Submitted:
14 August 2023
Posted:
16 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
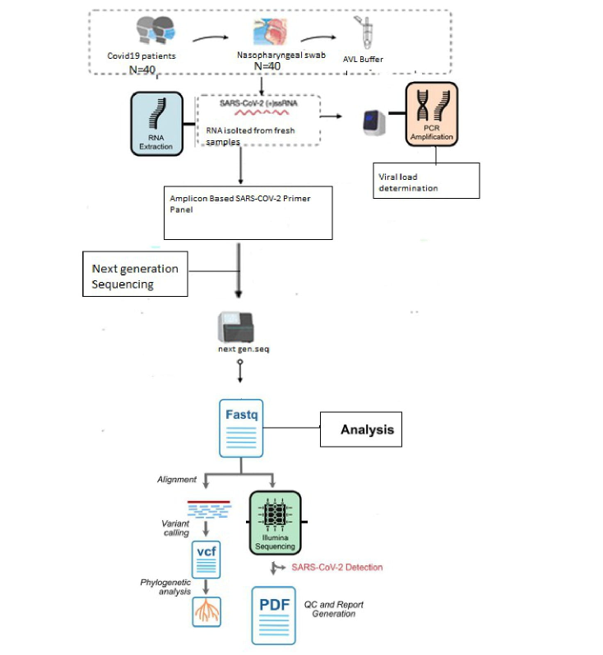
2. Materials and Methods
2.1. Setting
2.2. Clinical sample and processing
2.2.1. Sampling
- Viral RNA extraction and Real-Time PCR
- Complete Genome Sequencing of SARS-COV-2 Omicron Variant
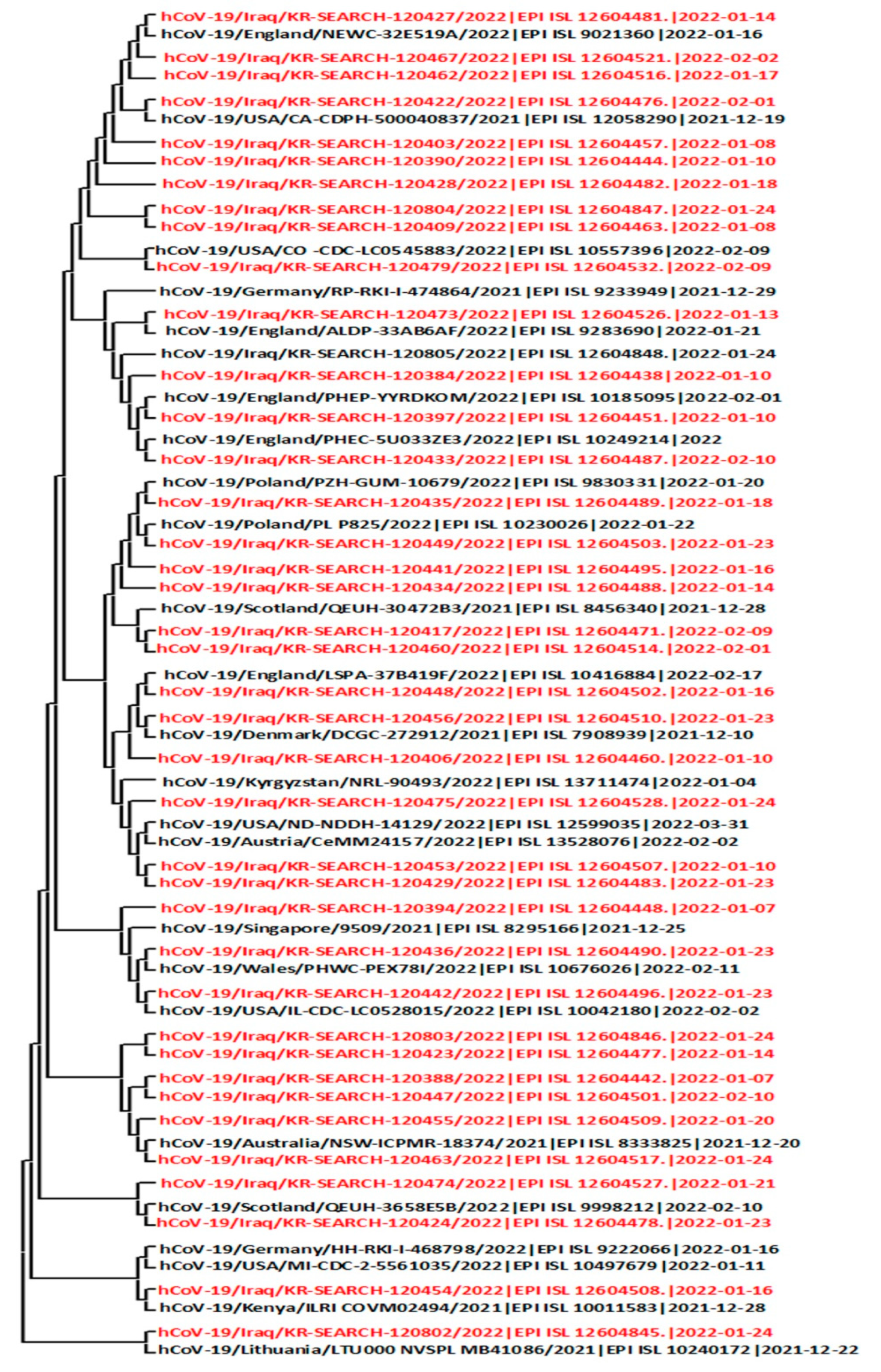
3. Genome Alignment, and Phylogenetic Analysis
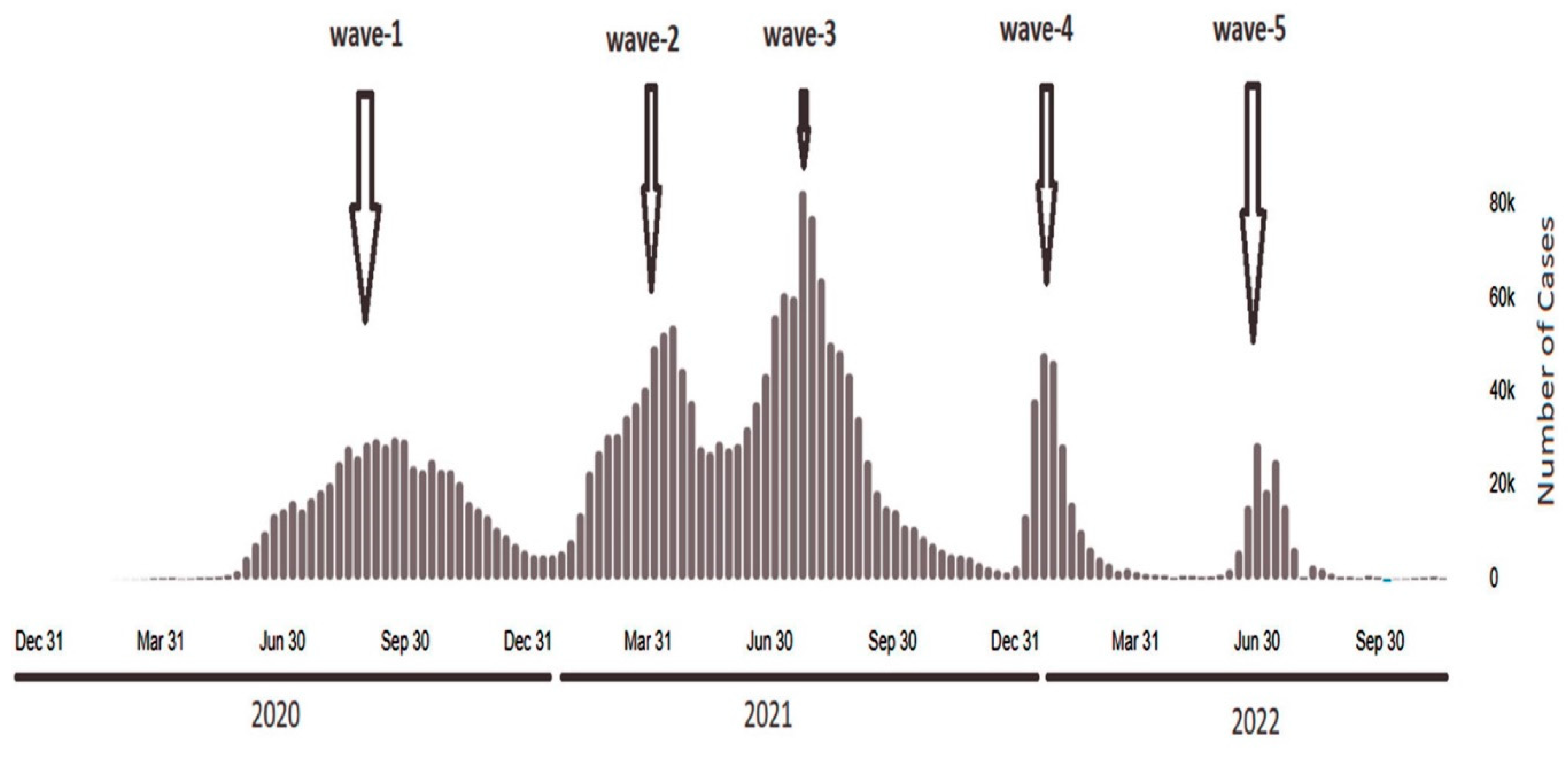
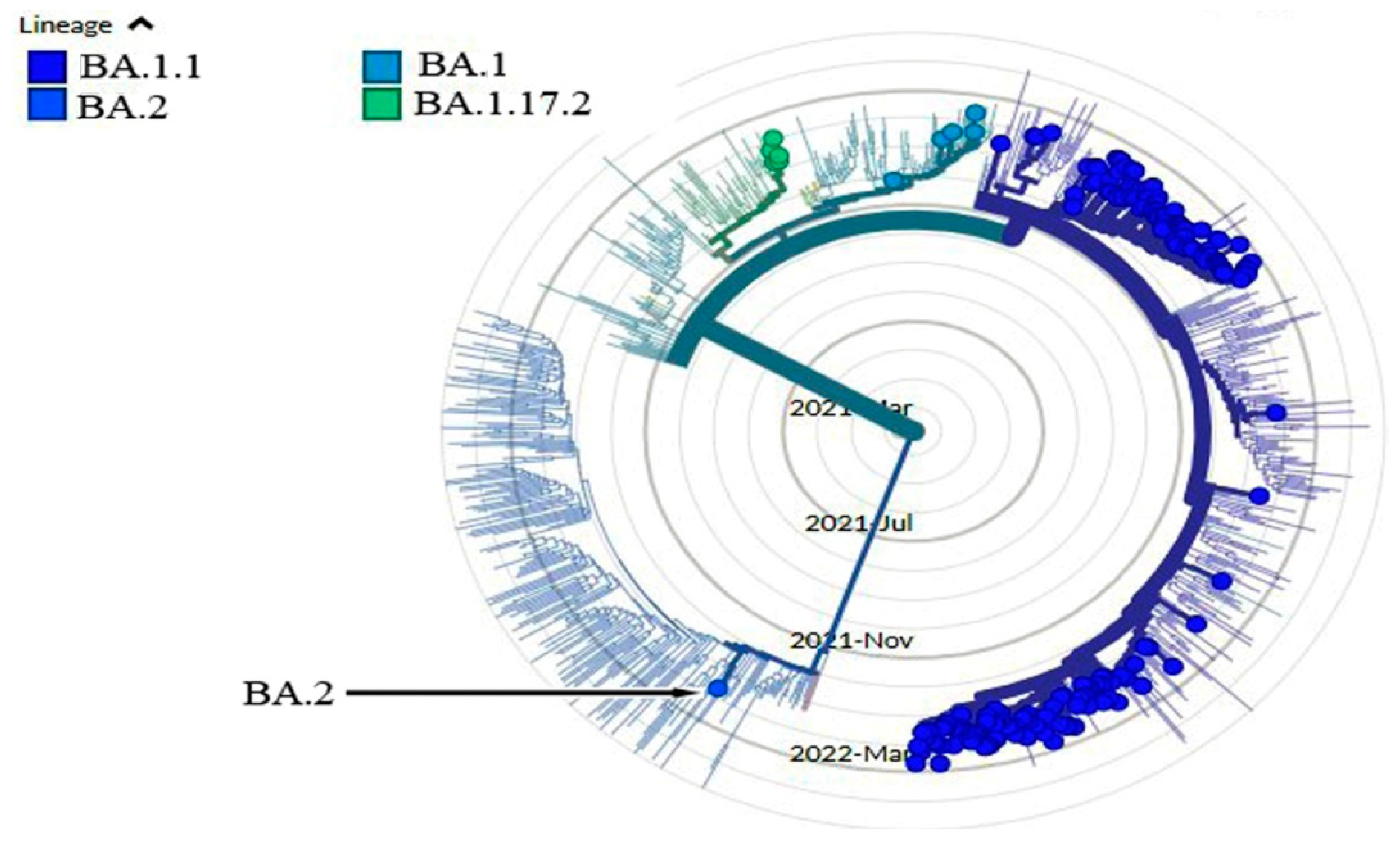
4. Results and Discussions
5. Conclusion
Author Contributions
Funding
Institutional Review Board Statement:
Informed Consent Statement:
Data Availability Statement
Conflicts of Interest
References
- Kim D, Lee J-Y, Yang J-S, Kim JW, Kim VN, Chang H, et al. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914-21. e10.
- Sidiqi KR, Sabir DK, Ali SM, Kodzius R, et al. Does early childhood vaccination protect against COVID-19?. Frontiers in molecular biosciences, 2020, 7: 120.
- Huremović D. Psychiatry of pandemics: a mental health response to infection outbreak: Springer; 2019.
- Shaibu JO, Onwuamah CK, James AB, Okwuraiwe AP, Amoo OS, Salu OB, et al. Full length genomic sanger sequencing and phylogenetic analysis of Severe Acute Respiratory Syndrome.
- Coronavirus 2 (SARS-CoV-2) in Nigeria. PloS one. 2021;16(1):e0243271.
- Aldiabat K, Kwekha Rashid A, Talafha H, Karajeh A,et al. The extent of smartphones users to adopt the use of cloud storage. J Comput Sci. 2018;14(12):1588-98.1.
- Alhayani B, Abbas ST, Mohammed HJ, Mahajan HB,et al. Intelligent secured two-way image transmission using corvus corone module over WSN. Wireless Personal Communications. 2021;120(1):665-700.
- Hasan HS, Abdallah AA, Khan I, Alosman HS, Kolemen A, Alhayani B,et al. Novel unilateral dental expander appliance (udex): a compound innovative materials. Computers, Materials and Continua. 2021:3499-511.
- Mostafaei S, Sayad B, Azar MEF, Doroudian M, Hadifar S, Behrouzi A, et al. The role of viral and bacterial infections in the pathogenesis of IPF: a systematic review and meta-analysis. Respiratory research. 2021;22(1):1-14.
- Hui EK-W. Reasons for the increase in emerging and re-emerging viral infectious diseases. Microbes and infection. 2006;8(3):905-16.
- Domingo E. Viruses at the edge of adaptation. Virology. 2000;270(2):251-3.
- Taheri M, Rad LM, Hussen BM, Nicknafs F, Sayad A, Ghafouri-Fard S,et al. Evaluation of expression of VDR-associated lncRNAs in COVID-19 patients. BMC Infectious Diseases. 2021;21(1):1-10.
- Ibrahim FM, Alkaim A, Kadhom M, Sabir DK, Salih N, Yousif E,et al. Chemistry of selected drugs for SARS-CoV-2 inhibition: tested in vitro and approved by the FDA. Chem Int. 2021;7(3):212-6.
- Khan S, Hussain A, Vahdani Y, Kooshki H, Hussen BM, Haghighat S, et al. Exploring the interaction of quercetin-3-O-sophoroside with SARS-CoV-2 main proteins by theoretical studies: A probable prelude to control some variants of coronavirus including Delta. Arabian Journal of Chemistry. 2021;14(10):103353.
- Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Global challenges. 2017;1(1):33-46.
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. others.(2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine.
- Chen, Z., Azman, A. S., Chen, X., Zou, J., Tian, Y., Sun, R., ... & Yu, H. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nature genetics, 2022, 54.4: 499-507.
- Khare S, Gurry C, Freitas L, Schultz MB, Bach G, Diallo A. . GISAID’s role in pandemic response. China CDC weekly, 2021, 3.49: 1049.
- Motayo BO, Oluwasemowo OO, Olusola BA, Akinduti PA, Arege OT, Obafemi YD. Evolution and genetic diversity of SARS-CoV-2 in Africa using whole genome sequences. International Journal of Infectious Diseases, 2021, 103: 282-287.
- World Health Organization. WHO Announces Simple, Easy-To-Say Labels for SARS-CoV-2 Variants of Interest and Concern.(2021). 2022.
- Elbe S, Buckland-Merrett G. Data, disease and diplomacy: Data, disease and diplomacy: GISAID's innovative contribution to global health. Global challenges, 2017, 1.1: 33-46.
- Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics, 2018, 34.23: 4121-4123.
- Rambaut A, Holmes EC, O’Toole A, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nature microbiology, 2020, 5.11: 1403-1407.
- Choi, J.Y.; Smith, D.M. SARS-CoV-2 variants of concern. Yonsei Med. J. 2021, 62, 961–968.
- Merza MA, Haleem Al Mezori AA, Mohammed HM, et al. COVID-19 outbreak in Iraqi Kurdistan: the first report characterizing epidemiological, clinical, laboratory, and radiological findings of the disease. Diabetes MetabSyndr. 2020;14(4):547-554. doi: 10.1016/j.dsx.2020.04.047.
- Merza MA, Aswad SM, Sulaiman HM, et al. Clinical and epidemiological characteristics and outcomes of Coronavirus disease-19 patients in a large longitudinal study. Int J Health Sci (Qassim). 2021;15(4):29-41.
- OCHA. Panel study IV: impact of COVID-19 on small- and medium-sized enterprises in Iraq - June 2020 to June 2021. https://reliefweb.int/report/iraq/panel-study-iv-impact-covid-19-small-and-medium sizedenterprisesiraq-june-2020-june. Accessed June 2022.
- Al-Rashedi NAM, Alburkat H, Hadi AO, et al. High prevalence of an alpha variant lineage with a premature stop codon in ORF7a in Iraq, winter 2020-2021. PLoS One. 2022;17(5):e0267295. Published May 26, 2022.
- Rudaw. Kurdistan region, Iraq record first cases of COVID-19 Omicron variant. Rudaw.net.https://www.rudaw.net/english/middleeast/iraq/060120222. Accessed January 6, 2022.
- Worldometers. Worldometer’s COVID-19 data. https://www.worldometers.info/coronavirus/country/iraq/. Accessed February 25, 2022.
- Ong SWX, Chiew CJ, Ang LW, et al. Clinical and virological features of SARS-CoV-2 variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta) [publishedonline ahead of print, Aug 23, 2021]. Clin Infect Dis. 2021;ciab721.
- Finlay C, Brett A, Henry L, Yuka J, Franck K, Neale B. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021; 26(30).
- Al-Rashedi NAM, Licastro D, Rajasekharan S, Monego D, Marcello A, Munahi MG, et al. Genome sequencing of a novel coronavirus SARS-CoV-2 isolate from Iraq. Microbiol Resour Announc. 2021; 10 (4):e01316–20. https://doi.org/10.1128/MRA.01316-20 PMID: 33509990.
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Challenges 2017, 1, 33–46.
- Aksamentov, I.; Roemer, C.; Hodcroft, E.B.; Neher, R.A. Nextclade: Clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021, 6, 3773.
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., ... & Cao, B., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The lancet, 2020, 395.10223: 497-506.
- Woo PC, Lau SK, Lam CS, Lai KK, Huang Y, Lee P, et al. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. Journal of virology. 2009;83(2):908-17.
- Xia X. Domains and functions of spike protein in Sars-Cov-2 in the context of vaccine design. Viruses. 2021;13(1):109.
- Gupta D, Sharma P, Singh M, Kumar M, Ethayathulla A, Kaur P,et al. Structural and functional insights into the spike protein mutations of emerging SARS-CoV-2 variants. Cellular and Molecular Life Sciences. 2021;78(24):7967-89.
- Mahajan S, Kode V, Bhojak K, Karunakaran C, Lee K, Manoharan M, et al. Immunodominant T-cell epitopes from the SARS-CoV-2 spike antigen reveal robust pre-existing T-cell immunity in unexposed individuals. Sci Rep. (2021) 11:13164. doi: 10.1038/s41598-021-92521-4.
- Wang C, Liu Z, Chen Z, Huang X, Xu M, He T, et al. The establishment of reference sequence for SARS-CoV-2 and variation analysis. Journal of medical virology. 2020;92(6):667-74.
- Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nature communications. 2020;11(1):19.
- Daniloski Z, Jordan TX, Ilmain JK, Guo X, Bhabha G, TenOever BR, et al. The Spike D614G mutation increases SARS-CoV-2 infection of multiple human cell types. Elife. 2021;10:e65365.
- Zhang J, Cai Y, Xiao T, Lu J, Peng H, Sterling SM, et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science. 2021;372(6541):525-30.
- Ahmad L. Implication of SARS-CoV-2 immune escape spike variants on secondary and vaccine breakthrough infections. Frontiers in immunology. 2021:4563.
- Korber, B., Fischer, W. M., Gnanakaran, S., Yoon, H., Theiler, J., Abfalterer, W., ... & Montefiori, D. C., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell, 2020, 182.4: 812-827. e19.
- Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284-94. e9.
- Shah M, Woo HG. Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Frontiers in immunology. 2022;12:6031.
- Barton MI, MacGowan SA, Kutuzov MA, Dushek O, Barton GJ, Van Der Merwe PA,et al. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. Elife. 2021;10:e70658.
- Greaney, A. J., Loes, A. N., Crawford, K. H., Starr, T. N., Malone, Greaney, A. J., Loes, A. N., Crawford, K. H., Starr, T. N., Malone, et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell host & microbe, 2021, 29.3: 463-476. e6.
- Khateeb J, Li Y, Zhang H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Critical Care. 2021;25(1):1-8.
- Barrett CT, Neal HE, Edmonds K, Moncman CL, Thompson R, Branttie JM, et al. Effect of mutations in the SARS-CoV-2 spike protein on protein stability, cleavage, and cell-cell fusion function. bioRxiv. 2021.
- Harvey, W. T., Carabelli, A. M., Jackson, B., Gupta, R. K., Thomson, E. C., Harrison, E. M., ... & Robertson, D. L.., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nature Reviews Microbiology, 2021, 19.7: 409-424.
- Mishra SK, Tripathi T. One year update on the COVID-19 pandemic: Where are we now? Acta tropica. 2021;214:105778.
- Guruprasad K. Geographical distribution of amino acid mutations in human SARS-CoV-2 orf1ab poly-proteins compared to the equivalent reference proteins from China. ChemRxiv. (2021). 10.33774/chemrxiv-2021-lf2zd-v2.
- Graham RL, Sparks JS, Eckerle LD, Sims AC, Denison MR,et al. SARS coronavirus replicase proteins in pathogenesis. Virus research. 2008;133(1):88-100.
- Thomas S. Mapping the nonstructural transmembrane proteins of severe acute respiratory syndrome coronavirus 2. Journal of Computational Biology. 2021;28(9):909-21.
- Wolff G, Melia CE, Snijder EJ, Bárcena M,et al. Double-membrane vesicles as platforms for viral replication. Trends in microbiology. 2020;28(12):1022-33.
- Diao B, Wen K, Zhang J, Chen J, Han C, Chen Y, et al. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clinical Microbiology and Infection. 2021;27(2):289. e1-. e4.
- Gao T, Gao Y, Liu X, Nie Z, Sun H, Lin K, et al. Identification and functional analysis of the SARS-COV-2 nucleocapsid protein. BMC microbiology. 2021;21(1):1-10.
- Ramesh S, Govindarajulu M, Parise RS, Neel L, Shankar T, Patel S, et al. Emerging SARS-CoV-2 variants: a review of its mutations, its implications and vaccine efficacy. Vaccines. 2021;9(10):1195.
- Washington NL, Gangavarapu K, Zeller M, Bolze A, Cirulli ET, Barrett KMS, et al. Emergence and rapid transmission of SARS-CoV-2 B. 1.1. 7 in the United States. Cell. 2021;184(10):2587-94. e7.
- Jung, C.; Kmiec, D.; Koepke, L.; Zech, F.; Jacob, T.; Sparrer, K.M.J.; Kirchhoff, F. Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning? J. Virol. 2022, 96, e02077-21.
- de Silva, T.I.; Liu, G.; Lindsey, B.B.; Dong, D.; Moore, S.C.; Hsu, N.S.; Shah, D.; Wellington, D.; Mentzer, A.J.; Angyal, A.; et al. The Impact of Viral Mutations on Recognition by SARS-CoV-2 Specific T Cells. iScience 2021, 24, 103353.
- Washington N.L., Gangavarapu K., Zeller M., Bolze A., Cirulli E.T., Schiabor Barrett K.M., Larsen B.B., Anderson C., White S., Cassens T., et al. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell. 2021;184:2587–2594.e7.
- Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370:eabe9403.
- Kannan, S. R., Spratt, A. N., Sharma, K., Chand, H. S., Byrareddy, S. N., & Singh, K..Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. J Autoimmun. 2022 Jan; 126: 102779.
| Mutation | Position | Nucleotide change | Code | Amino acid Change | Type of Mutation | ||
|---|---|---|---|---|---|---|---|
| ORF1a (266...13468) | |||||||
| 444 | GTT > GCT | V 60 A | Valin>Alanine | Non-synonymous SNV | |||
| 593 | CAT > TAT | H 110 Y | Histidine>Tyrosine | Non-synonymous SNV | |||
| 670 | AGT > AGG | S 135 R | Serine>Arginine | Non-synonymous SNV | |||
| 1415 | CTT > TTT | L 384 F | Leucine>Phenylalanine | Non-synonymous SNV | |||
| 2790 | ACT > ATT | T 842 I | Threonine>Isoleucine | Non-synonymous SNV | |||
| 2832 | AAG > AGG | K 856 R | Lysine>Arginine | Non-synonymous SNV | |||
| 2883 | TGT > TAT | C 873 Y | Cisteine>Tyrosine | Non-synonymous SNV | |||
| 3896 | GTT > TTT | V 1211 F | Valine>Phenylalanine | Non-synonymous SNV | |||
| 4184 | GGT > AGT | G 1307 S | Glycine>Serine | Non-synonymous SNV | |||
| 4893 | ACA > ATA | T 1543 I | Threonin>Isoleucine | Non-synonymous SNV | |||
| 5007 | ACG > ATG | T 1581 M | Threonin>Methionine | Non-synonymous SNV | |||
| 510 – 518 | ATG > -TG | del82/84 | del82/84 | Non-frame shift deletion | |||
| 519 | ATG > -TG | M 85 V | Methionine>Valine | Non-synonymous SNV | |||
| 6176 | GAT > AAT | D 1971 N | Aspartic acid>Asparagine | Non-synonymous SNV | |||
| 6513 - 6515 | del2083/2083 | del2083/2083 | Non-synonymous deletion | ||||
| 6516 | TTA > -TA | L 2084 I | Leucine>Isoleucine | Non-synonymous SNV | |||
| 7036 | TTA > TTT | L 2257 F | Leucine>Phenylalanine | Non-synonymous SNV | |||
| 7488 | ACT > ATT | T 2408 I | Threonine>Isoleucine | Non-synonymous SNV | |||
| 8393 | GCT > ACT | A 2710 T | Alanine>Threonin | Non-synonymous SNV | |||
| 9344 | CTT > TTT | L 3027 F | Leucine>Phenylalanine | Non-synonymous SNV | |||
| 9474 | GCT > GTT | A 3070 V | Alanine>Valine | Non-synonymous SNV | |||
| 9534 | ACT > ATT | T 3090 I | Threonine>Isoleucine | Non-synonymous SNV | |||
| 9866 | CTT > TTT | L 32201 I | Leucine>Isoleucine | Non-synonymous SNV | |||
| 10029 | ACC > ATC | T 3255 I | Threonin>Isoleucine | Non-synonymous SNV | |||
| 10323 | AAG > AGG | K 3353 R | Lysine>Arginine | Non-synonymous SNV | |||
| 10449 | CCC > CAC | P 3395 H | Proline>Histidine | Non-synonymous SNV | |||
| 11405 | GTC > TTC | V 3714 F | Valine>Phenylalanine | Non-synonymous SNV | |||
| 11285-11293 | del3674/3676 | del3674/3676 | Non-frame shift deletion | ||||
| 11537 | ATT > GTT | I 3758 V | Isoleucine>Valine | Non-synonymous SNV | |||
| 12534 | ACT > ATT | T 409 I | Threonine>Isoleucine | Non-synonymous SNV | |||
| ORF1b (13468...21555) | |||||||
| 13756 | ATA > GTA | I 97 V | Isoleucine>Valine | Non-synonymous SNV | |||
| 14408 | CCT > CTT | P 314 L | Proline>Leucine | Non-synonymous SNV | |||
| 14821 | CCA > TCA | P 452 S | Proline>Serine | Non-synonymous SNV | |||
| 15641 | AAT > AGT | N 725 S | Asparagine>Serine | Non-synonymous SNV | |||
| 15982 | GTA > ATA | V 839 I | Valine>Isoleucine | Non-synonymous SNV | |||
| 16744 | GGT > AGT | G 1093 S | Glycine>Serine | Non-synonymous SNV | |||
| 17410 | GGT > TGT | R 1315 C | Arginine>Cisteine | Non-synonymous SNV | |||
| 18163 | ATA > GTA | I 1566 V | Isoleucine>Valine | Non-synonymous SNV | |||
| 18433 | GAT > CAT | D 165 H | Aspartic acid>Histidine | Non-synonymous SNV | |||
| 19999 | GTT > TTT | V 2178 F | Valine>Phenylalanine | Non-synonymous SNV | |||
| 20003 | GAT > GGT | P 2179 G | Proline>Glycine | Non-synonymous SNV | |||
| S (21563...25384) | |||||||
| 21765 - 21770 | TACATG > - - - | del69/70 | del69/70 | Non-synonymous deletion | |||
| 21789 | ACT > ATT | T 76 I | Threonine>Isoleucine | Non-synonymous SNV | |||
| 21846 | ACT > ATT | T95I | Threonine>Isoleucine | Non-frame shift deletion | |||
| 21987 | GGT > GAT | G142D | Glycine>Aspartic acid | Non-synonymous SNV | |||
| 21987 - 21995 | del142/144 | del142/144 | Non-frame shift deletion | ||||
| 21996 | TAC > -AC | Y 145 D | Tyrosine>Aspartic acid | Non-synonymous SNV | |||
| 22194 - 22196 | AAT > A-- | del211/211 | del211/211 | Non-synonymous deletion | |||
| 22197 | TTA > -TA | L 212 I | Leucine>Isoleucine | Non-synonymous SNV | |||
| 222000 | GTG > GGG | V 213 G | Valine>Glycine | Non-synonymous SNV | |||
| 22578 | GCT > GAT | G339D | Glycine>Aspartic acid | Non-synonymous SNV | |||
| 22599 | AGA > AAA | R346K | Arginine>Lysine | Non-synonymous SNV | |||
| 22673 | T > C | S371L | Serine>Leucine | Non-synonymous SNV | |||
| 22674 | C > T | S 373 P | Serine>Proline | Non-synonymous SNV | |||
| 22686 | TCC > TTC | S 375 F | Serine>Phenylalanine | Non-synonymous SNV | |||
| 22688 | ACT > GCT | T 376 A | Threonine>Isoleucine | Non-synonymous SNV | |||
| 22786 | AGA > AGC | R408S | Arginine>Serine | Non-synonymous SNV | |||
| 22813 | AAG > AAT | K 417 N | Lysine>Asparagine | Non-synonymous SNV | |||
| 22882 | AAT > AAG | N440K | Asparagine>Lysine | Non-synonymous SNV | |||
| 22898 | GGT > AGT | G446S | Glycine>Serine | Non-synonymous SNV | |||
| 23013 | GAA > GCA | E 484 A | Glutamic acid > isoleucine | Non-synonymous SNV | |||
| 22992 | AGC > AAC | S477N | Serine>Asparagine | Non-synonymous SNV | |||
| 22995 | ACA > AAA | T478K | Threonine>Lysine | Non-synonymous SNV | |||
| 23040 | CAA > CGA | Q493R | Glutamine>Arginine | Non-synonymous SNV | |||
| 23048 | G > A | G496S | Glycine>Serine | Non-synonymous SNV | |||
| 23055 | A > G | Q498R | Glutamine>Arginine | Non-synonymous SNV | |||
| 23063 | AAT > TAT | N501Y | Asparagine>Tyrosine | Non-synonymous SNV | |||
| 23075 | TAC > CAC | Y505H | Tyrosine>Histidine | Non-synonymous SNV | |||
| 23202 | ACA > AAA | T547K | Threonine>Lysine | Non-synonymous SNV | |||
| 23403 | GAT > GGT | D614G | Aspartic acid>Glycine | Non-synonymous SNV | |||
| 23525 | CAT > TAT | H655Y | Histidine>Tyrosine | Non-synonymous SNV | |||
| 23599 | T > G | N679K | Asparagine>Lysine | Non-synonymous SNV | |||
| 23604 | CCT > CAT | P681H | Proline>Histidine | Non-synonymous SNV | |||
| 23854 | AAC > AAA | N764K | Asparagine>Lysine | Non-synonymous SNV | |||
| 23948 | GAT > TAT | D796Y | Aspartic acid>Tyrosine | Non-synonymous SNV | |||
| 24130 | ACC > AAA | N856K | Asparagine>Lysine | Non-synonymous SNV | |||
| 24424 | CAA > CAT | Q954H | Glutamine>Histidine | Non-synonymous SNV | |||
| 24469 | AAT > AAA | N969K | Asparagine>Lysine | Non-synonymous SNV | |||
| 24503 | CCT > TTT | L981F | Leucine>Phenylalanine | Non-synonymous SNV | |||
| ORF3a (25393…26220) | |||||||
| 25471 | GAT > TAT | D 27 Y | Aspartic acid>Tyrosine | Non-synonymous SNV | |||
| 26060 | ACT > ATT | T 223 I | Threonine>Isoleucine | Non-synonymous SNV | |||
| M (26523... 27191) | 26530 | GAT > GGT | D 3 G | Aspartic acid>Glycine | Non-synonymous SNV | ||
| 26577 | CAA > GAA | Q 19 E | Glutamine>Glutamic acid | Non-synonymous SNV | |||
| 26709 | GCT > ACT | A 63 T | Alanine>Threonin | Non-synonymous SNV | |||
| ORF6 (27202…27387) | 27269 | AAA > -AA | K 23 * | K23* | Non-synonymous SNV | ||
| 27266 - 27268 | TTA > - - - | del22/23 | del22/23 | Non-frame shift deletion | |||
| ORF9b (28284…28577) | 28311 | CCC > TCC | P 10 S | Proline>Serine | Non-synonymous SNV | ||
| N (28274…29533) | 28881 | AGG > AAA | R 203 K | Arginine>Lysine | Non-synonymous SNV | ||
| 28882 | AGG > AAA | R203 K | Arginine>Lysine | Non-synonymous SNV | |||
| 28883 | GGA > ACG | G 204 R | Glycine>Arginine | Non-synonymous SNV | |||
| 28311 | CCC > CTC | P 13 L | Proline>Leucine | Non-synonymous SNV | |||
| 28725 | CCT > CTT | P 151 L | Proline>Leucine | Non-synonymous SNV | |||
| 29000 | GGC > AGC | G 243 S | Glycine>Serine | Non-synonymous SNV | |||
| 29005 | CAA > CAC | Q 244 H | Glutamine>Histidine | Non-synonymous SNV | |||
| 29510 | AGT > CGT | S 413 R | Serine > Arginine | Non-synonymous SNV | |||
| Upload Acc. ID | Distance | Quality | Accession ID Of Close Related Genome | Collection date | Submission date | lineage | Country/State |
|---|---|---|---|---|---|---|---|
| EPI_ISL_12604438 | 1 | 0.959 | EPI_ISL_10185095 | 2/1/2022 | 2/22/2022 | BA.1.1 | United Kingdom / England |
| EPI_ISL_12604442 | 0 | 0.998 | EPI_ISL_12604518 | 2/10/2022 | 5/9/2022 | BA.1.1 | Iraq / Kurdistan / Duhok |
| EPI_ISL_12604444 | 0 | 0 | EPI_ISL_11163451 | 1/26/2022 | 3/18/2022 | BA.1.1 | USA / Tennessee |
| EPI_ISL_12604448 | 0 | 1 | EPI_ISL_11501531 | 2/18/2022 | 3/28/2022 | BA.1.1 | Canada / Saskatchewan |
| EPI_ISL_12604478 | 0 | 0.974 | EPI_ISL_9222066 | 1/16/2022 | 1/28/2022 | BA.1.1 | Germany / Hamburg |
| EPI_ISL_12604481 | 0 | 0.943 | EPI_ISL_9021360 | 1/16/2022 | 1/24/2022 | BA.1.1 | United Kingdom / England |
| EPI_ISL_12604482 | 0 | 1 | EPI_ISL_9041699 | 1/5/2022 | 1/24/2022 | BA.1.1 | USA / Maryland |
| EPI_ISL_12604483 | 0 | 0.999 | EPI_ISL_13528111 | 2/6/2022 | 6/28/2022 | BA.1.1 | Austria / Styria / Liezen |
| EPI_ISL_12604487 | 0 | 0.977 | EPI_ISL_9830331 | 1/20/2022 | 2/12/2022 | BA.1 | Poland /Swietokrzyskie Voivodeship |
| EPI_ISL_12604488 | 0 | 0.947 | EPI_ISL_8456340 | 12/28/2021 | 1/7/2022 | BA.1.1 | United Kingdom / Scotland |
| EPI_ISL_12604489 | 1 | 1 | EPI_ISL_17134891 | 1/13/2022 | 2/28/2022 | BA.1 | USA / Arkansas |
| EPI_ISL_12604490 | 0 | 0.997 | EPI_ISL_10676026 | 2/11/2022 | 3/4/2022 | BA.1.1 | United Kingdom / Wales |
| EPI_ISL_12604495 | 1 | 0.94 | EPI_ISL_11955489 | 1/12/2022 | 4/11/2022 | BA.1.1 | United Kingdom / England |
| EPI_ISL_12604496 | 1 | 0.942 | EPI_ISL_10042180 | 2/2/2022 | 2/17/2022 | BA.1.1 | USA / Illinois |
| EPI_ISL_12604501 | 0 | 1 | EPI_ISL_12604497 | 2/10/2022 | 5/9/2022 | BA.1.1 | Iraq / Kurdistan / Duhok |
| EPI_ISL_12604502 | 0 | 0.994 | EPI_ISL_12604412 | 2/5/2022 | 5/9/2022 | BA.1.1 | Iraq / Kurdistan / Duhok |
| EPI_ISL_12604503 | 0 | 1 | EPI_ISL_11252814 | 3/3/2022 | 3/21/2022 | BA.2 | USA / Ohio |
| EPI_ISL_12604507 | 1 | 0.999 | EPI_ISL_11041477 | 3/7/2022 | 3/15/2022 | BA.1.1 | United Kingdom / Scotland |
| EPI_ISL_12604508 | 0 | 1 | EPI_ISL_10011583 | 12/28/2021 | 2/17/2022 | BA.1.1 | Kenya / Migori |
| EPI_ISL_12604509 | 1 | 0.954 | EPI_ISL_9517295 | 1/14/2022 | 2/4/2022 | BA.1.1 | Iraq / Baghdad |
| EPI_ISL_12604510 | 0 | 0.991 | EPI_ISL_7908939 | 12/10/2021 | 12/21/2021 | BA.1.1 | Denmark / Syddanmark |
| EPI_ISL_12604514 | 1 | 0.903 | EPI_ISL_9835746 | 1/11/2022 | 2/13/2022 | BA.1.1 | United Kingdom / England |
| EPI_ISL_12604516 | 0 | 1 | EPI_ISL_10181069 | 2/2/2022 | 2/22/2022 | BA.1.1 | United Kingdom / Scotland |
| EPI_ISL_12604517 | 0 | 0.999 | EPI_ISL_10497679 | 1/11/2022 | 3/1/2022 | BA.1.1 | USA / Michigan |
| EPI_ISL_12604521 | 0 | 1 | EPI_ISL_17128823 | 1/20/2022 | 3/21/2022 | BA.1.1 | USA / Arkansas |
| EPI_ISL_12604527 | 1 | 0.986 | EPI_ISL_9998212 | 2/10/2022 | 2/17/2022 | BA.1.1 | United Kingdom / Scotland |
| EPI_ISL_12604528 | 1 | 1 | EPI_ISL_13674544 | 1/13/2022 | 7/6/2022 | BA.1.1 | USA / Minnesota |
| EPI_ISL_12604532 | 2 | 0.989 | EPI_ISL_9233949 | 12/29/2021 | 1/28/2022 | BA.1.1 | Germany / Rhineland-Palatinate |
| EPI_ISL_12604845 | 1 | 0.946 | EPI_ISL_10240172 | 12/22/2021 | 2/23/2022 | BA.1.1 | Lithuania / Kauno apskritis |
| EPI_ISL_12604846 | 0 | 1 | EPI_ISL_12604443 | 1/24/2022 | 5/9/2022 | BA.1.1 | Iraq / Kurdistan / Duhok |
| EPI_ISL_12604847 | 0 | 1 | EPI_ISL_9041699 | 1/5/2022 | 1/24/2022 | BA.1.1 | USA / Maryland |
| EPI_ISL_12604848 | 0 | 1 | EPI_ISL_9985416 | 2/4/2022 | 2/16/2022 | BA.1.1 | Poland / Lodzkie / Lodz |
| EPI_ISL_12604451 | 0 | 0.954 | EPI_ISL_10249214 | 2022 | 2/23/2022 | BA.1 | United Kingdom / England |
| EPI_ISL_12604457 | 1 | 0.94 | EPI_ISL_10557396 | 2/9/2022 | 3/2/2022 | BA.1.1 | USA / Colorado |
| EPI_ISL_12604460 | 2 | 0.971 | EPI_ISL_13711474 | 1/4/2022 | 7/8/2022 | BA.1.1 | Kyrgyzstan / Chui |
| EPI_ISL_12604463 | 1 | 1 | EPI_ISL_12604423 | 1/19/2022 | 5/9/2022 | BA.1.1 | Iraq / Kurdistan / Duhok |
| EPI_ISL_12604471 | 0 | 0.932 | EPI_ISL_9224622 | 12/30/2021 | 1/28/2022 | BA.1 | Germany / North Rhine-Westphalia |
| EPI_ISL_12604476 2 0.954 | 2 | 0.954 | EPI_ISL_12058290 | 12/19/2021 | 4/14/2022 | BA.1.1 | USA / California / Placer County |
| EPI_ISL_12604526 | 0 | 0.907 | EPI_ISL_11405543 | 2/27/2022 | 3/25/2022 | BA.1.1 | South Korea |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





