1. Introduction
Community-acquired meningoencephalitis (CAME) is a life-threatening condition characterized by acute central nervous system (CNS) inflammation, caused by microbial infection in 30-50% cases (ref). Viral and bacterial infection are the most causes of CNS diseases [
1], while fungal and parasites CNS prognosis are usually detected in immunocompromised patients [
2,
3,
4,
5]. Clinical manifestation of CAME depends on the causative pathogen and the inflammation level, including fever, headache, neck stiffness, confusion, seizures, as well as nausea, vomiting, photo-phonophobia and loss of consciousness and coma in specific situations [
6]. Current diagnosis of CAME is based on a combination of physical examination, CT scan or magnetic resonance imaging, and cerebrospinal fluid (CSF) analysis [
7]. The CSF culture remained a gold standard of CAME routine diagnosis, which failed in 10–56% cases [
8], and Multiplex RT-PCRs targeting a limited number of pathogens involved in CNS diseases [
9]. Real-time metagenomics next-generation sequencing (RT-mNGS) is a throughput method for the identification of pathogen genomes directly from CSF samples in real-time [
10,
11,
12]. With no prior amplification or specific target, RT-mNGS is a universal method able to diagnose mono and polymicrobial infections directly from clinical samples, as well as
in-silico prediction of their antibiotic susceptibility profiles and genotypes [
13]. Herein, we reported the first case of polymicrobial CAME in a pediatric patient in Niger, diagnosed by RT-mNGS applied directly on CSF sample.
2. Case Presentation
A 10-year-old female patient was admitted at the pediatric emergency unit of Amirou Boubacar Diallo National hospital, Niamey, Niger, with complains of fever and focal neurologic deficit. The patient is living with her family in Niamey, and no chronic disease or significant past medical history, as well as no specific medication or medical advice has been reported. At her arrival admission, the patient was awake, confused, and irresponsive. The Glasgow Coma Score (GCS) was 12 and the patient’s vital signs were as follows: temperature 38.2 °C, blood pressure 180/120 mmHg, pulse rate 121 beats/min, and respiratory rate 21 breaths/min with oxygen saturation (SpO2) of 96 % in ambient/room air. Neurological examination revealed a right hemiplegia without neck stiffness. Based on the scoring system proposed by the Medical Research Council (MRC), the patient had muscle power of 0/5 and 0/5 in the right upper and lower extremities, respectively. Deep tendon reflexes were symmetrical, and flexor plantar reflexes were present. Otherwise, other physical examinations were unremarkable. The CSF analysis after lumbar puncture showed blood cell count 46 /µL (97% lymphocytes), normal glycorrhachia level, protein level of 41 mg/dL. Blood analysis was significant for leukocytosis, profound hyponatremia (118 mmol/L), blood creatinine and urea level rises, C-reactive protein level (CRP) was increased (9.50 mg/dL), and the direct blood cultures were negatives. The brain computed tomography (CT) scan without contrast showed bright cortical sulci with asymmetric white matter lesions visible as areas of low attenuation (Figure S1). National Institute of Health Stroke Scale (NIHSS) score was 7 points. Empirical intravenous ceftriaxone and metronidazole were initiated, pending diagnostic confirmation.
The patient left the hospital after 10-days hospitalization, against medical advice due to financial constraints, at the request of her parents. Unfortunately, the patient died two days later. Although, all routine microbiological investigations including molecular assays and CSF culture were negative.
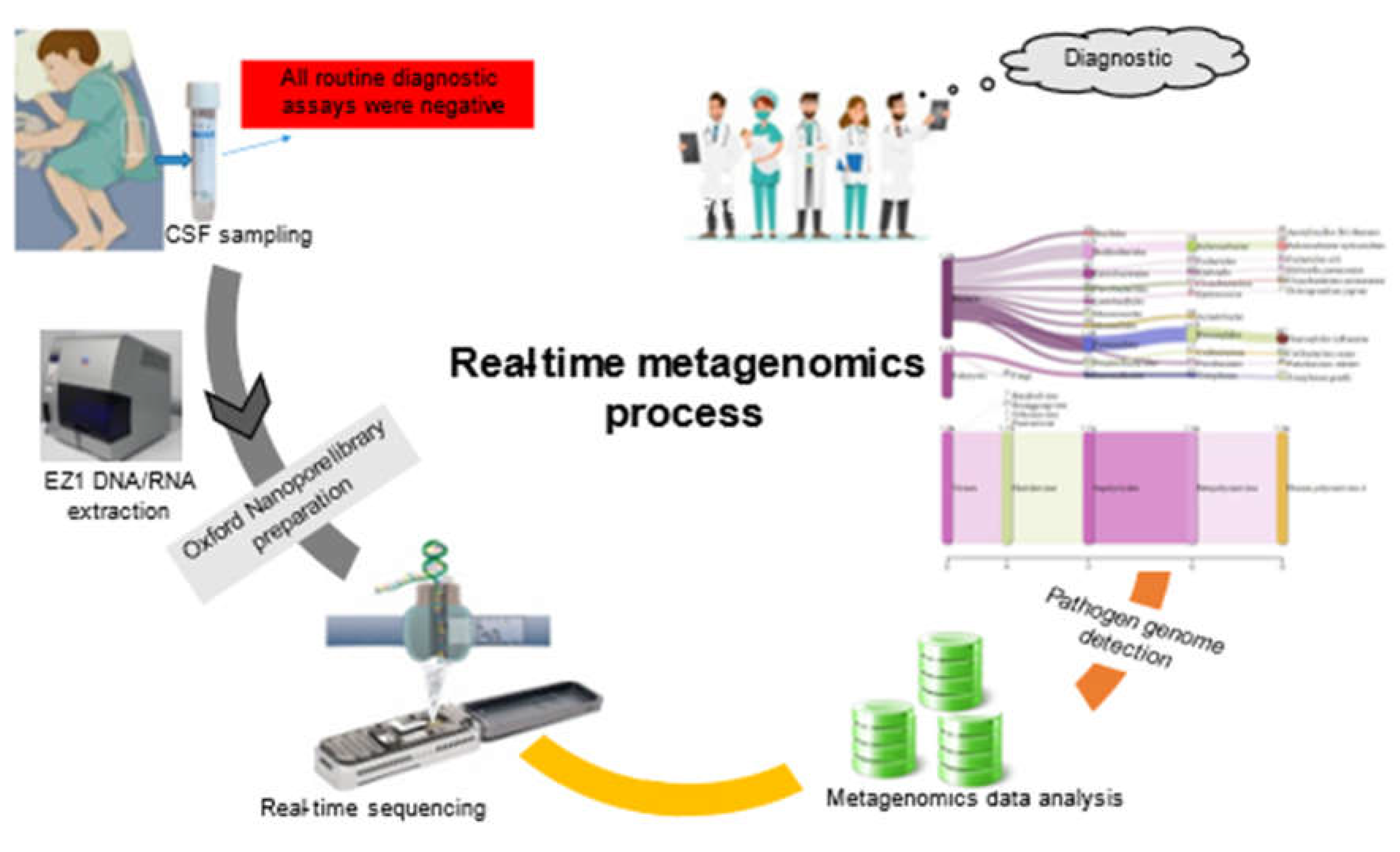
A few days letter, metagenomic-based on real-time sequencing was applied directly on DNA/RNA extracted from 50 µL leftover CSF using EZ1 Virus Mini Kit v2.0 (Qiagen, Courtaboeuf, France), then sequenced in eight hours run using an Oxford Nanopore MinION as previously described [
14,
15], to detect all potential pathogen genomes non-routinely targeted by RT-PCR assays. In parallel, 1 ng DNA/RNA was used for paired-end Illumina Nextera-XT library preparation as previously described (8), to confirm the MinION results.
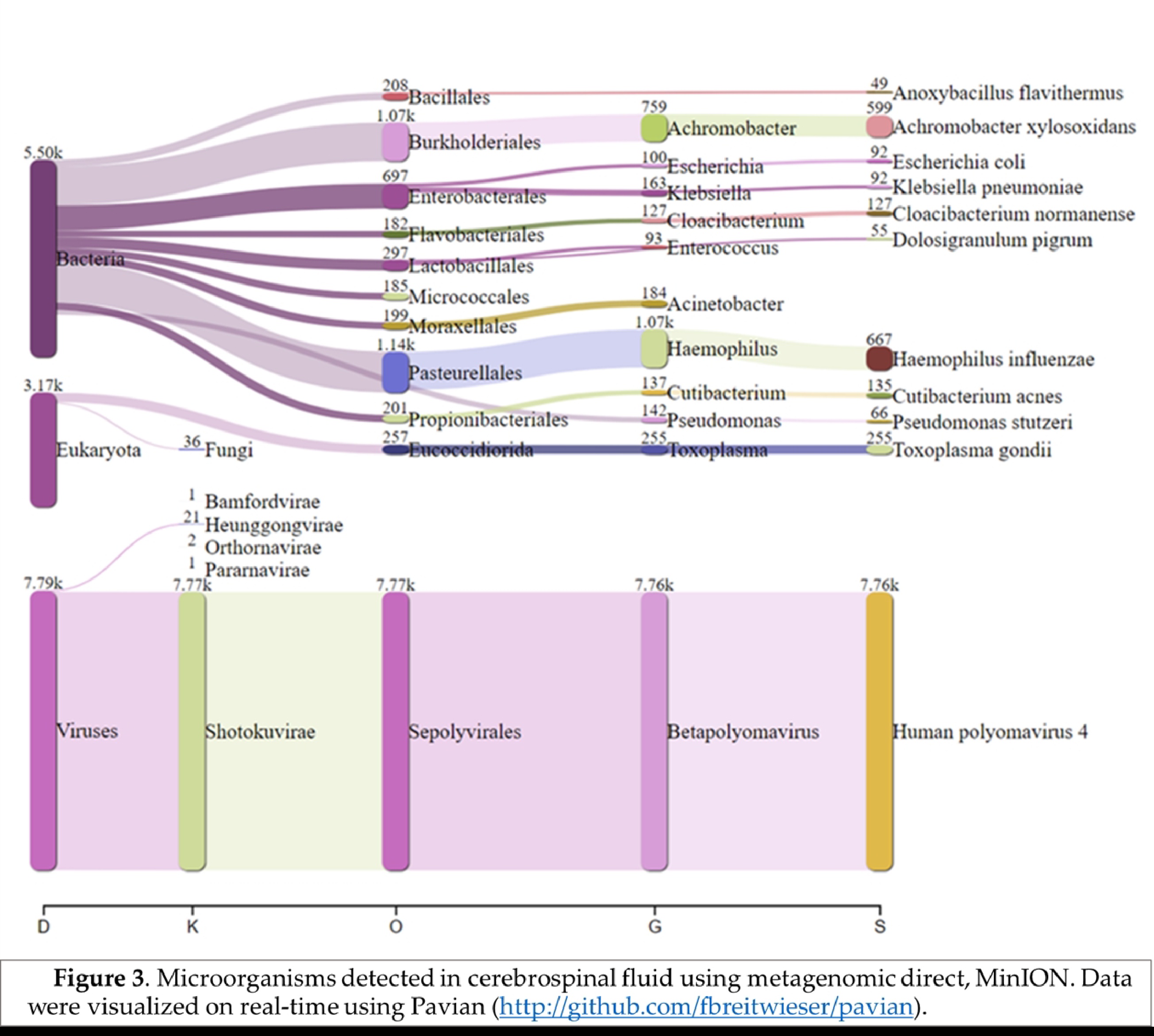
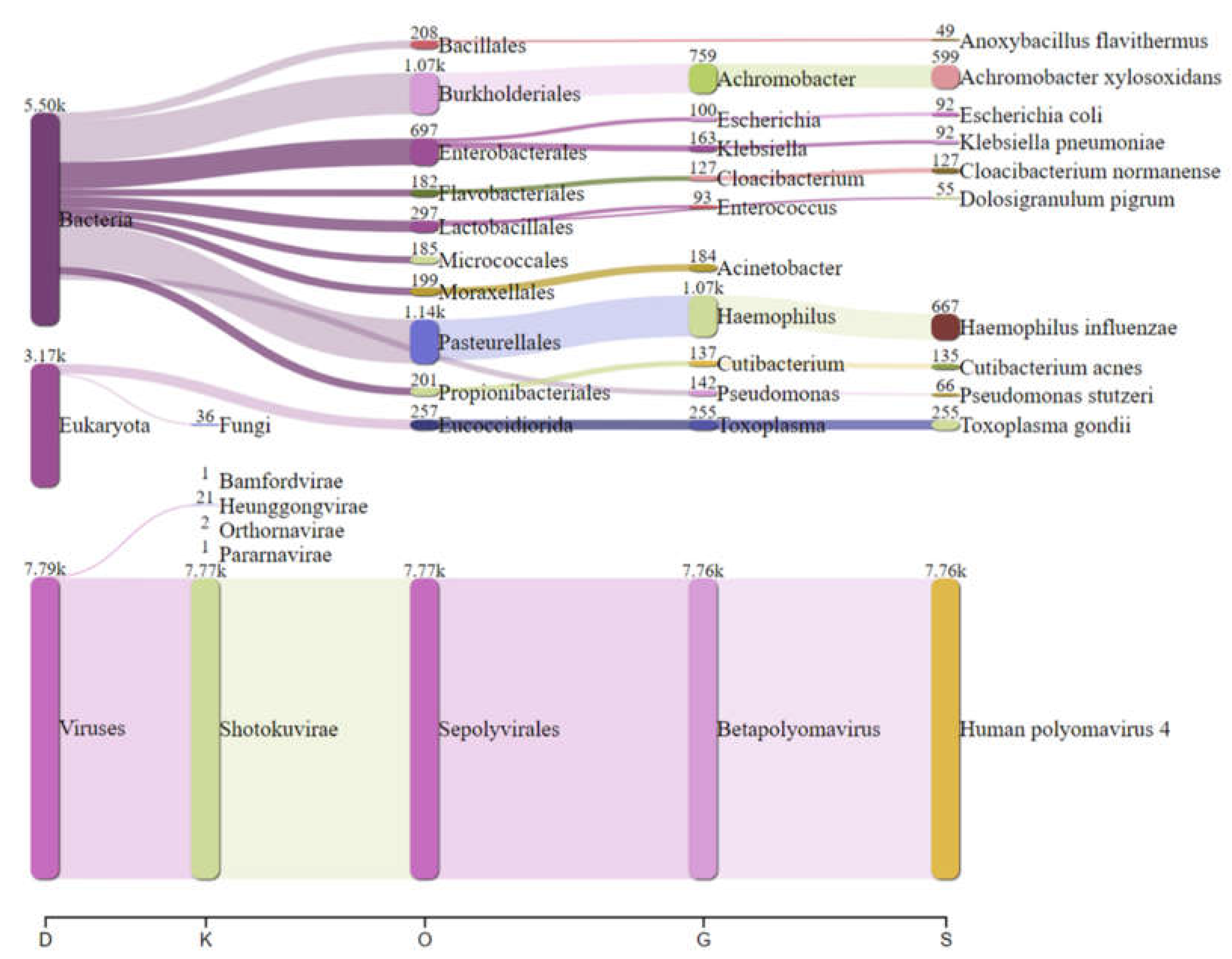
Real-time analysis of the MinION data was performed using EPI2ME software (version 2019.11.11-2920621). In parallel, mNGS data were analyzed by Kraken 2 on galaxy online software (
https://usegalaxy.eu/) and visualized using Pavian online software (
https://fbreitwieser.shinyapps.io/pavian/) (
Figure 1). Total MinION and Illumina reads were concatenated and assembled using spades assembler (version: 3.15.4) and blasted against NCBI GenBank database and mapped to the heat-blast reference genomes using CLC genomic workbench (Qiagen) (version: 21.1.1).
3. Results and Discussion
Real-time analysis of MinION data detected 132 WU Polyomavirus specific reads after one-hour run. Blast nucleotide of the fasta sequences after assembly of both Illumina and Nanopore reads against NCBI GenBank database identified WU polyomavirus strain W33 (GenBank accession no: GU296367.1), isolated from pediatric Australian patient [
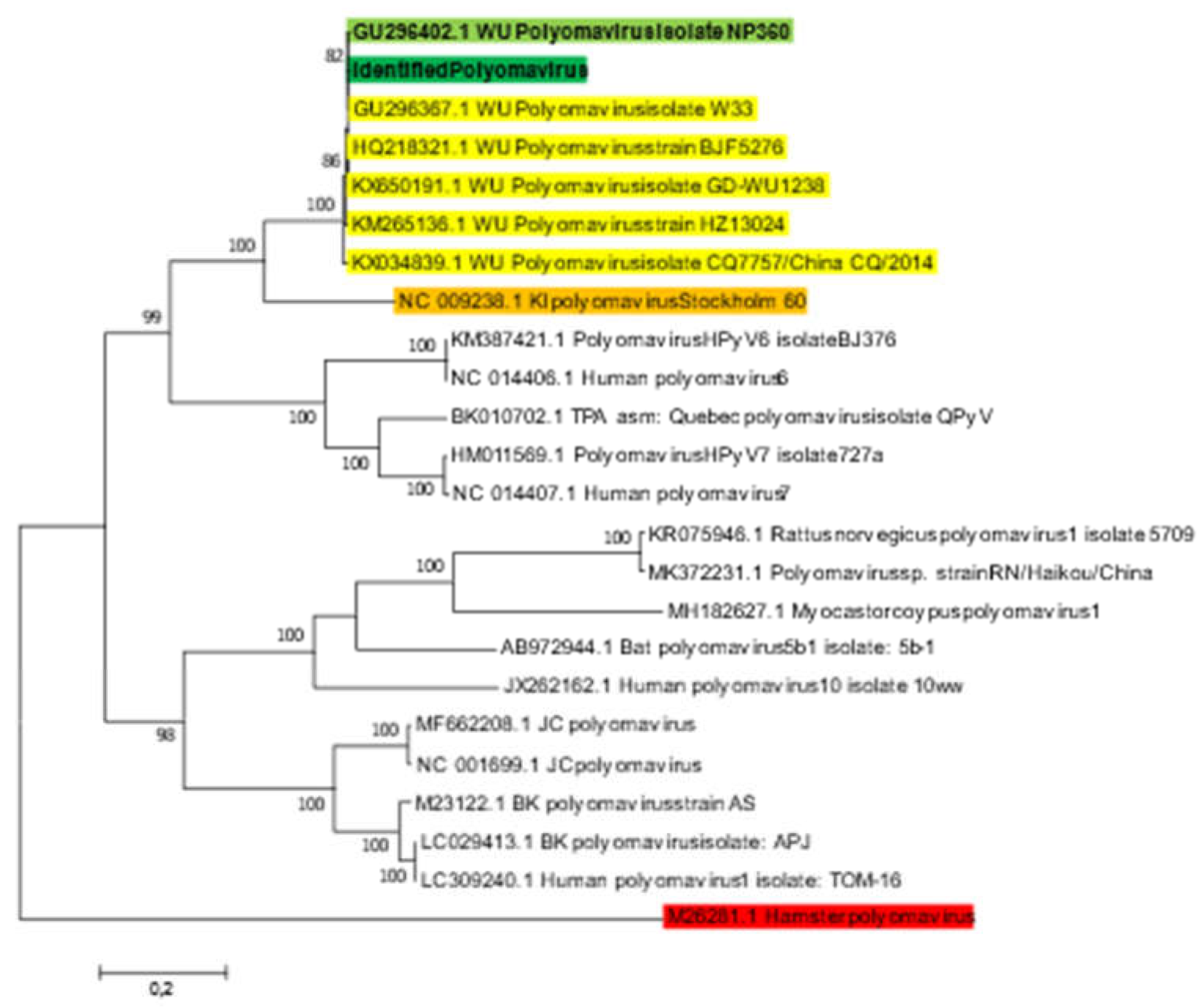
16]. The blast sequence alignment exhibited 100% coverage and 99.98% identity. Mapping of the total reads against the hit-blast sequence (GenBank accession no: GU296367.1) using CLC software showed that 11,632/1,048,634 (1.12%) of reads were matched with the reference genome, generating 5,228 bp corresponding to 100% polyomavirus genome coverage, with 165 mean depths. Phylogenetic analysis based on whole genome sequence of the identified Polyomavirus strain (BANPedHNABD), the six hit-blast WU Polyomavirus and other representative polyomavirus genomes recovered from GenBank database, showed that the identified Polyomavirus sequence clustered with other WU Polyomavirus isolates, and mainly with WU polyomavirus isolate NP360 (GenBank accession n°: GU296402) at 82% sequence similarity (
Figure 2), belonging to human polyomavirus 4 family, isolated from patients with acute respiratory infection [
16]. The pathogenicity of WU polyomavirus and its association with CSN prognosis remained unclear. Despite its previous detection in CSF samples [
17,
18], no cases of WU polyomavirus meningoencephalitis have been reported at present. It is far more likely that WU polyomavirus is an incidental finding related to virus reactivation or chronic shedding rather than being the etiologic cause of the patient's meningitis [
19]. WU polyomavirus is a persisting virus that is reactivated by an inflammatory process, or a predisposing or aggravating factor [
20]. Primary infection with WU polyomavirus probably occurs early in childhood, as suggested by the seroprevalence in young people [
21]. JC and Bk polyomaviruses are the most targeted polyomavirus in routine diagnostic, commonly associated to respiratory and CNS diseases [
22,
23], and WU polyomavirus is completely neglected in routine diagnosis.
Haemophilus influenzae specific reads (667 reads) were detected which explains the possible co-infection bacteria-DNA virus in this case. Blastn after total reads assembly identified
H. influenzae strain PittGG (GenBank accession n° CP000672), belonging to the non-typable
H. influenzae genotype by muti-locus-sequence-typing analysis. As
H. influenzae CNS infection had been reported in both young and elderly patients [
13].
Furthermore,
Achromobacter xylosoxidans (599 reads) were detected in this patient.
Achromobacter xylosoxidans is a Gram-negative bacterium that was first isolated from human ear discharge [
24]. Since,
Achromobacter xylosoxidans has been documented as an opportunistic pathogen in numerous health care-associated infections including meningitis [
25,
26].
In addition, 255 reads specific to
Toxoplasma gondii were identified in the sequenced CSF, could be the direct cause of CAME, as previously documented in CNS toxoplasmosis in young patient [
27] (
Figure 3), which can facilitate other bacterial and viral infections.
Based on our findings, we classified this case as a polymicrobial meningoencephalitis including viral, bacterial and protozoa infection. Using RT-mNGS, pathogen genome could be detected directly from CSF sample [
16]. With no specific target, RT-mNGS seems to be an adapted method to diagnose non-routinely detectable pathogen, as well as their genotype and antimicrobial susceptibility in reduced time [
28]. Faced on high prevalence of routinely undocumented CSF (≈90%) [
29] RT-mNGS given an opportunity for early documentation of CNS infection in pediatric population in endemic countries, that challenging several cases may be escaped current routine investigations. Universal detection of pathogen genome by RT-mNGS open new perspective for clinical microbiology laboratories to implement this strategy as an alternative emergency diagnosis of life-threatening CNS infections.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: The brain computed tomography (CT) scan without contrast showing bright cortical sulci with asymmetric white matter lesions visible as areas of low attenuation.
Author Contributions
A.Y contributed to experimental design, routine analyses, data analysis, data interpretation and writing the first draft. M.G. contributed to clinical diagnostic and CSF sampling. M.S.S and S.A collected samples and collected clinical information. M.B.S performed routine analyses. A.O, A.O-O, B.S, D.M, M.D, E.A and S.M contributed to critically reviewing the manuscript, data interpretation and coordinated and directed the work. M.M. contributed to experimental design, data analysis, performed bioinformatics data analysis, and data interpretation, and writing of the original draft of the manuscript. AY and MM ensured the critical reviewing and final validation of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
All procedures were approved by the Research Ethics Committee of Amirou Boubacar Diallo National Hospital (HNABD/2022/012, approved on 11 January 2022).
Informed Consent Statement
The patient provided a written consent for the publication.
Data Availability Statement
Real-time metagenomic next-generation sequencing data for this study are available from GenBank under GenBank accession: OP852794.1.
Acknowledgments
We thank the Niger Republic National reference laboratory for HIV, Tuberculosis, and antimicrobial resistance for performing routine microbiological investigations including molecular assays and CSF culture used in this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hasbun, R.; Rosenthal, N.; Balada-Llasat, J.M.; Chung, J.; Duff, S.; Bozzette, S.; Zimmer, L.; Ginocchio, C.C. Epidemiology of Meningitis and Encephalitis in the United States, 2011–2014. Clinical Infectious Diseases 2017, 65, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, H.; Sivasubramanian, G. Cryptococcus Neoformans Meningoencephalitis in an Immunocompetent Patient after COVID-19 Infection. Case Reports in Infectious Diseases 2021, 2021, e5597473. [Google Scholar] [CrossRef]
- Güémez, A.; García, E. Primary Amoebic Meningoencephalitis by Naegleria Fowleri: Pathogenesis and Treatments. Biomolecules 2021, 11, 1320. [Google Scholar] [CrossRef]
- Pham Thu, H.; Đao Huu, N.; Thi Thu, T.L.; Van, L.N. Case Report: Angiostrongylus Cantonensis Meningoencephalitis in a 9-Month-Old Baby in Vietnam. Am J Trop Med Hyg 2020, 103, 723–726. [Google Scholar] [CrossRef]
- Saraya, A.; Mahavihakanont, A.; Shuangshoti, S.; Sittidetboripat, N.; Deesudchit, T.; Callahan, M.; Wacharapluesadee, S.; Wilde, H.; Hemachudha, T. Autoimmune Causes of Encephalitis Syndrome in Thailand: Prospective Study of 103 Patients. BMC Neurology 2013, 13, 150. [Google Scholar] [CrossRef] [PubMed]
- Fitch, M.T.; Abrahamian, F.M.; Moran, G.J.; Talan, D.A. Emergency Department Management of Meningitis and Encephalitis. Infect Dis Clin North Am 2008, 22, 33–52. [Google Scholar] [CrossRef] [PubMed]
- Poplin, V.; Boulware, D.R.; Bahr, N.C. Methods for Rapid Diagnosis of Meningitis Etiology in Adults. Biomarkers in Medicine 2020, 14, 459–479. [Google Scholar] [CrossRef]
- Kanu, O.O.; Ojo, O.; Esezobor, C.; Bankole, O.; Olatosi, J.; Ogunleye, E.; Asoegwu, C.; Eghosa, M.; Adebayo, B.; Oladele, R.; et al. Pediatric Brain Abscess – Etiology, Management Challenges and Outcome in Lagos Nigeria. Surg Neurol Int 2021, 12, 592. [Google Scholar] [CrossRef]
- Vincent, J.-J.; Zandotti, C.; Baron, S.; Kandil, C.; Levy, P.-Y.; Drancourt, M.; Raoult, D.; Ninove, L. Point-of-Care Multiplexed Diagnosis of Meningitis Using the FilmArray® ME Panel Technology. Eur J Clin Microbiol Infect Dis 2020, 39, 1573–1580. [Google Scholar] [CrossRef]
- Peng, J.-M.; Du, B.; Qin, H.-Y.; Wang, Q.; Shi, Y. Metagenomic Next-Generation Sequencing for the Diagnosis of Suspected Pneumonia in Immunocompromised Patients. Journal of Infection 2021, 82, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, H.; Chen, H.; Li, Z.; Ding, W.; Wang, J.; Yin, Y.; Jin, L.; Sun, S.; Jing, C.; et al. Metagenomic Next-Generation Sequencing to Identify Pathogens and Cancer in Lung Biopsy Tissue. EBioMedicine 2021, 73, 103639. [Google Scholar] [CrossRef] [PubMed]
- Graff, K.; Dominguez, S.R.; Messacar, K. Metagenomic Next-Generation Sequencing for Diagnosis of Pediatric Meningitis and Encephalitis: A Review. Journal of the Pediatric Infectious Diseases Society 2021, 10, S78–S87. [Google Scholar] [CrossRef] [PubMed]
- Morsli, M.; Boudet, A.; Kerharo, Q.; Stephan, R.; Salipante, F.; Dunyach-Remy, C.; Houhamdi, L.; Fournier, P.-E.; Lavigne, J.P.; Drancourt, M. Real-Time Metagenomics-Based Diagnosis of Community-Acquired Meningitis: A Prospective Series, Southern France. eBioMedicine 2022, 84, 104247. [Google Scholar] [CrossRef] [PubMed]
- Morsli, M.; Vincent, J.-J.; Milliere, L.; Colson, P.; Drancourt, M. Direct Next-Generation Sequencing Diagnosis of Echovirus 9 Meningitis, France. Eur J Clin Microbiol Infect Dis 2021, 40, 2037–2039. [Google Scholar] [CrossRef] [PubMed]
- Morsli, M.; Bodet, A.; Kerharo, Q.; Stéphane, R.; Saliponte, F.; Remy, C.D.; Houhamdi, L.; Fournier, P.-E.; Lavigne, J.-P.; Drancourt, M. Real-Time Metagenomics-Based Diagnosis of Community-Acquired Meningitis: A Prospective Series, Southern France. Southern France.
- Bialasiewicz, S.; Rockett, R.; Whiley, D.W.; Abed, Y.; Allander, T.; Binks, M.; Boivin, G.; Cheng, A.C.; Chung, J.-Y.; Ferguson, P.E.; et al. Whole-Genome Characterization and Genotyping of Global WU Polyomavirus Strains. Journal of Virology 2010, 84, 6229–6234. [Google Scholar] [CrossRef] [PubMed]
- Paticheep, S.; Kongsaengdao, S. Viral Infection of Central Nervous System in Children: One Year Prospective Study. J Med Assoc Thai 2011, 94 Suppl 7, S24–31. [Google Scholar]
- Barzon, L.; Squarzon, L.; Pacenti, M.; Scotton, P.G.; Palù, G. Detection of WU Polyomavirus in Cerebrospinal Fluid Specimen from a Patient with AIDS and Suspected Progressive Multifocal Leukoencephalopathy. The Journal of Infectious Diseases 2009, 200, 314–315. [Google Scholar] [CrossRef]
- Olausson, J.; Brunet, S.; Vracar, D.; Tian, Y.; Abrahamsson, S.; Meghadri, S.H.; Sikora, P.; Lind Karlberg, M.; Jakobsson, H.E.; Tang, K.-W. Optimization of Cerebrospinal Fluid Microbial DNA Metagenomic Sequencing Diagnostics. Sci Rep 2022, 12, 3378. [Google Scholar] [CrossRef]
- Neske, F.; Blessing, K.; Ullrich, F.; Pröttel, A.; Kreth, H.W.; Weissbrich, B. WU Polyomavirus Infection in Children, Germany. Emerg Infect Dis 2008, 14, 680–681. [Google Scholar] [CrossRef]
- Lazim, H.H.; Al-Obaidi, A.B. WU and KI Polyomaviruses. Microbes and Infectious Diseases 2022. [Google Scholar] [CrossRef]
- Elsner, C.; Dörries, K. Evidence of Human Polyomavirus BK and JC Infection in Normal Brain Tissue. Virology 1992, 191, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Del Valle, L.; Enam, S.; Lara, C.; Miklossy, J.; Khalili, K.; Gordon, J. Primary Central Nervous System Lymphoma Expressing the Human Neurotropic Polyomavirus, JC Virus, Genome. Journal of Virology 2004, 78, 3462–3469. [Google Scholar] [CrossRef]
- Yabuuchi, E.; Ohyama, A. Achromobacter Xylosoxidans n. Sp. from Human Ear Discharge. Japanese Journal of Microbiology 1971, 15, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Bellissimo, F.; Pinzone, M.R.; Tosto, S.; Nunnari, G.; Cacopardo, B. Achromobacter Xylosoxidans Meningitis in an Immunosuppressed Patient. QJM 2014, 107, 65–66. [Google Scholar] [CrossRef]
- Namnyak, S.S.; Holmes, B.; Fathalla, S.E. Neonatal Meningitis Caused by Achromobacter Xylosoxidans. J Clin Microbiol 1985, 22, 470–471. [Google Scholar] [CrossRef] [PubMed]
- Prandota, J.; Gryglas, A.; Fuglewicz, A.; Żesławska-Faleńczyk, A.; Ujma-Czapska, B.; Szenborn, L.; Mierzwa, J. Recurrent Headaches May Be Caused by Cerebral Toxoplasmosis. World J Clin Pediatr 2014, 3, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Morsli, M.; Bechah, Y.; Coulibaly, O.; Toro, A.; Fournier, P.E.; Houhamdi, L.; Drancourt, M. Direct Diagnosis of Pasteurella Multocida Meningitis Using Next-Generation Sequencing. The Lancet Microbe 2022, 3, e6. [Google Scholar] [CrossRef] [PubMed]
- Morsli, M.; Salipante, F.; Kerharo, Q.; Boudet, A.; Stephan, R.; Dunyach-Remy, C.; Zandotti, C.; Lavigne, J.-P.; Drancourt, M. Dynamics of Community-Acquired Meningitis Syndrome Outbreaks in Southern France. Frontiers in Microbiology 2023, 13. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).