Submitted:
19 August 2023
Posted:
22 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Mass-Spectrometry screening, isolation, and purification of cysteine-rich peptides in aqueous extracts of C. canephora and C. liberica
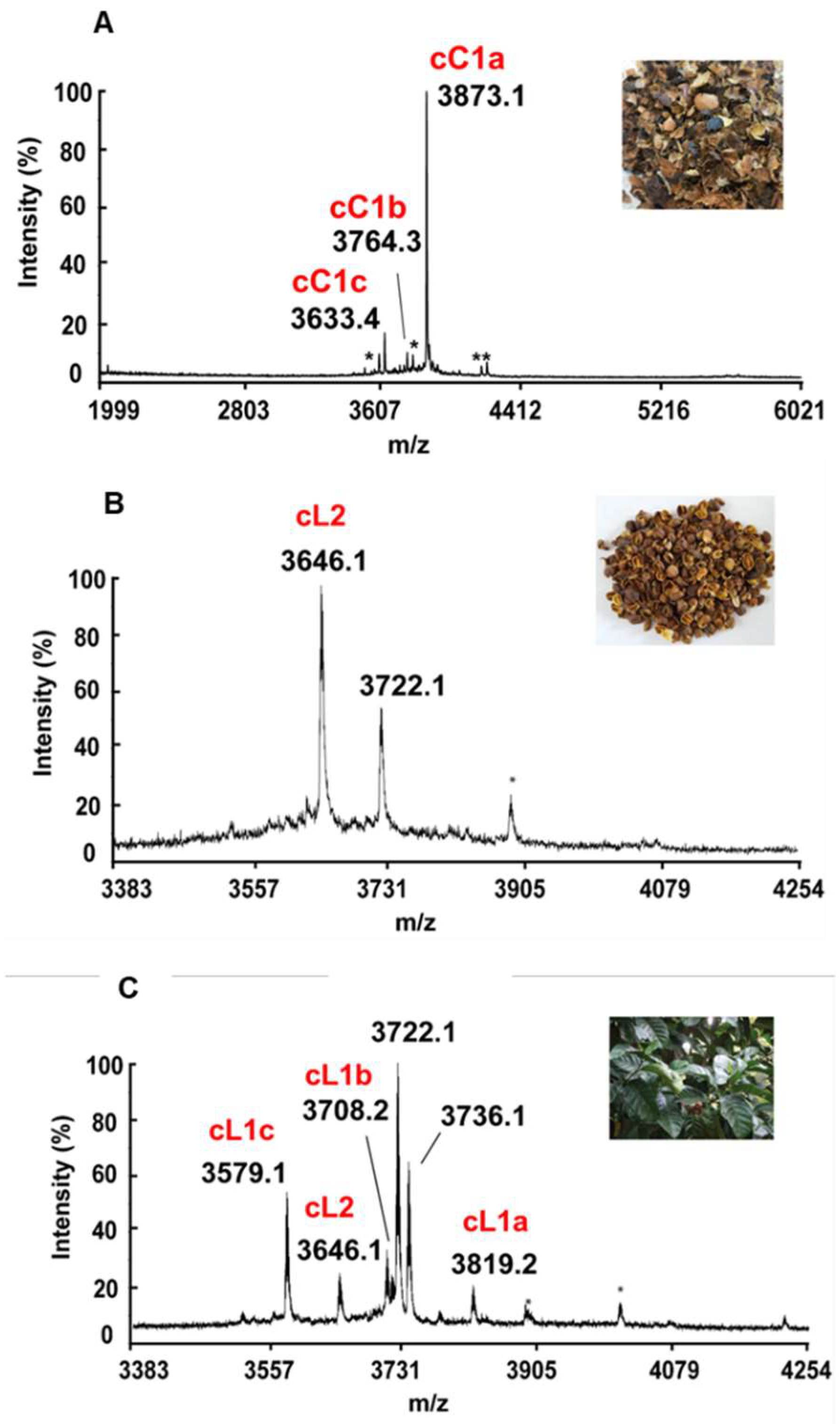
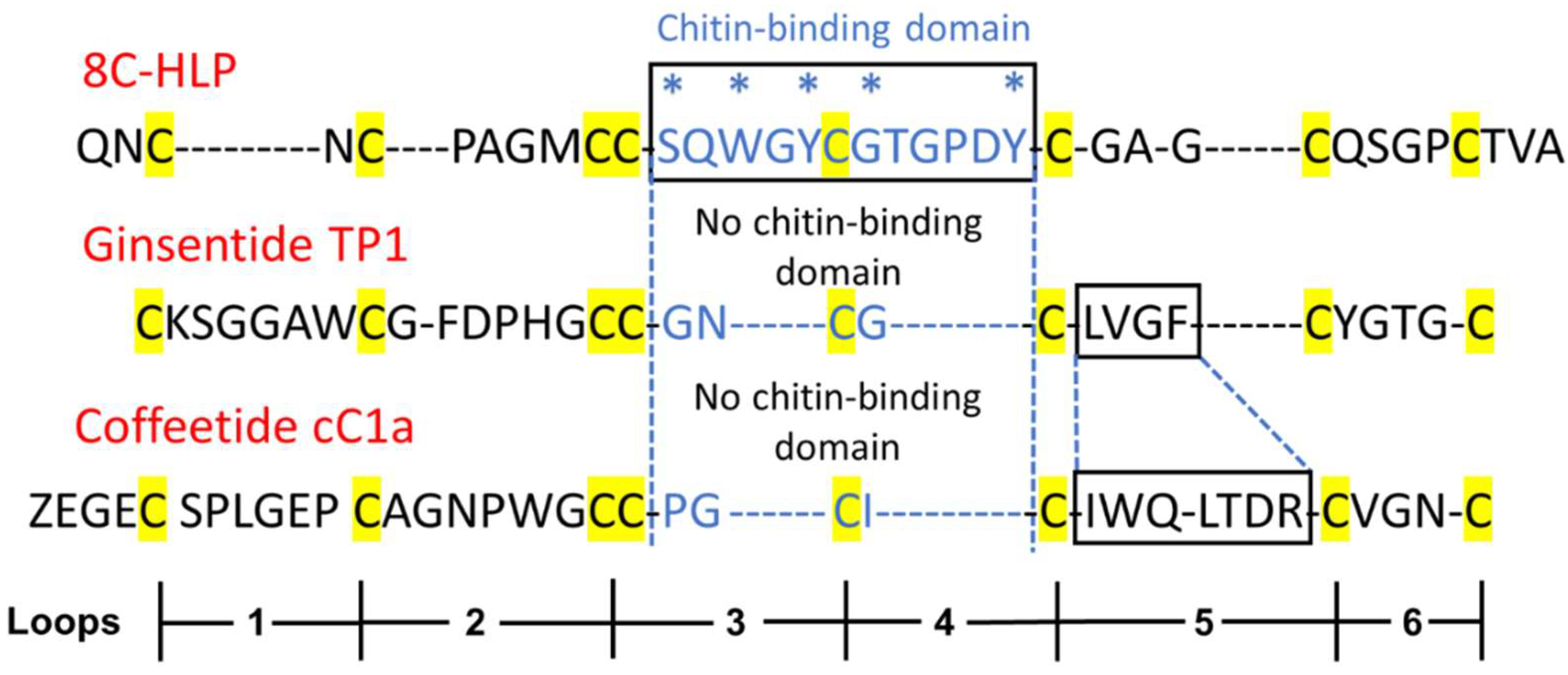
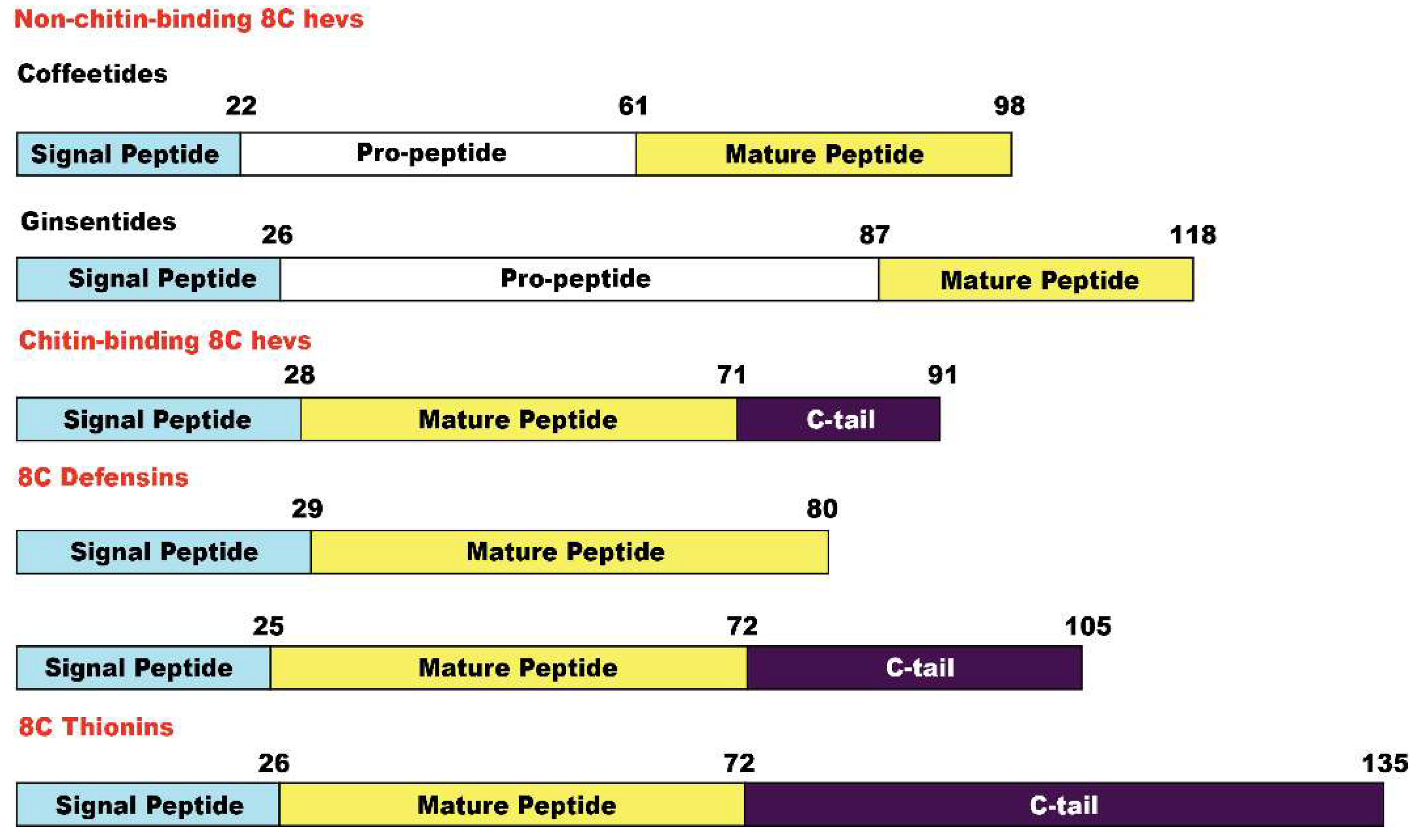
2.2. Sequencing, database search and transcriptomic analysis of coffeetides

| Peptide | Species | Amino acid sequence | Mass (Da) 1 | Charge 2 | PI | Approach 3 |
|---|---|---|---|---|---|---|
| Loops | 1 2 3 4 5 6 | |||||
| cC1a | C. canephora | ZEGECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CVGNC | 3873.1 | -3 | 4.00 | T, P |
| cC1b | C. canephora | -EGECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CVGNC | 3764.3 | -3 | 4.00 | T, P |
| cC1c | C. canephora | --GECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CVGNC | 3633.4 | -2 | 4.14 | T, P |
| cC2 | C. canephora | QEGECSPFGKPCRYNPWGCCDSCVCVAT-PADE-GRCLGNC | 4145.6 | -2 | 4.51 | T |
| cC3 | C. canephora | QEGECSPLGKPCKYNPWGCCGSCLCIVDQP-THEGTCVGNC | 4206.7 | -2 | 4.83 | T |
| cC4 | C. canephora | QEGECSPLGKPCRYNPWGCCGSCLCIVDQP-THEGTCVGNC | 4234.7 | -2 | 4.83 | T |
| cC5 | C. canephora | QEGECSALGKPCRYNPSGCCGLCVCVIPDPTDE-GSCIGIC | 4067.7 | -3 | 4.18 | T |
| cC6 | C. canephora | QEVECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CVGNC | 3931.6 | -3 | 4.00 | T |
| cC7 | C. canephora | QEPSCIPVLGSCVGNPWGCCPGCMCIRQ-LTDR---CHGYC | 3976.6 | 0 | 6.69 | T |
| cL1a | C. liberica | ZEGECSPAGKPCR--PLGCCGACLCIVDHP-THEGTCVGNC | 3819.2 | -2 | 5.36 | T, P |
| cL1b | C. liberica | -EGECSPAGKPCR--PLGCCGACLCIVDHP-THEGTCVGNC | 3708.2 | -2 | 5.36 | T, P |
| cL1c | C. liberica | --GECSPAGKPCR--PLGCCGACLCIVDHP-THEGTCVGNC | 3579.1 | -1 | 6.01 | T, P |
| cL2 | C. liberica | --GECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CIGNC | 3649.2 | -2 | 4.14 | T, P |
| cL3 | C. liberica | GKDTCIGLLESCKDDPWGCCFGCVCLWP--GDL---CRGSC | 3834.4 | -2 | 4.36 | T |
| cL4 | C. liberica | QEGECSPAGKSCR--PVRCCDFCVCVVDYP-THVGTCRGNC | 4069.7 | -2 | 4.36 | T |
| cL5 | C. liberica | QEGECSPAGKPCR--PVRCCDFCVCVVDYP-THVGTCRGNC | 4082.7 | 0 | 6.70 | T |
| cL6 | C. liberica | QEPSCIPVLGSCVGNPWGCCPGCMCIRQ-LTDR---CHGYC | 3976.7 | 0 | 6.69 | T |
| cA1 | C. arabica | QEGECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CIGNC | 3903.5 | -3 | 4.00 | T |
| cA2 | C. arabica | QEGECSPLGEACAGNPWGCCPGCICIWQ-LTDR---CVGNC | 3863.5 | -3 | 4.00 | T |
| cA3 | C. arabica | QEPSCLPAGESCTGNPWGCCPGCICIWQ-LTER---CVGNC | 3905.6 | -2 | 4.25 | T |
| cA4 | C. arabica | QEPSCIPVGEPCAGNPGGCCDGCICIWQ-LTDR---CAGSC | 3733.5 | -3 | 3.92 | T |
| cA5 | C. arabica | QEGECSPLGKPCRYNPRGCCDFCVCVVADVTDEEGSCRGNC | 4389.7 | -3 | 4.44 | T |
| cA6 | C. arabica | RKDTCIGLLESCKDDPYGCCPGCVCLWP--GDL---CRGDC | 3884.6 | -2 | 4.46 | T |
| cA7 | C. arabica | QEGECSPAGKPCR--PVRCCDSCLCIVDYP-THVGTCRGNC | 4047.7 | 0 | 6.70 | T |
| cA8 | C. arabica | QEGECSPLGKPCAGNPWGCCPGCICIWQ-LTDR---CIGNC | 3902.5 | -1 | 4.68 | T |
| cA9 | C. arabica | QEPSCIPVGEPCAGNPGGCCDGCICIWQ-LTDR---CAGSC | 3733.3 | -3 | 3.92 | T |
| cA10 | C. arabica | QEGECSPLGEPCAGNPWGCCPGCICIWQ-LTDR---CVGNC | 3890.1 | -3 | 4.00 | T |
| cR1 | C. racemosa | QEPRCIPALGSCVGNPWGCCFGCMCIRQ-LTDR---CLGYC | 4043.7 | +1 | 7.70 | T |
| cR2 | C. racemosa | QEPRCIPVFGSCVGNPWGCCFGCMCIRQ-HTNR---CLGYC | 4128.7 | +2 | 8.22 | T |
| cR3 | C. racemosa | QEGECSPFGKPCRYNPWGCCGSCLCVVDHP-THEGTCVGNC | 4263.7 | -2 | 5.36 | T |
| cR4 | C. racemosa | QEGECSPFGKPCRYNPWGCCGSCVCVVDHP-THEGTCLGNC | 4263.7 | -2 | 5.36 | T |
| cR5 | C. racemosa | QEGKCSPAGKPC--DPWGCCDFCVCVVDFPGGE-GRCAGNC | 3885.5 | -2 | 4.44 | T |
| cR6 | C. racemosa | QEEKCSPAGKPCRYNPRGCCDFCVCVVDFPGGE-GSCLGNC | 4218.7 | -1 | 4.94 | T |
| cR7 | C. racemosa | GKDTCIGLLESCKDDPWGCCPGCVCLWP--GDL---CRGSC | 3780.5 | -2 | 4.36 | T |
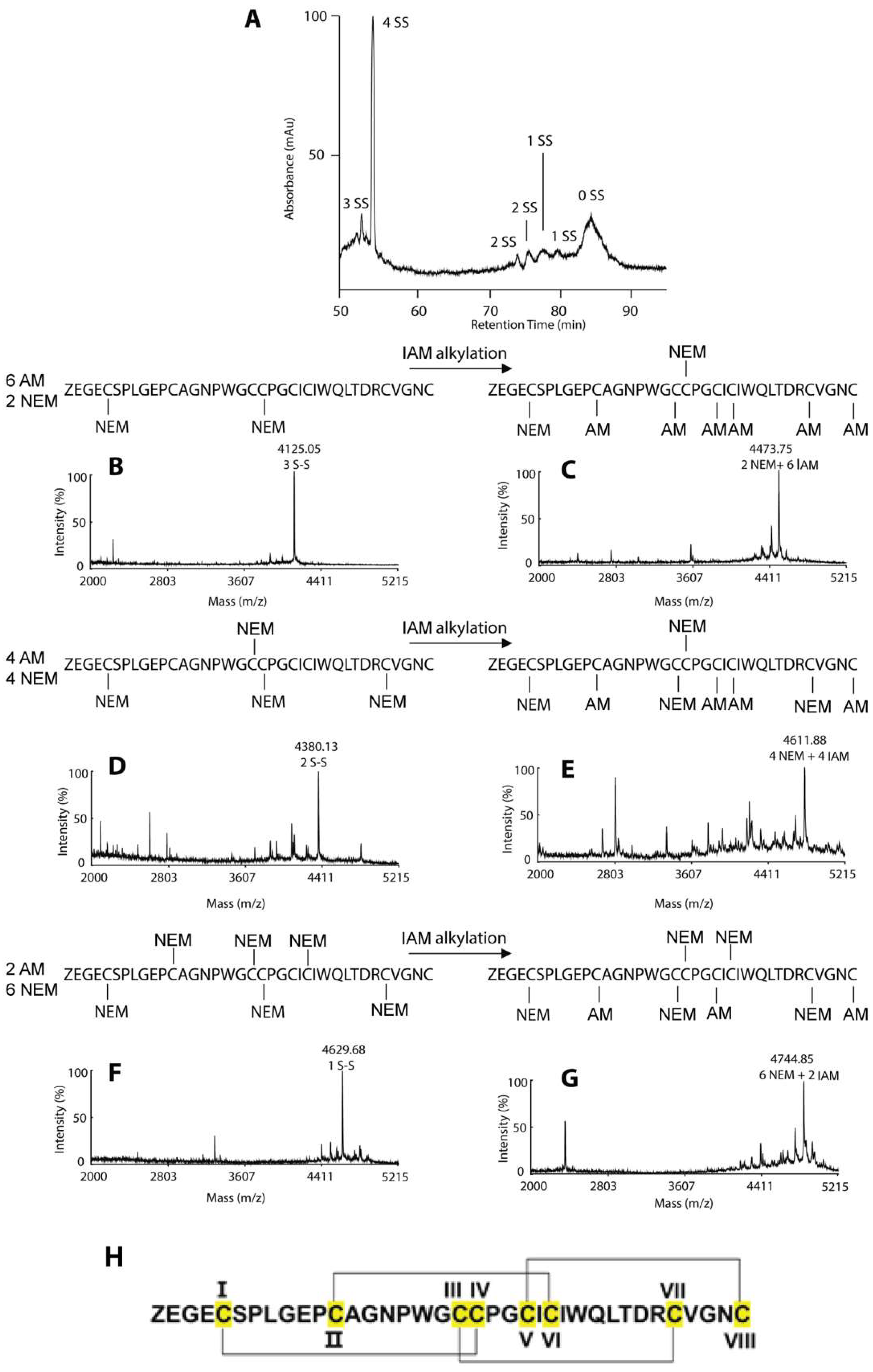
2.3. Disulfide mapping of coffeetide cC1a
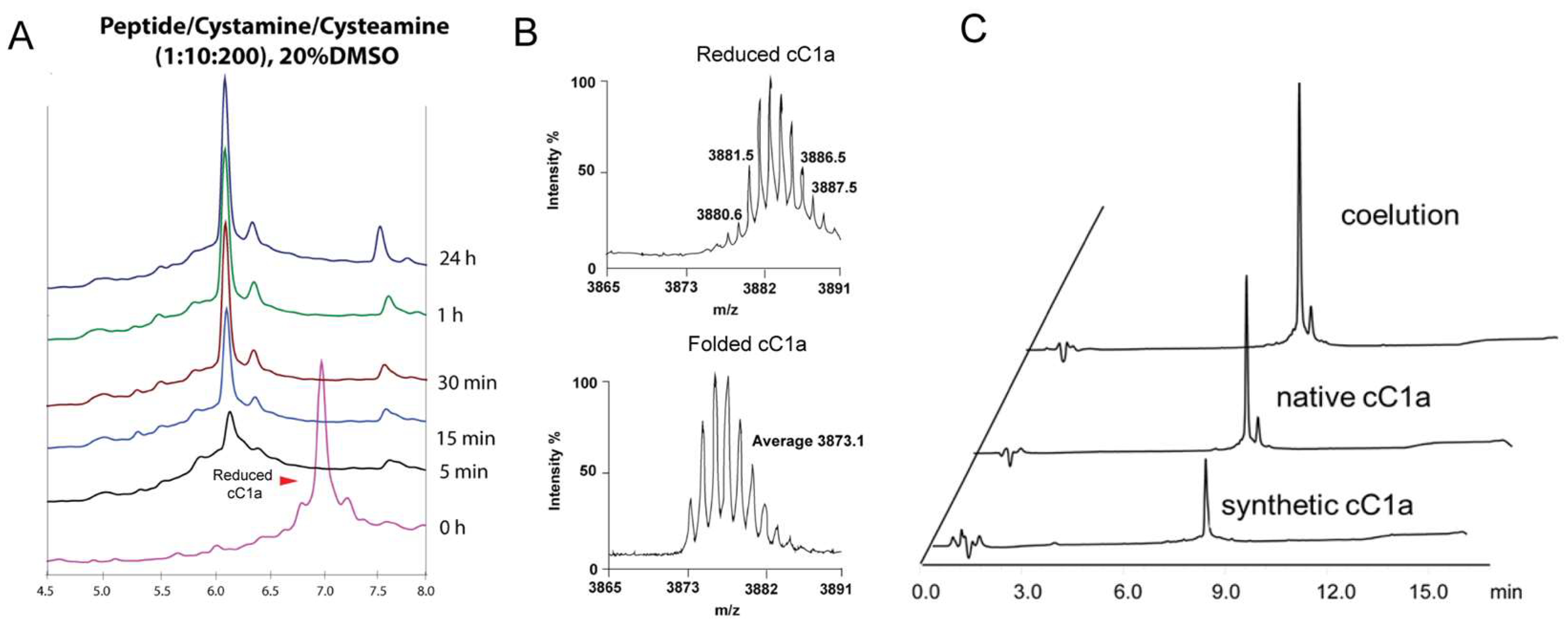
2.4. Chemical synthesis and oxidative folding of coffeetide cC1a
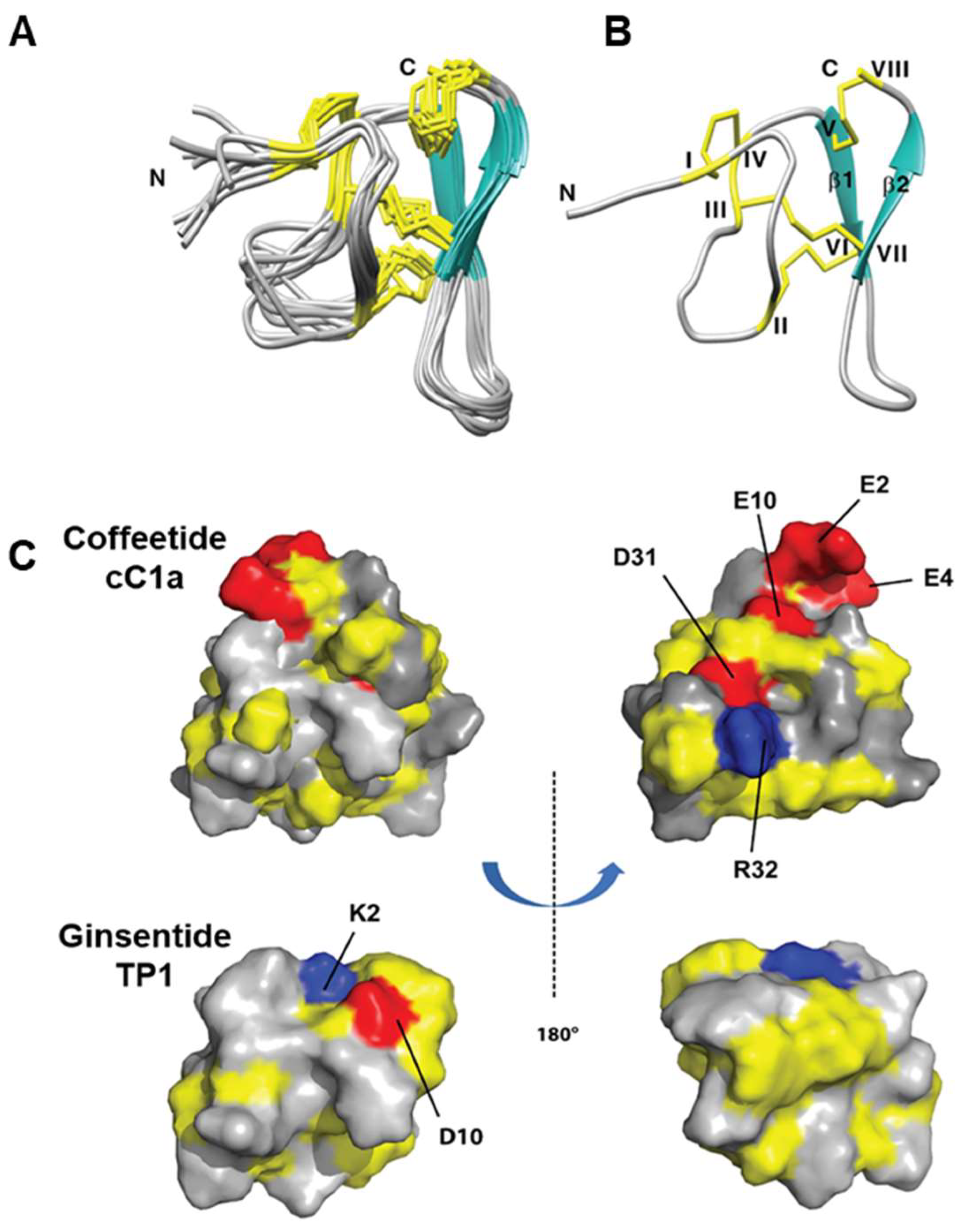
2.5. Solution NMR Structure of cC1a
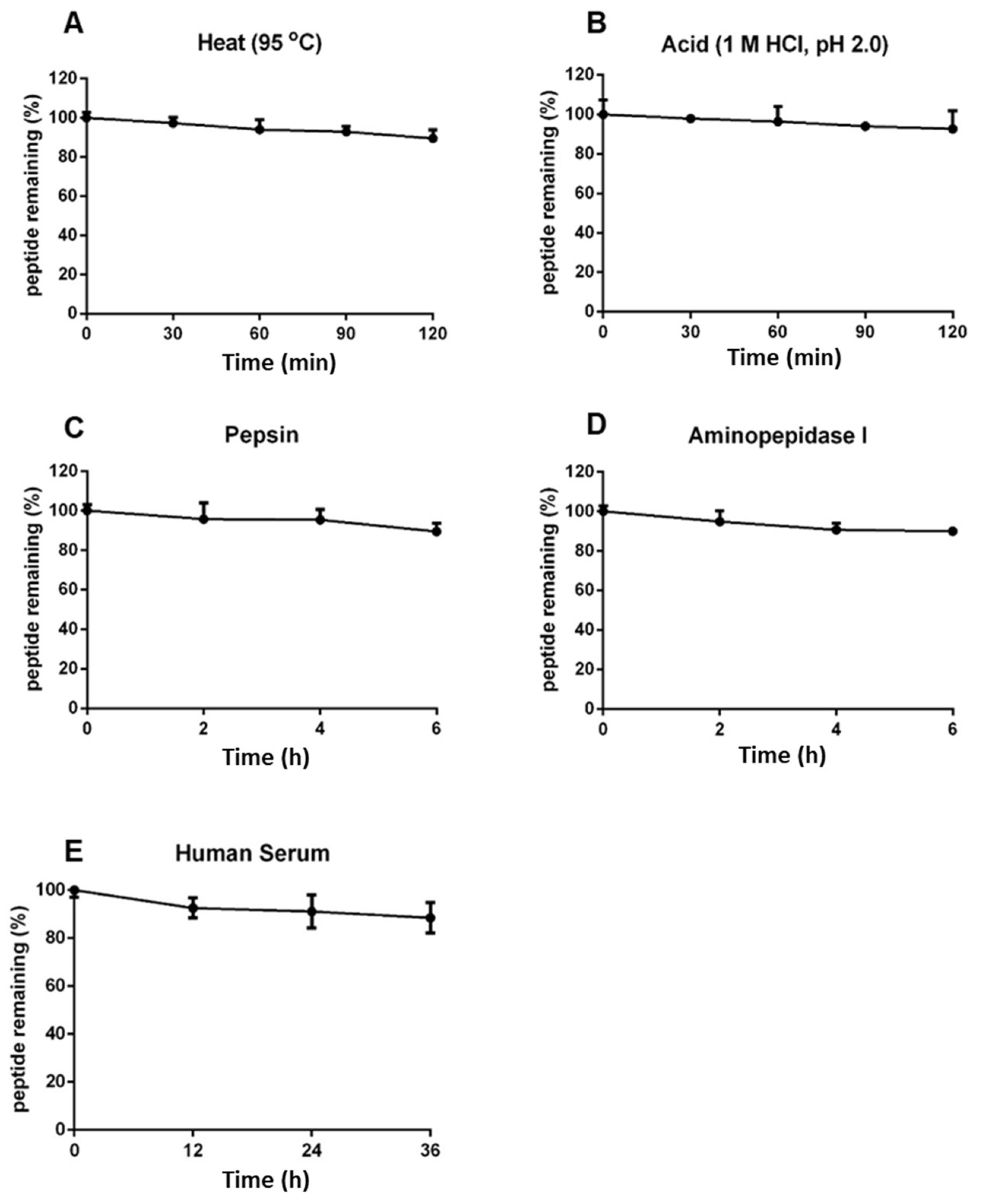
2.6. Coffeetide cC1a is resistant to heat, acid, proteolytic, and human serum-mediated degradation
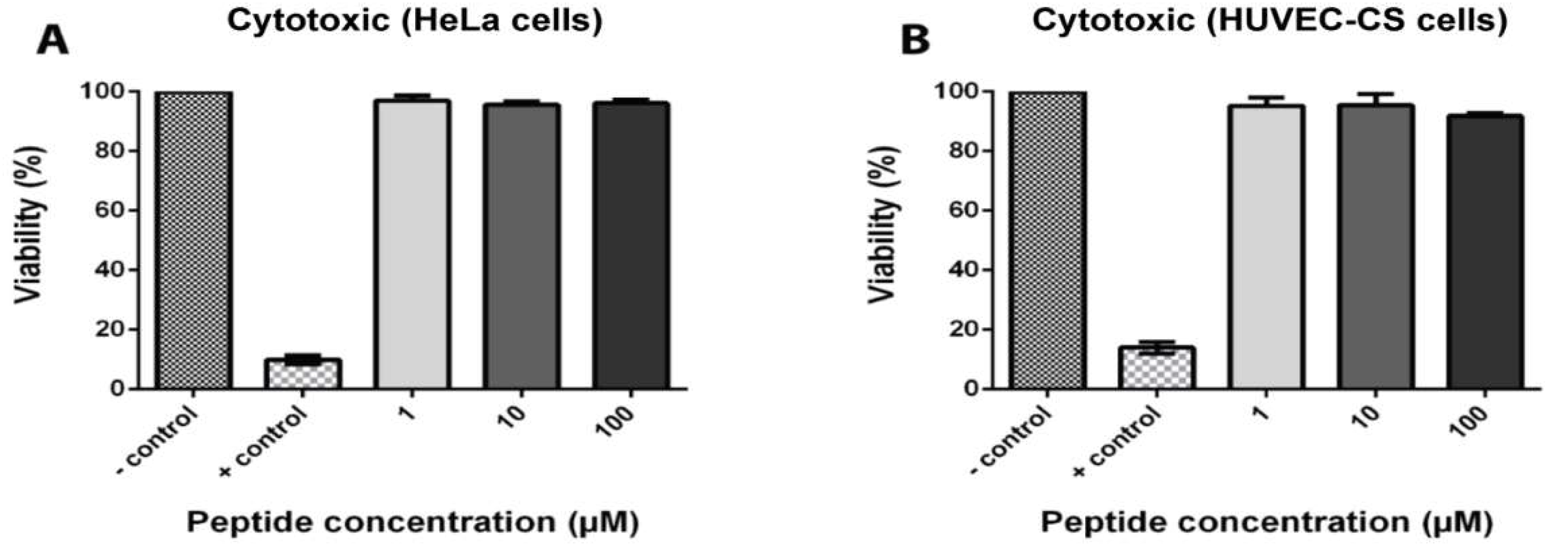
2.7. Coffeetide cC1a is non-cytotoxic
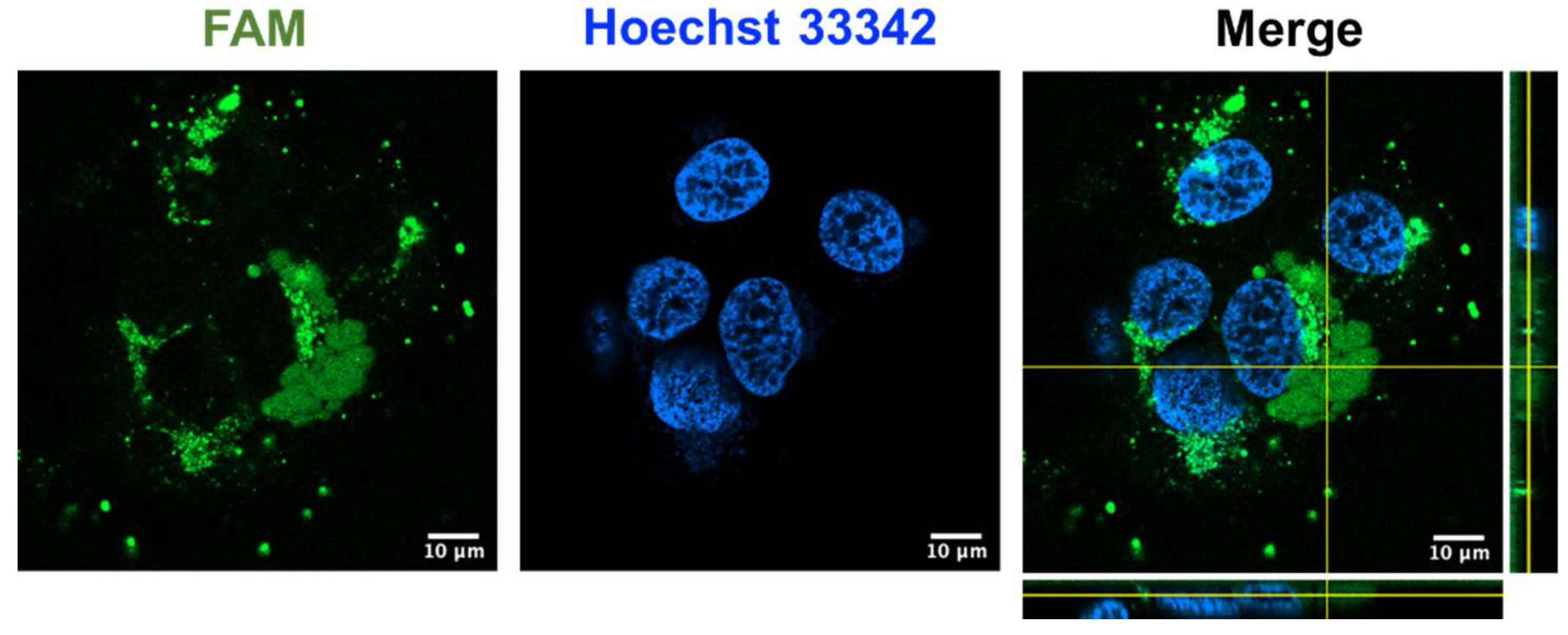
2.8. Coffeetide cC1a is cell-penetrating
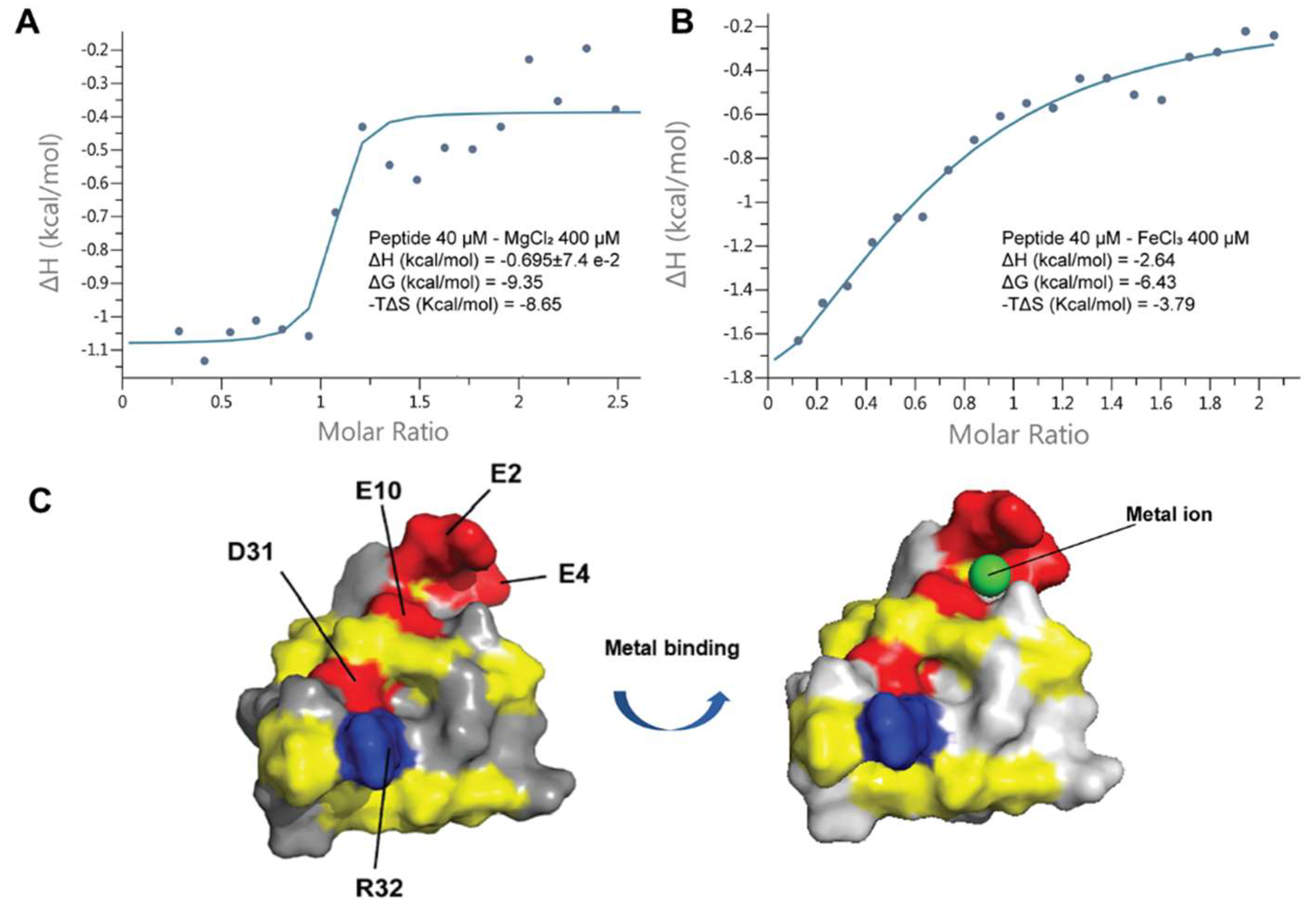
2.9. Coffeetide cC1a is metal binding
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Plant materials
4.3. Extraction and screening of Coffea canephora and Coffea liberica
4.4. Isolation and purification of coffeetides
4.5. Data mining and bioinformatics analysis
4.6. Sequence determination of coffeetides
4.7. Disulfide mapping
4.8. NMR solution structure
4.9. Chemical synthesis of coffeetide
4.10. Oxidative folding of coffeetide cC1a
4.11. Stability assays
4.12. Cell culture
4.13. Toxicity assay
4.14. Cell penetrating assay
4.15. Isothermal Titration Calorimetry (ITC) assay
4.16. Statistical analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
References
- Hu, G.L.; Wang, X.; Zhang, L.; Qiu, M.H. The sources and mechanisms of bioactive ingredients in coffee. Food & function 2019, 10, 3113–3126. [Google Scholar] [CrossRef]
- Matosinhos, R.C.; Bezerra, J.P.; Barros, C.H.; Fernandes Pereira Ferreira Bernardes, A.C.; Coelho, G.B.; Carolina de Paula Michel Araújo, M.; Dian de Oliveira Aguiar Soares, R.; Sachs, D.; Saúde-Guimarães, D.A. Coffea arabica extracts and their chemical constituents in a murine model of gouty arthritis: How they modulate pain and inflammation. Journal of ethnopharmacology 2022, 284, 114778. [Google Scholar] [CrossRef]
- Veltri, C.; Grundmann, O. Current perspectives on the impact of Kratom use. Substance abuse and rehabilitation 2019, 10, 23–31. [Google Scholar] [CrossRef]
- Bisht, S.; Sisodia, S. Coffea arabica: A wonder gift to medical science. J. Nat. Pharmaceut 2010, 1, 58–65. [Google Scholar] [CrossRef]
- Bisht, S.; Sisodia, S.S. Coffea arabica: A wonder gift to medical science. Journal of Natural Pharmaceuticals 2010, 1, 58. [Google Scholar] [CrossRef]
- Gökcen, B.B.; Şanlier, N. Coffee consumption and disease correlations. Critical Reviews in Food Science and Nutrition 2019, 59, 336–348. [Google Scholar] [CrossRef]
- Ali, A.; Zahid, H.F.; Cottrell, J.J.; Dunshea, F.R. A Comparative Study for Nutritional and Phytochemical Profiling of Coffea arabica (C. arabica) from Different Origins and Their Antioxidant Potential and Molecular Docking. Molecules 2022, 27, 5126. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Bhupathiraju, S.N.; Satija, A.; van Dam, R.M.; Hu, F.B. Long-term coffee consumption and risk of cardiovascular disease: A systematic review and a dose-response meta-analysis of prospective cohort studies. Circulation 2014, 129, 643–659. [Google Scholar] [CrossRef]
- Chieng, D.; Kistler, P.M. Coffee and tea on cardiovascular disease (CVD) prevention. Trends in cardiovascular medicine 2022, 32, 399–405. [Google Scholar] [CrossRef]
- James, J.E. Critical review of dietary caffeine and blood pressure: A relationship that should be taken more seriously. Psychosomatic medicine 2004, 66, 63–71. [Google Scholar] [CrossRef]
- Geraets, L.; Moonen, H.J.; Wouters, E.F.; Bast, A.; Hageman, G.J. Caffeine metabolites are inhibitors of the nuclear enzyme poly (ADP-ribose) polymerase-1 at physiological concentrations. Biochemical pharmacology 2006, 72, 902–910. [Google Scholar] [CrossRef]
- Gonzalez de Mejia, E.; Ramirez-Mares, M.V. Impact of caffeine and coffee on our health. Trends in endocrinology and metabolism: TEM 2014, 25, 489–492. [Google Scholar] [CrossRef]
- Bonita, J.S.; Mandarano, M.; Shuta, D.; Vinson, J. Coffee and cardiovascular disease: In vitro, cellular, animal, and human studies. Pharmacol Res 2007, 55, 187–198. [Google Scholar] [CrossRef]
- Socała, K.; Szopa, A.; Serefko, A.; Poleszak, E.; Wlaź, P. Neuroprotective Effects of Coffee Bioactive Compounds: A Review. International journal of molecular sciences 2020, 22, 107. [Google Scholar] [CrossRef]
- Mansour, A.; Mohajeri-Tehrani, M.R.; Samadi, M.; Qorbani, M.; Merat, S.; Adibi, H.; Poustchi, H.; Hekmatdoost, A. Effects of supplementation with main coffee components including caffeine and/or chlorogenic acid on hepatic, metabolic, and inflammatory indices in patients with non-alcoholic fatty liver disease and type 2 diabetes: A randomized, double-blind, placebo-controlled, clinical trial. Nutrition journal 2021, 20, 35. [Google Scholar] [CrossRef]
- Muchtaridi, M.; Lestari, D.; Khairul Ikram, N.K.; Gazzali, A.M.; Hariono, M.; Wahab, H.A. Decaffeination and Neuraminidase Inhibitory Activity of Arabica Green Coffee (Coffea arabica) Beans: Chlorogenic Acid as a Potential Bioactive Compound. Molecules 2021, 26, 3402. [Google Scholar] [CrossRef]
- Stefanello, N.; Spanevello, R.M.; Passamonti, S.; Porciúncula, L.; Bonan, C.D.; Olabiyi, A.A.; Teixeira da Rocha, J.B.; Assmann, C.E.; Morsch, V.M.; Schetinger, M.R.C. Coffee, caffeine, chlorogenic acid, and the purinergic system. Food and chemical toxicology: An international journal published for the British Industrial Biological Research Association 2019, 123, 298–313. [Google Scholar] [CrossRef]
- Murthy, P.S.; Madhava Naidu, M. Sustainable management of coffee industry by-products and value addition—A review. Resources, Conservation and Recycling 2012, 66, 45–58. [Google Scholar] [CrossRef]
- Pagett, M.; Teng, K.S.; Sullivan, G.; Zhang, W. Reusing Waste Coffee Grounds as Electrode Materials: Recent Advances and Future Opportunities. Global challenges 2023, 7, 2200093. [Google Scholar] [CrossRef]
- Qi, S.; Huang, H.; Huang, J.; Wang, Q.; Wei, Q. Lychee (Litchi chinensis Sonn.) seed water extract as potential antioxidant and anti-obese natural additive in meat products. Food Control 2015, 50, 195–201. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, M.; Su, G.; Cai, M.; Zhou, C.; Huang, J.; Lin, L. The antioxidant activities and the xanthine oxidase inhibition effects of walnut (Juglans regia L.) fruit, stem and leaf. International Journal of Food Science & Technology 2015, 50, 233–239. [Google Scholar] [CrossRef]
- Lin, L.; Huang, J.; Sun-Waterhouse, D.; Zhao, M.; Zhao, K.; Que, J. Maca (Lepidium meyenii) as a source of macamides and polysaccharide in combating of oxidative stress and damage in human erythrocytes. International Journal of Food Science & Technology 2017. [Google Scholar] [CrossRef]
- Huang, J.; Wong, K.H.; Tay, S.V.; How, A.; Tam, J.P. Cysteine-Rich Peptide Fingerprinting as a General Method for Herbal Analysis to Differentiate Radix Astragali and Radix Hedysarum. Frontiers in plant science 2019, 10, 973. [Google Scholar] [CrossRef]
- Tam, J.P.; Nguyen, G.K.T.; Loo, S.; Wang, S.; Yang, D.; Kam, A. Ginsentides: Cysteine and Glycine-rich Peptides from the Ginseng Family with Unusual Disulfide Connectivity. Sci Rep 2018, 8, 16201. [Google Scholar] [CrossRef] [PubMed]
- Lay, F.; Anderson, M. Defensins-components of the innate immune system in plants. Current Protein and Peptide Science 2005, 6, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Heitz, A.; Avrutina, O.; Le-Nguyen, D.; Diederichsen, U.; Hernandez, J.-F.; Gracy, J.; Kolmar, H.; Chiche, L. Knottin cyclization: Impact on structure and dynamics. BMC structural biology 2008, 8, 54. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial Peptides from Plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef]
- Nguyen, P.Q.T.; Ooi, J.S.G.; Nguyen, N.T.K.; Wang, S.; Huang, M.; Liu, D.X.; Tam, J.P. Antiviral Cystine Knot α-Amylase Inhibitors from Alstonia scholaris*. Journal of Biological Chemistry 2015, 290, 31138–31150. [Google Scholar] [CrossRef]
- Loo, S.; Tay, S.V.; Kam, A.; Lee, W.; Tam, J.P. Hololectin Interdomain Linker Determines Asparaginyl Endopeptidase-Mediated Maturation of Antifungal Hevein-Like Peptides in Oats. Frontiers in plant science 2022, 13, 899740. [Google Scholar] [CrossRef]
- Kini, S.G.; Wong, K.H.; Tan, W.L.; Xiao, T.; Tam, J.P. Morintides: Cargo-free chitin-binding peptides from Moringa oleifera. BMC plant biology 2017, 17, 68. [Google Scholar] [CrossRef]
- Wong, K.H.; Tan, W.L.; Serra, A.; Xiao, T.; Sze, S.K.; Yang, D.; Tam, J.P. Ginkgotides: Proline-Rich Hevein-Like Peptides from Gymnosperm Ginkgo biloba. Frontiers in plant science 2016, 7, 1639. [Google Scholar] [CrossRef]
- Wong, K.H.; Tan, W.L.; Kini, S.G.; Xiao, T.; Serra, A.; Sze, S.K.; Tam, J.P. Vaccatides: Antifungal glutamine-rich hevein-like peptides from Vaccaria hispanica. Frontiers in plant science 2017, 8, 1100. [Google Scholar] [CrossRef]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Progress in Polymer Science 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Loo, S.; Kam, A.; Dutta, B.; Zhang, X.; Feng, N.; Sze, S.K.; Liu, C.-F.; Wang, X.; Tam, J.P. Decoding the Cure-all Effects of Ginseng. bioRxiv 2023. [Google Scholar] [CrossRef]
- Gruber, C.W.; Elliott, A.G.; Ireland, D.C.; Delprete, P.G.; Dessein, S.; Göransson, U.; Trabi, M.; Wang, C.K.; Kinghorn, A.B.; Robbrecht, E. Distribution and evolution of circular miniproteins in flowering plants. The Plant Cell 2008, 20, 2471–2483. [Google Scholar] [CrossRef] [PubMed]
- Kini, S.G.; Wong, K.H.; Tan, W.L.; Xiao, T.; Tam, J.P. Morintides: Cargo-free chitin-binding peptides from Moringa oleifera. BMC plant biology 2017, 17, 68–68. [Google Scholar] [CrossRef]
- Porto, W.F.; Souza, V.A.; Nolasco, D.O.; Franco, O.L. In silico identification of novel hevein-like peptide precursors. Peptides 2012, 38, 127–136. [Google Scholar] [CrossRef]
- Kam, A.; Loo, S.; Fan, J.S.; Sze, S.K.; Yang, D.; Tam, J.P. Roseltide rT7 is a disulfide-rich, anionic, and cell-penetrating peptide that inhibits proteasomal degradation. The Journal of biological chemistry 2019, 294, 19604–19615. [Google Scholar] [CrossRef]
- Tam, J.P.; Wu, C.R.; Liu, W.; Zhang, J.W. Disulfide bond formation in peptides by dimethyl sulfoxide. Scope and applications. Journal of the American Chemical Society 1991, 113, 6657–6662. [Google Scholar] [CrossRef]
- Tam, J.P.; Wong, C.T.T. Chemical Synthesis of Circular Proteins. Journal of Biological Chemistry 2012, 287, 27020–27025. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.T.T.; Taichi, M.; Nishio, H.; Nishiuchi, Y.; Tam, J.P. Optimal Oxidative Folding of the Novel Antimicrobial Cyclotide from Hedyotis biflora Requires High Alcohol Concentrations. Biochemistry 2011, 50, 7275–7283. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.S.; Huang, J.Y.; Wong, K.H.; Tay, S.V. Coffeetides: Iron-binding cysteine rich peptides from coffee waste. 2020. [Google Scholar]
- Wang, S.; Nguyen, K.T.G.; Luo, S.-n.; Tam, J.P.; Yang, D. Ginsentides: Characterization, Structure and Application of a New Class of Highly Stable Cystine Knot Peptides in Ginseng. Journal of Back and Musculoskeletal Rehabilitation 2015. [Google Scholar]
- Loo, S.; Kam, A.; Xiao, T.; Nguyen, G.K.; Liu, C.F.; Tam, J.P. Identification and Characterization of Roseltide, a Knottin-type Neutrophil Elastase Inhibitor Derived from Hibiscus sabdariffa. Sci Rep 2016, 6, 39401. [Google Scholar] [CrossRef]
- Loo, S.; Kam, A.; Xiao, T.; Tam, J.P. Bleogens: Cactus-Derived Anti-Candida Cysteine-Rich Peptides with Three Different Precursor Arrangements. Frontiers in plant science 2017, 8, 2162. [Google Scholar] [CrossRef]
- Kam, A.; Loo, S.; Dutta, B.; Sze, S.K.; Tam, J.P. Plant-derived mitochondria-targeting cysteine-rich peptide modulates cellular bioenergetics. The Journal of biological chemistry 2019, 294, 4000–4011. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-C.; Son, Y.-O.; Pratheeshkumar, P.; Shi, X. Oxidative stress and metal carcinogenesis. Free Radical Biology and Medicine 2012, 53, 742–757. [Google Scholar] [CrossRef]
- Castillo, M.; Fernández-Gómez, B.; Martínez Sáez, N.; Iriondo-DeHond, A.; Mesa, M.D. Coffee by-products. 2019. [Google Scholar]
- Twardzik, D.R.; Peterkofsky, A. Glutamic acid as a precursor to N-terminal pyroglutamic acid in mouse plasmacytoma protein. Proceedings of the National Academy of Sciences 1972, 69, 274–277. [Google Scholar] [CrossRef]
- Kini, S.G.; Nguyen, P.Q.; Weissbach, S.; Mallagaray, A.; Shin, J.; Yoon, H.S.; Tam, J.P. Studies on the Chitin Binding Property of Novel Cysteine-Rich Peptides from Alternanthera sessilis. Biochemistry 2015, 54, 6639–6649. [Google Scholar] [CrossRef]
- Nguyen, P.Q.; Luu, T.T.; Bai, Y.; Nguyen, G.K.; Pervushin, K.; Tam, J.P. Allotides: Proline-Rich Cystine Knot alpha-Amylase Inhibitors from Allamanda cathartica. Journal of natural products 2015, 78, 695–704. [Google Scholar] [CrossRef]
- Jiayi, H. Discovery, characterization, and applications of cysteine-rich peptides from medicinal plants. Nanyang Technological University, 2019. [Google Scholar]
- Nguyen, G.K.T.; Zhang, S.; Nguyen, N.T.K.; Nguyen, P.Q.T.; Chiu, M.S.; Hardjojo, A.; Tam, J.P. Discovery and Characterization of Novel Cyclotides Originated from Chimeric Precursors Consisting of Albumin-1 Chain a and Cyclotide Domains in the Fabaceae Family *. Journal of Biological Chemistry 2011, 286, 24275–24287. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Taichi, M.; Wei, N.; Yang, H.; Luo, K.Q.; Tam, J.P. An Orally Active Bradykinin B(1) Receptor Antagonist Engineered as a Bifunctional Chimera of Sunflower Trypsin Inhibitor. J Med Chem 2017, 60, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Verdine, G.L. Drugging the undruggable using stapled peptides. In Abstracts of Papers of The American Chemical Society; 2010. [Google Scholar]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic acids research 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nature methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The proteomics protocols handbook; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar]
- Hall, T. BioEdit: An important software for molecular biology. GERF Bull Biosci 2011, 2, 60–61. [Google Scholar]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome research 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Jeener, J.; Meier, B.; Bachmann, P.; Ernst, R. Investigation of exchange processes by two-dimensional NMR spectroscopy. The Journal of chemical physics 1979, 71, 4546–4553. [Google Scholar] [CrossRef]
- Kumar, A.; Ernst, R.; Wüthrich, K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochemical and biophysical research communications 1980, 95, 1–6. [Google Scholar] [CrossRef]
- Bax, A.; Davis, D.G. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. Journal of Magnetic Resonance (1969) 1985, 65, 355–360. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Wüthrich, K.; Billeter, M.; Braun, W. Pseudo-structures for the 20 common amino acids for use in studies of protein conformations by measurements of intramolecular proton-proton distance constraints with nuclear magnetic resonance. Journal of molecular biology 1983, 169, 949–961. [Google Scholar] [CrossRef]
- Brünger, A.T.; Adams, P.D.; Clore, G.M.; DeLano, W.L.; Gros, P.; Grosse-Kunstleve, R.W.; Jiang, J.S.; Kuszewski, J.; Nilges, M.; Pannu, N.S. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D 1998, 54, 905–921. [Google Scholar]
- DeLano, W.L. PyMOL. 2002. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





