1. Introduction
The emergence of SARS-CoV-2 infection presented a novel and significant clinical challenge, imposing a substantial burden on healthcare systems worldwide [
1,
2,
3]. This unique and complex form of hypoxemic acute respiratory failure is frequently accompanied by multiorgan failure, requiring organ support, and, consequently, admission to an intensive care unit (ICU) [
4]. Moreover, these patients are highly susceptible to ICU-acquired infections due to prolonged invasive support, long length of ICU stay, and finally several immunosuppressive therapies [
3,
5].
ICU-acquired infections are a common complication among critically ill patients, with an estimated mortality rate reaching >40% [
6]. However, diagnosing ICU-acquired infections relies heavily on maintaining a high clinical suspicion and conducting a comprehensive evaluation of radiological signs, laboratory results, and microbiological sampling, while facing the challenge of distinguishing between high rates of tissue colonization and actual infection [
7,
8,
9,
10]. Nonetheless, the importance of its recognition is paramount for timely prescription of empiric antimicrobial therapy.
These daily clinical challenges are further exacerbated in COVID-19 patients, characterized by a significant pro-inflammatory cytokine profile, extensive pulmonary involvement, and exceptionally high mortality rates [
4,
11,
12,
13,
14]. Frequently, there is difficulty to distinguishing the clinical, laboratory and radiology manifestations of the underlying viral disease from a new ICU-acquired infection [
15]. Currently, there is limited and conflicting evidence supporting the use of C-reactive protein (CRP) [
12,
16,
17,
18] or Procalcitonin (PCT) [
6,
11,
19,
20,
21] for diagnosing ICU-acquired infections in COVID-19 patients, usually limited to the evaluation of their potential role in diagnosing ventilator-associated pneumonia. Moreover, published data often focus on the use of these biomarkers at ICU admission or ICU-acquired infection diagnosis, neglecting their behavior over time, its kinetics, and their potential predictive value [
22,
23,
24,
25,
26,
27].
The objective of our study is to evaluate the value of daily measurements of CRP and PCT in early identification of ICU-acquired infections in COVID-19 patients. We hypothesize that CRP and PCT kinetics could be useful in predicting ICU-acquired infections in COVID-19 patients prior to infection diagnosis.
2. Materials and Methods
2.1. Study Cohort
A prospective observational cohort study was performed at an ICU of Centro Hospitalar Lisboa Ocidental. The study was approved by the National Ethics Committee for Clinical Research (reference REC: 2020_EO_02).
All adult mechanically ventilated patients admitted consecutively to the ICU between January 1st 2020 and March 31st 2021 were considered for the study. Patients were included if they had an admission to the ICU with a COVID19 respiratory infection diagnosis and a length of stay of at least 72h. COVID19 respiratory infection was diagnosed using clinical and radiological criteria confirming pulmonary involvement with a SARS-CoV-2 positive RT-PCR test.
Within the study population, patients were stratified into two distinct groups: the Infected group, comprising all patients with a documented ICU-acquired infection during their stay in the intensive care unit (ICU), and the non-Infected group, which included all patients without a diagnosis of an intercurrent ICU-acquired infection during their ICU length of stay.
An ICU-acquired infection was defined as the presence of new clinical signs of sepsis, a change in baseline Sequential Organ Failure Assessment (SOFA) score of 2 points or more, and a positive culture that warranted the initiation of empiric antibiotic therapy, as determined through comprehensive clinical evaluation conducted by the attending physicians. These assessment criteria were considered only if they manifested at least 48 hours after the patients' admission to the ICU and if the patient was not receiving antibiotics for at least 5 days before infection diagnosis. The non-Infected group comprehended all patients without criteria for an ICU-acquired infection.
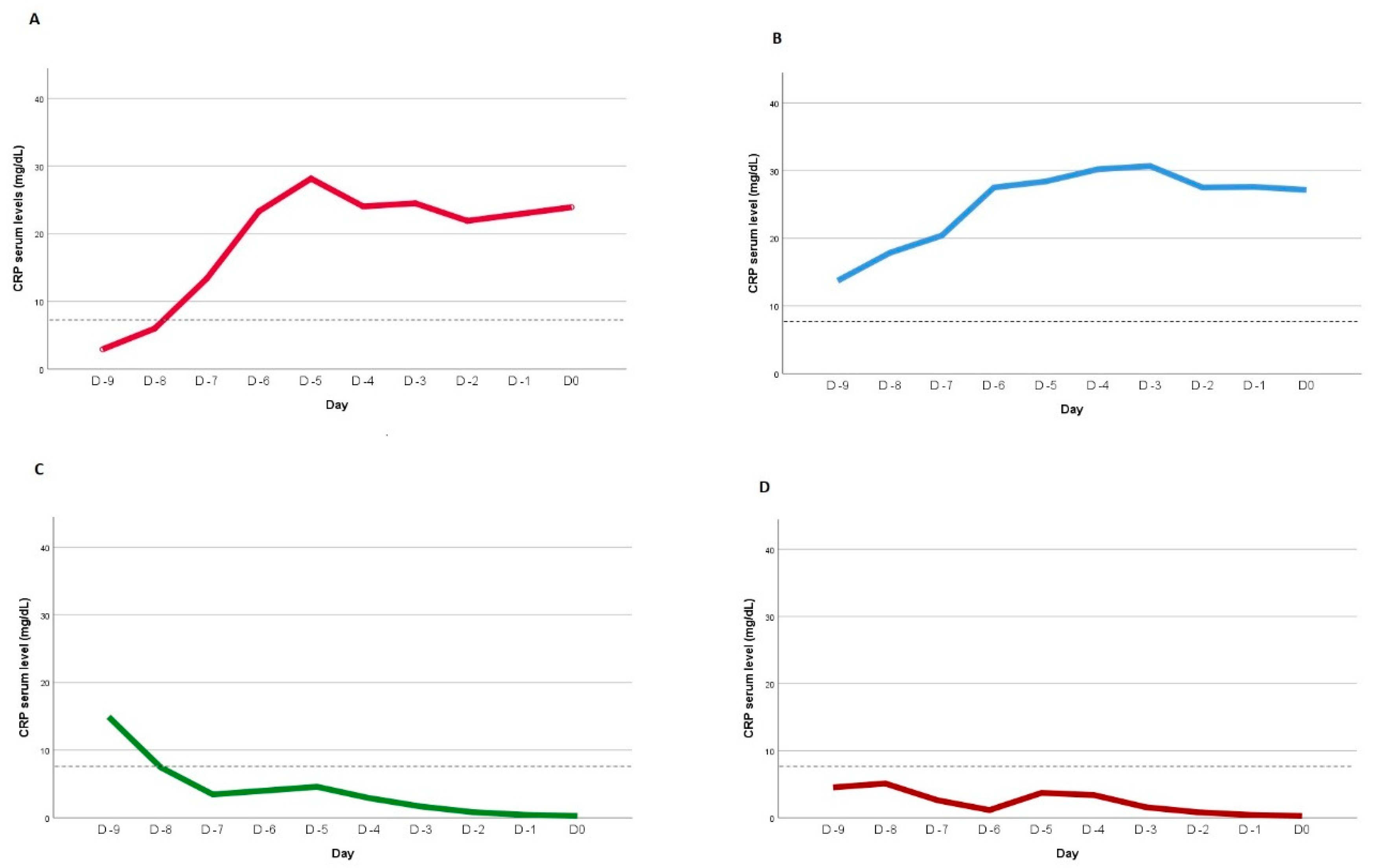
Four CRP kinetic patterns were defined after non-linear mixed-effects modeling to the individual CRP kinetic profiles, using as cut-off value point for infection a previously identified value of 8.7mg/dL (Figure1) [
28]. Pattern A was defined by a day 0 CRP serum level > 8.7mg/dL and, in the previous days, it was at least once below that cut-off value. Pattern B was assumed if serum CRP level was consistently above 8.7mg/dL until day 0. Pattern C was defined when the serum CRP level was <8.7mg/dL at day 0 and at least once above that value in the previous days. Finally, pattern D occurred when CRP serum level was persistently <8.7mg/dL at all evaluations.
2.2. Sampling and Data Collection
To detect ICU-acquired infection, a positive microbiologic culture was considered on the following biological cultures: endotracheal aspirate, bronchoalveolar lavage or blood. For the first a cutoff of 105 CFU/ml in culture was considered for positivity, and for the bronchoalveolar lavage a cutoff of 104 CFU/ml was considered for positivity. For the blood samples, a microorganism documentation was used to document culture positivity. This is consistent with the recommendations provided by the Centers for Disease Control and International ERS/ESICM/ESCMID/ALAT guidelines [
29,
30].
Clinical data were prospectively collected from patient’s electronic health records and included demographics, comorbidities, daily laboratory values, Acute Physiology and Chronic Health Evaluation II (APACHE II) score, SOFA scores, therapies administered and data related to microbiological samples, from patients’ electronic health records. Additionally, data concerning ongoing or previous antibacterial therapy at the day of ICU-acquired infection diagnosis were also recorded to ensure patient eligibility. CRP and Procalcitonin daily values were registered in both groups. Data was stored in a pseudo-anonymized database.
Biomarkers serum levels were analyzed comparing the infected and non-infected groups. All identified patterns were also compared, assessing their differences in mortality and clinical course outcomes.
2.3. Statistical Analysis Plan
All Gaussian distributed variables were expressed as mean (SD), and nonnormally distributed variables as median (interquartile range [IQR]). Categorical variables were expressed as numbers and percentages. To assess differences between the two main groups the student t test and the Mann-Whitney U test were used for continuous variables and the χ2 test was used for categorical variables.
Receiver operating characteristics curves were plotted for the maximum CRP and maximum PCT registered. The accuracy of these variables was assessed by calculating the area under the curve (AUC).
Time-dependent analysis of different variables was performed with general linear model, unmatched, univariate, repeated-measures analysis using a split-plot design approach, using a similar approach to a previous study [
28]. The day of ICU-acquired infection diagnosis was considered day 0 in the Infected group. In the non-infected group, the day 10 after ICU admission was considered as day 0 after its identification as median day of ICU-acquired infection diagnoses in the Infected group. In the non-infected group, the patients were only selected if they didn’t receive any antibiotic therapy in the previous 5 days before the selected day 0. CRP and Procalcitonin daily values were registered from day -10 to day 10 of ICU stay in both groups.
Multivariate logistic regression model was used to assess potential variables contributing to predict the occurrence of ICU-acquired infection. The variables considered were age, gender, SOFA score at admission and maximum registered CRP and PCT serum levels, before day 0 in ICU. They were considered to the model if statistically significant in a bivariate analysis and if they had an attributable odds ratio above 1.2. Multicollinearity was checked by computing the correlation coefficient between selected variables, and a coefficient below 0.4 was considered for exclusion of correlation. Model calibration and discrimination were accessed by the Hosmer-Lemeshow goodness-of fit test and c statistic, respectively. Results were reported as odds ratio with the 95% confidence interval.
Non-linear mixed-effects modeling was done to the individual CRP kinetic profiles, using as cut-off value point for infection a previously identified value of 8.7mg/dL. The model’s returned four distinct patterns, as previously described and as depicted in
Figure 1. Goodness-of-fit was assessed using Akaike information criterion with values of 126.5 indicating the model’s suitability.
All calculations will be performed in SPSS interface (version 26.0.0.0) and R (version 4.0.3). P-values <0.05 were considered statistically significant.
3. Results
136 patients were eligible for the study. Of these patients, 18 patients did not stay at ICU for 72h or longer and were excluded for the statistical analysis. The remaining 118 patients were included, 83 (70.3%) patients in the non-Infected group and 35 (29.7%) in the Infected group. Patients’ baseline demographic and primary clinical characteristics are summarized in
Table 1. The cultures and microbiological identification rates are depicted in
Table 1 of the Electronic Supplementary Material (ESM).
IQR denotes Interquartile range; SOFA denotes Sequential Organ Failure Assessment; SAPS denotes Simplified Acute Physiology Score; CRP denotes C-Reactive Protein; PCT denotes Procalcitonin.
The groups were not different in gender distribution, age or SAPS III at admission, although the group with ICU-acquired infection had significantly more respiratory, renal and hemodynamic organ support requirements, along with a higher SOFA score in the first 24h after admission (SOFA score at admission, p<0.001). Similarly, ICU length of stay and in-hospital length of stay were different between groups, albeit no differences in in-hospital mortality rates were identified in infected versus non-infected patients (
Table 2).
The median (interquartile range) of CRP and PCT were not different between non-infected and Infected groups at ICU admission. However, the Infected group presented significantly higher maximum values of CRP and PCT, during ICU stay, before day 0 (
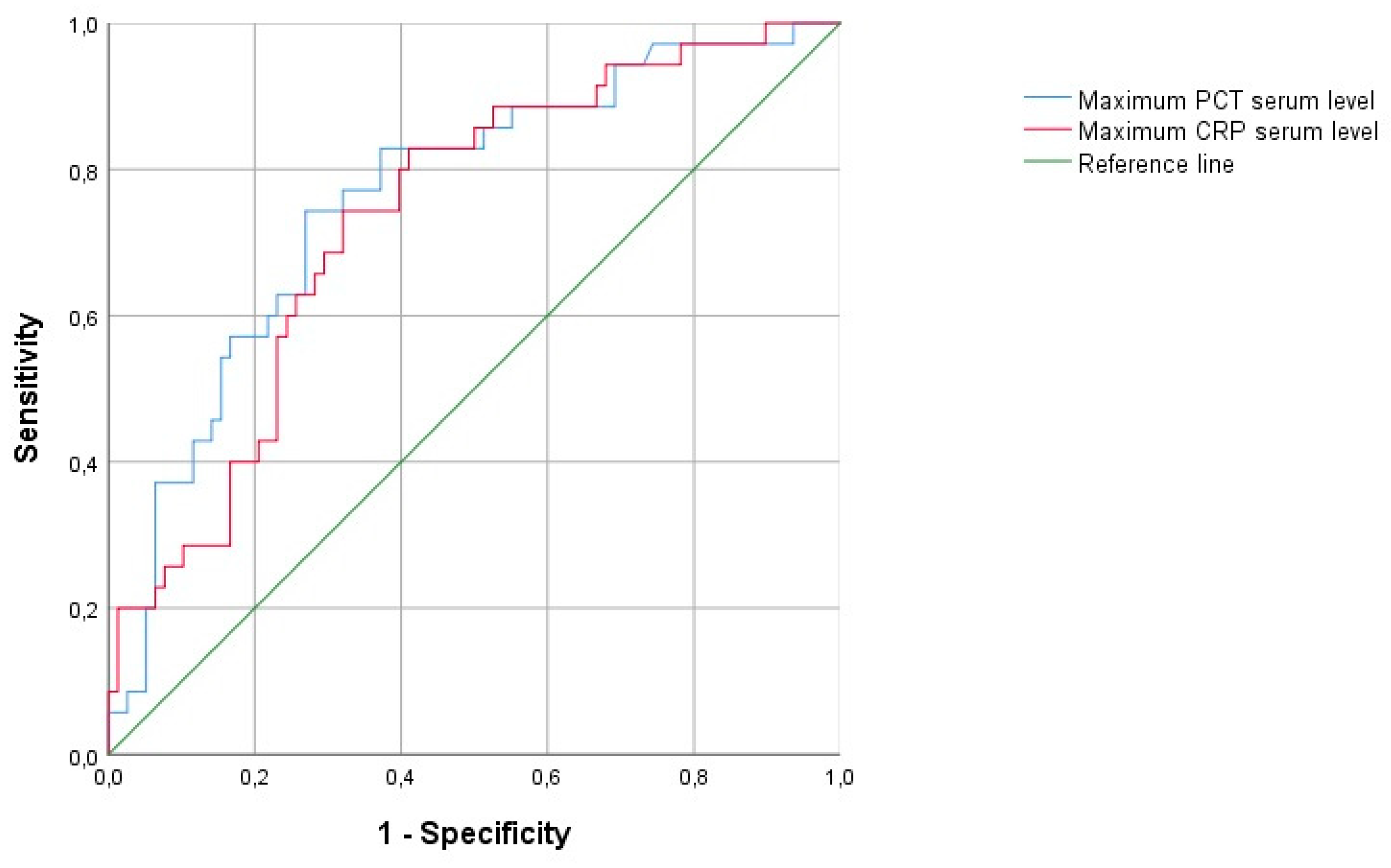
Table 2). The AUC of maximum CRP and PCT as predictors of infection were 0.734 and 0.762, respectively, with an overall similar quality of the model (0.64 vs 0.67, respectively. (
Figure 2).
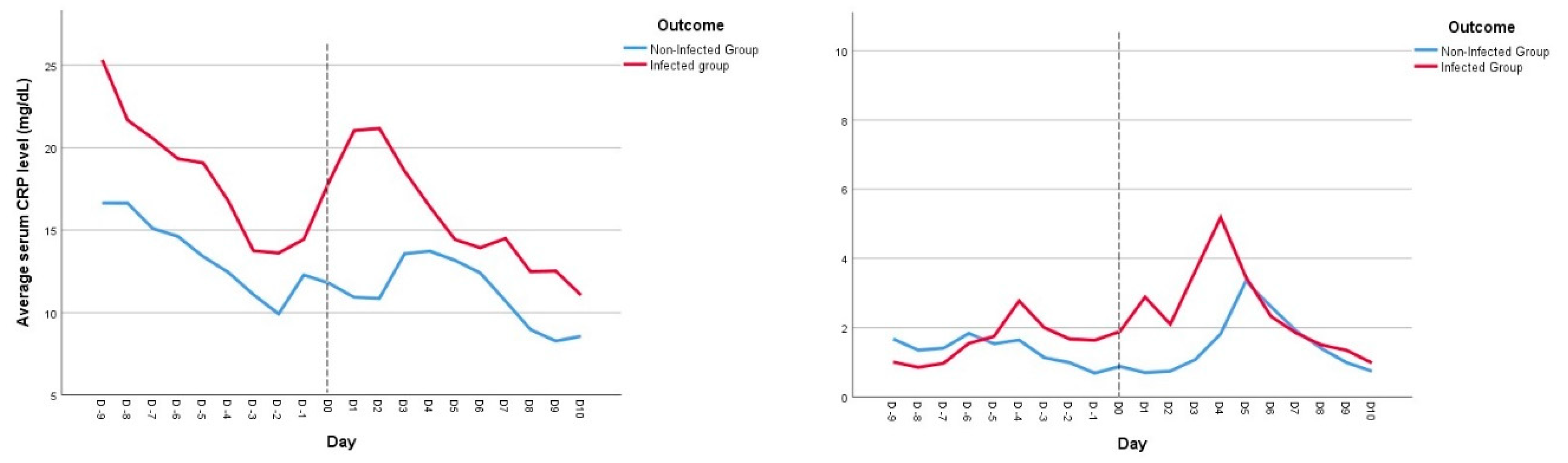
Time-dependent analysis of CRP (
Figure 3, Image A) showed a significant increase of this biomarker in the 48h before the day of infection diagnosis, whereas the CRP level in noninfected patients remained almost unchanged and steady, during the days before the event of interest (p=0.009). On the other hand, PCT serum levels were not different between the two groups (p=0.857) (
Figure 3, Image B).
For the multivariate logistic regression model, only SOFA score at admission, maximum CRP and PCT before day 0 were considered to the model and no multicollinearity was observed. The table 3 depicts the multivariable logistic regression analysis with identification of maximum CRP registered before day 0 as an independent predictor of ICU-acquired infection (model n = 89, comprehending all patients with ICU-acquired infection (n = 35), Hosmer-Lemeshow chi-squared 2.535 (p=0.96), AUC = 0.846).
Patients were classified based on previously defined CRP kinetic patterns observed prior to day 0. The majority of patients demonstrated CRP kinetic profiles corresponding to pattern B (51 patients, 43.2%) and pattern C (34 patients, 28.8%). Pattern A encompassed 9 patients (7.6%), while pattern D included 24 patients (20.4%). Key demographic and primary clinical characteristics of patients, grouped according to their respective patterns, are presented in
Table 2 of ESM. In terms of the distribution of CRP kinetic patterns between the Non-Infected and Infected groups, patterns A and B were more prevalent in the infected group, while patterns C and D were more prominent in the non-Infected groups (
Table 4). An analysis over time of the distinct CRP kinetics has revealed statistically significant differences in these evolving patterns (p < 0.001). However, no discernible differences were noted in terms of clinical severity upon admission, maximum CRP levels recorded during the ICU stay, or ICU and in-hospital mortality rates among the patients.
4. Discussion
Our study presents compelling evidence of distinct CRP and PCT kinetics in severe COVID19 patients with and without ICU-acquired infections. While no differences were observed between the two groups in the levels of biomarkers at ICU admission, there was a significant difference in their maximum values before the day of infection diagnosis. These findings are in accordance with previously published evidence highlighting the potential prediction value of PCT and CRP in the identification of ICU-acquired infections in COVID19 patients [
6,
12,
17]. However, our study advances this evaluation by emphasizing that, as single determination, these biomarkers only offer a moderately reasonable predictive value (AUC 0.73-0.76), precluding a more reasonable approach in their dynamic interpretation rather than a static value, especially considering the still unpredictable nature of these infections’ appearance. In fact, our results provide compelling evidence of the higher predictive value for infection diagnosis of CRP over PCT, using a longitudinal, time-dependent analysis of these biomarkers. The daily CRP values had a significant increase in the 48-hour period preceding the diagnosis of infection, whereas non-infected patients exhibited relatively stable CRP levels. On the other hand, no significant differences were found in daily PCT levels over time between the groups.
Contrary to the findings of Farrel-Cortês et. al [
11] and Richards, et. Al [
19] studies, our results from the multivariate logistic regression model establish CRP, rather than PCT, as an independent predictor of ICU-acquired infection with a proposed model with a more reliable prediction value, irrespective of the focus of the superimposed infection. On the other hand, our time-dependent analysis findings strongly challenge previous collected evidence, based on smaller cohorts [
11,
19], supporting the association of this biomarker with the development of acquired infections in the ICU. Moreover, it firmly challenges previous studies proposing the use of this biomarker as a helpful tool for antibiotic withdrawal and as a stewardship tool [
5].
Interestingly, our findings derived from the analysis of CRP kinetic patterns, prior to infection diagnosis, also challenge the prevailing paradigm of commonly accepted patterns as prognostic indicators for ICU-acquired infections. Within the analyzed cohort of critically ill COVID-19 patients, a persistent pro-inflammatory profile characterized by persistently elevated serum CRP levels over time was associated with a risk of developing an infection during the ICU stay, as delineated by pattern A and B. Notably, patients exhibiting these CRP kinetic patterns also displayed a discernible trend towards elevated mortality rates, suggesting a potential association between sustained elevation of serum CRP and heightened rates of organ dysfunction and infection, consequently translating to poorer clinical outcomes. Concretely, almost two thirds of infected patients presented the expected A and B patterns (22 patients, 62.9%). Similarly, 54.2% of patients without ICU-acquired infection exhibit predominantly C and D patterns. Nevertheless, a subset of patients who acquired infections during their ICU stay also displayed an alternative pattern characterized by a decline in CRP serum levels in the days preceding infection diagnosis (Pattern C).
To our knowledge, our results show a novel clinical scenario of biomarker profiling and its predictive value in COVID19 patients before the diagnosis of ICU-acquired infection. They further stand that CRP kinetics retains an acceptable infection predictive value and should be promptly considered in conjunction with relevant clinical assessment and microbiological culture collection in septic COVID-19 patients. Therefore, our results provide evidence that continuous monitoring of CRP may have a useful role in critical care setting in these patients and has the potential to refine and compose predictor models to expedite the identification of these infections and mitigate their impact on patients' survivability.
As strengths, our study represents a longitudinal analysis of a reasonably large cohort of critical care COVID-19 patients, reflecting real-world clinical decision-making. Importantly, biomarker sampling was performed irrespective of clinical suspicion of ICU-acquired infection, and a high overall rate of microbiological sampling was achieved. Furthermore, none of the eligible patients in this study were treated with Interleukin-6 antagonists which could potentially bias our results.
However, we acknowledge some limitations in our study. It is retrospective in nature, and we assumed the median day of infection diagnosis as the corresponding day 0 in the non-infected group, which may introduce determination bias due to the heterogeneous courses of COVID-19 disease over time. Furthermore, we observed an overall higher incidence of positive bacterial isolation than previously documented estimates [Langford], and we recognize the inherent limitations of microbiological culture methods in detecting the presence of infection. We also acknowledge that co-infection diagnosis at ICU admission in the analyzed COVID19 patients were not considered in the kinetic patterns modeling and outcome analysis, although no difference was found in those co-infection rates between both groups (4 patients (4.8%) in non-infection group versus 2 patients (5.7%) in the infection group). Additionally, the independent evaluation of the attending physician was used as a key element for ICU-acquired infection diagnosis and distinguishing infection from colonization in patients with microbiological bacterial isolates. While it was not corroborated by another expert's review, the majority of diagnoses were considered clinically relevant and treated accordingly. Nonetheless, this approach also reflects the real-world challenges faced by physicians in managing critically ill COVID-19 patients.
5. Conclusions
Our study presents a novel clinical scenario of biomarker profiling and its value in predicting ICU-acquired infections in COVID-19 patients. The kinetics of CRP may play a useful role in critical care settings and has the potential to refine and compose predictor models to expeditiously predict these infections, ultimately reducing their impact on patients' survivability. In addition, the identification of the CRP patterns could increase our ability to identify patients with ICU-acquired infection. The CRP patterns A and B were significantly more present in COVID19 patients with ICU-acquired infection. These findings contribute to the ongoing efforts to improve the management of ICU-acquired infections in critically ill COVID-19 patients and warrant further research to optimize the use of biomarkers in this context.
Supplementary Materials
The following supporting information can be downloaded at:
www.mdpi.com/xxx/s1,
Table 1 ESM – Characterization of microbiological cultures (A) and Isolated microorganisms (B) in the analyzed population;
Table 2 ESM – Demographic and primary clinical characteristics according to CRP kinetic patterns.
Author Contributions
JPC, LC and PP were responsible for conceptualization, methodology, validation, investigation, data curation and project administration. JPC was responsible for formal analysis and writing—original draft preparation. LC and PP were responsible for writing—review and editing, visualization, and supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Portuguese Ethics Committee for Clinical Investigation in Lisbon (REC: 2020_EO_02).
Informed Consent Statement
Considering the observational nature of study, the COVID19 pandemic state and the anonymity of the data collected, the ethic committee waived the need of written informed consent. All details and data collected that might disclose the subjects under the study were omitted or anonymized.
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available due to privacy issues, but are available from the corresponding author on reasonable request.
Conflicts of Interest
JPC and LC declare no conflict of interest. PP received honoraria for lectures and advisory boards from Abionic, Merck Sharp & Dohme, Sanofi, Gilead and Pfizer.
References
- Russo A, Olivadese V, Trecarichi EM, et al. Bacterial Ventilator-Associated Pneumonia in COVID-19 Patients: Data from the Second and Third Waves of the Pandemic. J Clin Med 2022;11(9). [CrossRef]
- Boyd S, Nseir S, Rodriguez A, et al. Ventilator-associated pneumonia in critically ill patients with COVID-19 infection: a narrative review. ERJ Open Res 2022;8(3). [CrossRef]
- Langford BJ, So M, Raybardhan S, et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect 2020;26(12):1622-9. [CrossRef]
- Chamorro-de-Vega E, Rodriguez-Gonzalez CG, Manrique-Rodríguez S, et al. Clinical course of severe patients with COVID-19 treated with tocilizumab: report from a cohort study in Spain. Expert Rev Clin Pharmacol 2021;14(2):249-60. [CrossRef]
- Roy A, Powers HR, Craver EC, et al. Antibiotic stewardship: Early discontinuation of antibiotics based on procalcitonin level in COVID-19 pneumonia. J Clin Pharm Ther 2022;47(2):243-7. [CrossRef]
- Ming DK, Myall AC, Hernandez B, et al. Informing antimicrobial management in the context of COVID-19: understanding the longitudinal dynamics of C-reactive protein and procalcitonin. BMC Infectious Diseases 2021;21(1):932. [CrossRef]
- Moreno J, Carvelli J, Lesaux A, et al. Ventilator Acquired Pneumonia in COVID-19 ICU Patients: A Retrospective Cohort Study during Pandemia in France. J Clin Med 2023;12(2). [CrossRef]
- Maes M, Higginson E, Pereira-Dias J, et al. Ventilator-associated pneumonia in critically ill patients with COVID-19. Crit Care 2021;25(1):25. [CrossRef]
- Khan W, Safi A, Muneeb M, et al. Complications of invasive mechanical ventilation in critically Ill Covid-19 patients - A narrative review. Ann Med Surg (Lond) 2022;80:104201. [CrossRef]
- Lingscheid T, Lippert LJ, Hillus D, et al. Characterization of antimicrobial use and co-infections among hospitalized patients with COVID-19: a prospective observational cohort study. Infection 2022;50(6):1441-52. [CrossRef]
- Côrtes MF, de Almeida BL, Espinoza EPS, et al. Procalcitonin as a biomarker for ventilator associated pneumonia in COVID-19 patients: Is it an useful stewardship tool? Diagn Microbiol Infect Dis 2021;101(2):115344. [CrossRef]
- Pink I, Raupach D, Fuge J, et al. C-reactive protein and procalcitonin for antimicrobial stewardship in COVID-19. Infection 2021;49(5):935-43. [CrossRef]
- Luan YY, Yin CH, Yao YM. Update Advances on C-Reactive Protein in COVID-19 and Other Viral Infections. Front Immunol 2021;12:720363. [CrossRef]
- Gragueb-Chatti I, Lopez A, Hamidi D, et al. Impact of dexamethasone on the incidence of ventilator-associated pneumonia and blood stream infections in COVID-19 patients requiring invasive mechanical ventilation: a multicenter retrospective study. Ann Intensive Care 2021;11(1):87. [CrossRef]
- Mason CY, Kanitkar T, Richardson CJ, et al. Exclusion of bacterial co-infection in COVID-19 using baseline inflammatory markers and their response to antibiotics. J Antimicrob Chemother 2021;76(5):1323-31. [CrossRef]
- Stringer D, Braude P, Myint PK, et al. The role of C-reactive protein as a prognostic marker in COVID-19. Int J Epidemiol 2021;50(2):420-9. [CrossRef]
- Houghton R, Moore N, Williams R, et al. C-reactive protein-guided use of procalcitonin in COVID-19. JAC Antimicrob Resist 2021;3(4):dlab180. [CrossRef]
- Potempa LA, Rajab IM, Hart PC, et al. Insights into the Use of C-Reactive Protein as a Diagnostic Index of Disease Severity in COVID-19 Infections. Am J Trop Med Hyg 2020;103(2):561-3. [CrossRef]
- Richards O, Pallmann P, King C, et al. Procalcitonin Increase Is Associated with the Development of Critical Care-Acquired Infections in COVID-19 ARDS. Antibiotics (Basel) 2021;10(11). [CrossRef]
- Hu R, Han C, Pei S, et al. Procalcitonin levels in COVID-19 patients. Int J Antimicrob Agents 2020;56(2):106051. [CrossRef]
- Rubio-Rivas M, Mora-Luján JM, Montero A, et al. The Use of Corticosteroids or Tocilizumab in COVID-19 Based on Inflammatory Markers. J Gen Intern Med 2022;37(1):168-75. [CrossRef]
- Malik P, Patel U, Mehta D, et al. Biomarkers and outcomes of COVID-19 hospitalisations: systematic review and meta-analysis. BMJ Evid Based Med 2021;26(3):107-8. [CrossRef]
- Alhazzani W, Møller MH, Arabi YM, et al. Surviving Sepsis Campaign: guidelines on the management of critically ill adults with Coronavirus Disease 2019 (COVID-19). Intensive Care Med 2020;46(5):854-87. [CrossRef]
- Del Sole F, Farcomeni A, Loffredo L, et al. Features of severe COVID-19: A systematic review and meta-analysis. Eur J Clin Invest 2020;50(10):e13378. [CrossRef]
- Ali N. Elevated level of C-reactive protein may be an early marker to predict risk for severity of COVID-19. J Med Virol 2020;92(11):2409-11. [CrossRef]
- Del Valle DM, Kim-Schulze S, Huang HH, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med 2020;26(10):1636-43. [CrossRef]
- Group ICC, Kartsonaki C. Characteristics and outcomes of an international cohort of 400,000 hospitalised patients with Covid-19. medRxiv 2021:2021.09.11.21263419. [CrossRef]
- Póvoa P, Coelho L, Almeida E, et al. Early identification of intensive care unit-acquired infections with daily monitoring of C-reactive protein: a prospective observational study. Crit Care 2006;10(2):R63. [CrossRef]
- Torres A, Niederman MS, Chastre J, et al. International ERS/ESICM/ESCMID/ALAT guidelines for the management of hospital-acquired pneumonia and ventilator-associated pneumonia: Guidelines for the management of hospital-acquired pneumonia (HAP)/ventilator-associated pneumonia (VAP) of the European Respiratory Society (ERS), European Society of Intensive Care Medicine (ESICM), European Society of Clinical Microbiology and Infectious Diseases (ESCMID) and Asociación Latinoamericana del Tórax (ALAT). Eur Respir J 2017;50(3). [CrossRef]
- Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med 2005;171(4):388-416. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).