 | 0.56 |
| 5-Fluorouracil | 0.7 | 0.95 | 0.71 | 0.87 | 0.94 | 0.51 |
| Halofuginone | 0.78 | 0.88 | 0.53 | 0.82 | 0.73 | 0.93 |
| NCT-503 | 0.68 | 0.88 | 0.74 | 0.86 | 0.95 | 1.11 |
| PLX8394 | 0.91 | 0.6 | 0.88 | 0.88 | 1.16 | 0.97 |
| Rapamycin | 1.29 | 1.05 | 1.56 | 0.97 | 1.57 | 2.38 |
| SCH772984 | 0.23 | 1.4 | 0.29 | 0.86 | 0.53 | 0.88 |
| Seliciclib | 0.47 | 0.85 | 0.51 | 0.96 | 0.87 | 0.88 |
| Selumetinib | 0.3 | 0.63 | 1.21 | 0.7 | 0.71 | 0.63 |
| Thapsigargin | 0.87 | 0.95 | 0.57 | 1.01 | 0.69 | 0.87 |
| THZ-1 | 0.71 | n.d. | 0.12 | n.d. | 0.4 | n.d. |
| V-9302 | 0.63 | 0.96 | 0.44 | 0.84 | 0.93 | 0.77 |
| YH16899 | 0.9 | 0.75 | 0.55 | 0.94 | 0.93 | 0.76 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





