Submitted:
12 August 2023
Posted:
14 August 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction

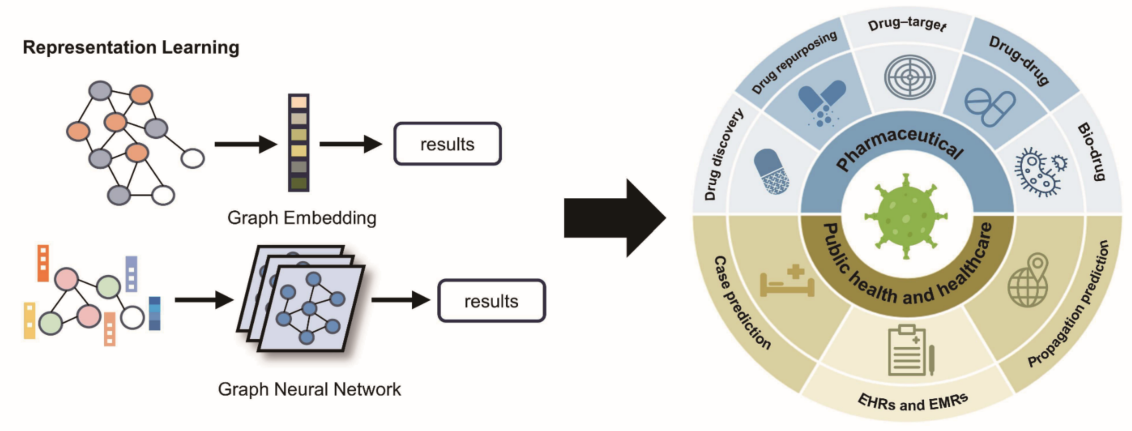
2. Representation Learning
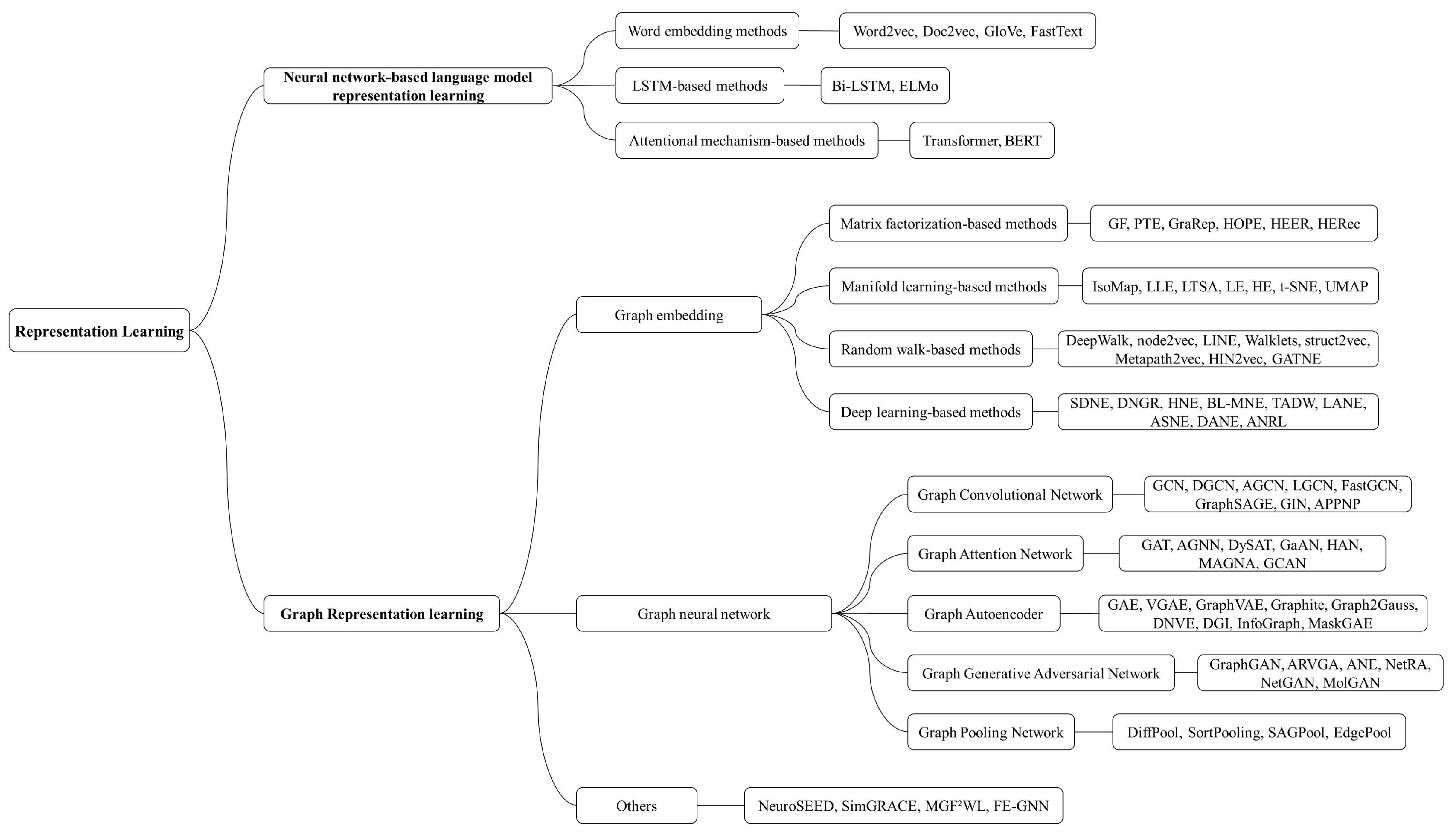
3. Overview of Representation Learning Methods
3.1. Neural Network-Based Language Model Representation Learning
3.2. Graph Representation Learning
3.2.1. Graph Embedding
3.2.2. Graph Neural Network-Based Methods
| Category | Method Name | Code Link |
|---|---|---|
|
Neural network-based language model representation learning |
Word2vec [37] | https://code.google.com/archive/p/word2vec/ |
| Doc2vec [38] | https://nbviewer.org/github/danielfrg/ word2vec/blob/main/examples/doc2vec.ipynb |
|
| GloVe [39] | https://nlp.stanford.edu/projects/glove/ | |
| FastText [40] | https://github.com/facebookresearch/fastText | |
| Bi-LSTM [42] | - | |
| ELMo [43] | https://allenai.org/allennlp/software/elmo | |
| Transformer [46] | https://github.com/tensorflow/tensorflow | |
| BERT [50] | https://github.com/google-research/bert | |
|
Graph representation learning |
||
| Graph embedding | GF [52] | - |
| PTE [53] | https://github.com/mnqu/PTE | |
| GraRep [54] | https://github.com/ShelsonCao/GraRep | |
| HOPE [55] | http://git.thumedia.org/embedding/HOPE | |
| HEER [56] | https://github.com/GentleZhu/HEER | |
| HERec [57] | https://github.com/librahu/HERec | |
| IsoMap [58] | https://github.com/scikit-learn/scikit-learn/blob/ main/sklearn/manifold/_isomap.py |
|
| LLE [59] | - | |
| LTSA [60] | - | |
| LE [61] | - | |
| HE [62] | - | |
| t-SNE [63] | https://lvdmaaten.github.io/tsne/ | |
| UMAP [64] | https://github.com/lmcinnes/umap | |
| DeepWalk [65] | https://github.com/phanein/deepwalk | |
| node2vec [66] | https://github.com/aditya-grover/node2vec | |
| LINE [67] | https://github.com/tangjianpku/LINE | |
| Walklets [68] | https://github.com/benedekrozemberczki/ walklets |
|
| struct2vec [69] | https://github.com/leoribeiro/struc2vec | |
| Metapath2vec [70] | https://ericdongyx.github.io/metapath2vec/ m2v.html |
|
| HIN2vec [71] | https://github.com/csiesheep/hin2vec | |
| GATNE [72] | https://github.com/THUDM/GATNE | |
| SDNE [73] | https://github.com/suanrong/SDNE | |
| DNGR [74] | https://github.com/ShelsonCao/DNGR | |
| HNE [75] | - | |
| BL-MNE [76] | - | |
| TADW [77] | https://github.com/thunlp/tadw | |
| LANE [78] | https://github.com/xhuang31/LANE | |
| ASNE [79] | https://github.com/lizi-git/ASNE | |
| DANE [80] | https://github.com/gaoghc/DANE | |
| ANRL [81] | https://github.com/cszhangzhen/ANRL | |
| Graph neural network | GCN [84] | https://github.com/tkipf/gcn |
| DGCN [85] | https://github.com/ZhuangCY/DGCN | |
| AGCN [86] | https://github.com/yimutianyang/AGCN | |
| LGCN [87] | https://github.com/divelab/lgcn | |
| FastGCN [88] | https://github.com/matenure/FastGCN | |
| GraphSAGE [89] | https://github.com/williamleif/GraphSAGE | |
| GIN [90] | https://github.com/weihua916/powerful-gnns | |
| APPNP [91] | https://github.com/gasteigerjo/ppnp | |
| GAT [92] | https://github.com/PetarV-/GAT | |
| AGNN [93] | - | |
| DySAT [94] | https://github.com/aravindsankar28/DySAT | |
| GaAN [95] | https://github.com/jennyzhang0215/GaAN | |
| HAN [96] | https://github.com/Jhy1993/HAN | |
| MAGNA [97] | https://github.com/xjtuwgt/GNN-MAGNA | |
| GCAN [98] | - | |
| GAE [99] | https://github.com/tkipf/gae | |
| VGAE [99] | https://github.com/tkipf/gae | |
| Graph neural network | GraphVAE [100] | https://github.com/snap-stanford/GraphRNN/ tree/master/baselines/graphvae |
| Graphite [101] | https://github.com/ermongroup/graphite | |
| Graph2Gauss [102] | https://github.com/abojchevski/gra ph2gauss |
|
| DNVE [103] | - | |
| DGI [104] | https://github.com/PetarV-/DGI | |
| InfoGraph [105] | https://github.com/fanyun-sun/Info Graph |
|
| MaskGAE [106] | https://github.com/EdisonLeeeee/Mas kGAE |
|
| GraphGAN [108] | https://github.com/hwwang55/GraphGAN | |
| ARVGA [109] | - | |
| ANE [110] | - | |
| NetRA [111] | https://github.com/chengw07/NetRA | |
| NetGAN [112] | https://github.com/danielzuegner/ne tgan |
|
| MolGAN [113] | https://github.com/nicola-decao/Mol GAN |
|
| DiffPool [114] | https://github.com/RexYing/diffpool | |
| SortPooling [115] | https://github.com/muhanzhang/DGCNN | |
| SAGPool [116] | https://github.com/inyeoplee77/SAG Pool |
|
| EdgePool [117] | - | |
| Others | NeuroSEED [118] | https://github.com/gcorso/NeuroSEED |
| SimGRACE [119] | https://github.com/mpanpan/SimGRACE | |
| MGF²WL [120] | - | |
| FE-GNN [121] | https://github.com/sajqavril/Featur e-Extension-Graph-Neural-Networks |
4. Representation Learning Methods for COVID-19
4.1. Pharmaceutical
4.1.1. Drug Discovery
4.1.2. Drug Repurposing
4.1.3. Drug–Target Interaction Prediction
4.1.4. Drug–Drug Interaction Prediction
4.1.5. Bio-Drug Interaction Prediction
4.2. Public Health and Healthcare
4.2.1. Case Prediction
4.2.2. Propagation Prediction
4.2.3. Analysis of Ehrs and Emrs
5. Challenges and Prospects
5.1. Data Quality
5.2. Hyperparameters and Labels
5.3. Interpretability and Extensibility
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- William T Harvey, Alessandro M Carabelli, Ben Jackson, Ravindra K Gupta, Emma C Thomson, Ewan M Harrison, Catherine Ludden, Richard Reeve, Andrew Rambaut, COVID-19 Genomics UK (COG-UK) Consortium, et al. Sars-cov-2 variants, spike mutations and immune escape. Nature Reviews Microbiology, 19(7):409–424, 2021.
- James Hadfield, Colin Megill, Sidney M Bell, John Huddleston, Barney Potter, Charlton Callender, Pavel Sagulenko, Trevor Bedford, and Richard A Neher. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics, 34(23):4121–4123, 2018. [CrossRef]
- David M Morens, Gregory K Folkers, and Anthony S Fauci. The concept of classical herd immunity may not apply to covid-19. The Journal of infectious diseases, 226(2):195–198, 2022. [CrossRef]
- Leiwen Fu, Bingyi Wang, Tanwei Yuan, Xiaoting Chen, Yunlong Ao, Thomas Fitzpatrick, Peiyang Li, Yiguo Zhou, Yi-fan Lin, Qibin Duan, et al. Clinical characteristics of coronavirus disease 2019 (covid-19) in china: a systematic review and meta-analysis. Journal of Infection, 80(6):656–665, 2020. [CrossRef]
- Elizabeth J Williamson, Alex J Walker, Krishnan Bhaskaran, Seb Bacon, Chris Bates, Caroline E Morton, Helen J Curtis, Amir Mehrkar, David Evans, Peter Inglesby, et al. Opensafely: factors associated with covid-19 death in 17 million patients. Nature, 584(7821):430, 2020.
- Yan-Rong Guo, Qing-Dong Cao, Zhong-Si Hong, Yuan-Yang Tan, Shou-Deng Chen, Hong-Jun Jin, Kai-Sen Tan, De-Yun Wang, and Yan Yan. The origin, transmission and clinical therapies on coronavirus disease 2019 (covid-19) outbreak–an update on the status. Military medical research, 7:1–10, 2020.
- Chaolin Huang, Yeming Wang, Xingwang Li, Lili Ren, Jianping Zhao, Yi Hu, Li Zhang, Guohui Fan, Jiuyang Xu, Xiaoying Gu, et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, china. The lancet, 395(10223):497–506, 2020.
- Thanh Thi Nguyen, Mohamed Abdelrazek, Dung Tien Nguyen, Sunil Aryal, Duc Thanh Nguyen, Sandeep Reddy, Quoc Viet Hung Nguyen, Amin Khatami, Edbert B Hsu, and Samuel Yang. Origin of novel coronavirus (covid-19): a computational biology study using artificial intelligence. BioRxiv, pages 2020–05, 2020. [CrossRef]
- Marco Cascella, Michael Rajnik, Abdul Aleem, Scott C Dulebohn, and Raffaela Di Napoli. Features, evaluation, and treatment of coronavirus (covid-19). Statpearls [internet], 2022.
- Shibo Jiang, Christopher Hillyer, and Lanying Du. Neutralizing antibodies against sars-cov-2 and other human coronaviruses. Trends in immunology, 41(5):355–359, 2020.
- Lok Bahadur Shrestha, Charles Foster, William Rawlinson, Nicodemus Tedla, and Rowena A Bull. Evolution of the sars-cov-2 omicron variants ba. 1 to ba. 5: Implications for immune escape and transmission. Reviews in Medical Virology, 32(5):e2381, 2022. [CrossRef]
- Bette Korber, Will M Fischer, Sandrasegaram Gnanakaran, Hyejin Yoon, James Theiler, Werner Abfalterer, Nick Hengartner, Elena E Giorgi, Tanmoy Bhattacharya, Brian Foley, et al. Tracking changes in sars-cov-2 spike: evidence that d614g increases infectivity of the covid-19 virus. Cell, 182(4):812–827, 2020.
- Yunlong Cao, Jing Wang, Fanchong Jian, Tianhe Xiao, Weiliang Song, Ayijiang Yisimayi, Weijin Huang, Qianqian Li, Peng Wang, Ran An, et al. Omicron escapes the majority of existing sars-cov-2 neutralizing antibodies. Nature, 602(7898):657–663, 2022.
- Manojit Bhattacharya, Ashish Ranjan Sharma, Kuldeep Dhama, Govindasamy Agoramoorthy, and Chiranjib Chakraborty. Omicron variant (b. 1.1. 529) of sars-cov-2: understanding mutations in the genome, s-glycoprotein, and antibody-binding regions. GeroScience, 44(2):619–637, 2022.
- Dhiraj Mannar, James W Saville, Xing Zhu, Shanti S Srivastava, Alison M Berezuk, Katharine S Tuttle, Ana Citlali Marquez, Inna Sekirov, and Sriram Subramaniam. Sars-cov-2 omicron variant: Antibody evasion and cryo-em structure of spike protein–ace2 complex. Science, 375(6582):760–764, 2022.
- Dinah V Parums. The xbb. 1.5 (‘kraken’) subvariant of omicron sars-cov-2 and its rapid global spread. Medical Science Monitor, 29, 2023.
- Imad A Basheer and Maha Hajmeer. Artificial neural networks: fundamentals, computing, design, and application. Journal of microbiological methods, 43(1):3–31, 2000. [CrossRef]
- Yifei Chen, Yi Li, Rajiv Narayan, Aravind Subramanian, and Xiaohui Xie. Gene expression inference with deep learning. Bioinformatics, 32(12):1832–1839, 2016. [CrossRef]
- Hongming Chen, Ola Engkvist, Yinhai Wang, Marcus Olivecrona, and Thomas Blaschke. The rise of deep learning in drug discovery. Drug discovery today, 23(6):1241–1250, 2018. [CrossRef]
- Mihalj Bakator and Dragica Radosav. Deep learning and medical diagnosis: A review of literature. Multimodal Technologies and Interaction, 2(3):47, 2018. [CrossRef]
- Jie Zhou, Ganqu Cui, Shengding Hu, Zhengyan Zhang, Cheng Yang, Zhiyuan Liu, Lifeng Wang, Changcheng Li, and Maosong Sun. Graph neural networks: A review of methods and applications. AI open, 1:57–81, 2020. [CrossRef]
- Jiacheng Xiong, Zhaoping Xiong, Kaixian Chen, Hualiang Jiang, and Mingyue Zheng. Graph neural networks for automated de novo drug design. Drug Discovery Today, 26(6):1382–1393, 2021. [CrossRef]
- Fang Yang, Kunjie Fan, Dandan Song, and Huakang Lin. Graph-based prediction of protein-protein interactions with attributed signed graph embedding. BMC bioinformatics, 21(1):1–16, 2020. [CrossRef]
- Xiao-Meng Zhang, Li Liang, Lin Liu, and Ming-Jing Tang. Graph neural networks and their current applications in bioinformatics. Frontiers in genetics, 12:690049, 2021. [CrossRef]
- Daniele Mercatelli, Laura Scalambra, Luca Triboli, Forest Ray, and Federico M Giorgi. Gene regulatory network inference resources: A practical overview. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 1863(6):194430, 2020. [CrossRef]
- Hongyun Cai, Vincent W Zheng, and Kevin Chen-Chuan Chang. A comprehensive survey of graph embedding: Problems, techniques, and applications. IEEE transactions on knowledge and data engineering, 30(9):1616–1637, 2018. [CrossRef]
- Mengjia Xu. Understanding graph embedding methods and their applications. SIAM Review, 63(4):825–853, 2021. [CrossRef]
- James Kotary, Ferdinando Fioretto, Pascal Van Hentenryck, and Bryan Wilder. End-to-end constrained optimization learning: A survey. arXiv preprint arXiv:2103.16378, 2021. [CrossRef]
- Xiao Wang, Deyu Bo, Chuan Shi, Shaohua Fan, Yanfang Ye, and S Yu Philip. A survey on heterogeneous graph embedding: methods, techniques, applications and sources. IEEE Transactions on Big Data, 2022. [CrossRef]
- Giulia Muzio, Leslie O’Bray, and Karsten Borgwardt. Biological network analysis with deep learning. Briefings in bioinformatics, 22(2):1515–1530, 2021.
- Zehong Zhang, Lifan Chen, Feisheng Zhong, Dingyan Wang, Jiaxin Jiang, Sulin Zhang, Hualiang Jiang, Mingyue Zheng, and Xutong Li. Graph neural network approaches for drug-target interactions. Current Opinion in Structural Biology, 73:102327, 2022. [CrossRef]
- Sezin Kircali Ata, Min Wu, Yuan Fang, Le Ou-Yang, Chee Keong Kwoh, and Xiao-Li Li. Recent advances in network-based methods for disease gene prediction. Briefings in bioinformatics, 22(4):bbaa303, 2021. [CrossRef]
- Oliver Wieder, Stefan Kohlbacher, Mélaine Kuenemann, Arthur Garon, Pierre Ducrot, Thomas Seidel, and Thierry Langer. A compact review of molecular property prediction with graph neural networks. Drug Discovery Today: Technologies, 37:1–12, 2020. [CrossRef]
- World. Statement on the fifteenth meeting of the ihr (2005) emergency committee on the covid-19 pandemic, May 2023.
- Ruiwei Feng, Yufeng Xie, Minshan Lai, Danny Z Chen, Ji Cao, and Jian Wu. Agmi: Attention-guided multi-omics integration for drug response prediction with graph neural networks. In 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pages 1295–1298. IEEE, 2021. [CrossRef]
- Yaxin Zhu, Peisheng Qian, Ziyuan Zhao, and Zeng Zeng. Deep feature fusion via graph convolutional network for intracranial artery labeling. In 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), pages 467–470. IEEE, 2022.
- Tomas Mikolov, Ilya Sutskever, Kai Chen, Greg S Corrado, and Jeff Dean. Distributed representations of words and phrases and their compositionality. Advances in neural information processing systems, 26, 2013.
- Quoc Le and Tomas Mikolov. Distributed representations of sentences and documents. In International conference on machine learning, pages 1188–1196. PMLR, 2014.
- Jeffrey Pennington, Richard Socher, and Christopher D Manning. Glove: Global vectors for word representation. In Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP), pages 1532–1543, 2014.
- Piotr Bojanowski, Edouard Grave, Armand Joulin, and Tomas Mikolov. Enriching word vectors with subword information. Transactions of the association for computational linguistics, 5:135–146, 2017.
- Sepp Hochreiter and Jürgen Schmidhuber. Long short-term memory. Neural computation, 9(8):1735–1780, 1997.
- Zhiheng Huang, Wei Xu, and Kai Yu. Bidirectional lstm-crf models for sequence tagging. arXiv preprint arXiv:1508.01991, 2015.
- Matthew E. Peters, Mark Neumann, Mohit Iyyer, Matt Gardner, Christopher Clark, Kenton Lee, and Luke Zettlemoyer. Deep contextualized word representations, 2018.
- Dzmitry Bahdanau, Kyunghyun Cho, and Yoshua Bengio. Neural machine translation by jointly learning to align and translate. arXiv preprint arXiv:1409.0473, 2014.
- Yoon Kim, Carl Denton, Luong Hoang, and Alexander M Rush. Structured attention networks. arXiv preprint arXiv:1702.00887, 2017.
- Ashish Vaswani, Noam Shazeer, Niki Parmar, Jakob Uszkoreit, Llion Jones, Aidan N Gomez, ukasz Kaiser, and Illia Polosukhin. Attention is all you need. Advances in neural information processing systems, 30, 2017.
- Jonas Gehring, Michael Auli, David Grangier, Denis Yarats, and Yann N Dauphin. Convolutional sequence to sequence learning. In International conference on machine learning, pages 1243–1252. PMLR, 2017.
- Sainbayar Sukhbaatar, Jason Weston, Rob Fergus, et al. End-to-end memory networks. Advances in neural information processing systems, 28, 2015.
- Alexander Miller, Adam Fisch, Jesse Dodge, Amir-Hossein Karimi, Antoine Bordes, and Jason Weston. Key-value memory networks for directly reading documents. arXiv preprint arXiv:1606.03126, 2016.
- Jacob Devlin, Ming-Wei Chang, Kenton Lee, and Kristina Toutanova. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805, 2018.
- Minh-Thang Luong, Hieu Pham, and Christopher D Manning. Effective approaches to attention-based neural machine translation. arXiv preprint arXiv:1508.04025, 2015.
- Amr Ahmed, Nino Shervashidze, Shravan Narayanamurthy, Vanja Josifovski, and Alexander J Smola. Distributed large-scale natural graph factorization. In Proceedings of the 22nd international conference on World Wide Web, pages 37–48, 2013.
- Jian Tang, Meng Qu, and Qiaozhu Mei. Pte: Predictive text embedding through large-scale heterogeneous text networks. In Proceedings of the 21th ACM SIGKDD international conference on knowledge discovery and data mining, pages 1165–1174, 2015.
- Shaosheng Cao, Wei Lu, and Qiongkai Xu. Grarep: Learning graph representations with global structural information. In Proceedings of the 24th ACM international on conference on information and knowledge management, pages 891–900, 2015.
- Mingdong Ou, Peng Cui, Jian Pei, Ziwei Zhang, and Wenwu Zhu. Asymmetric transitivity preserving graph embedding. In Proceedings of the 22nd ACM SIGKDD international conference on Knowledge discovery and data mining, pages 1105–1114, 2016.
- Yu Shi, Qi Zhu, Fang Guo, Chao Zhang, and Jiawei Han. Easing embedding learning by comprehensive transcription of heterogeneous information networks. In Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, pages 2190–2199, 2018.
- Chuan Shi, Binbin Hu, Wayne Xin Zhao, and S Yu Philip. Heterogeneous information network embedding for recommendation. IEEE Transactions on Knowledge and Data Engineering, 31(2):357–370, 2018. [CrossRef]
- Joshua B Tenenbaum, Vin de Silva, and John C Langford. A global geometric framework for nonlinear dimensionality reduction. science, 290(5500):2319–2323, 2000. [CrossRef]
- Sam T Roweis and Lawrence K Saul. Nonlinear dimensionality reduction by locally linear embedding. science, 290(5500):2323–2326, 2000. [CrossRef]
- Zhenyue Zhang and Hongyuan Zha. Principal manifolds and nonlinear dimensionality reduction via tangent space alignment. SIAM journal on scientific computing, 26(1):313–338, 2004. [CrossRef]
- Mikhail Belkin and Partha Niyogi. Laplacian eigenmaps for dimensionality reduction and data representation. Neural computation, 15(6):1373–1396, 2003. [CrossRef]
- David L Donoho and Carrie Grimes. Hessian eigenmaps: Locally linear embedding techniques for high-dimensional data. Proceedings of the National Academy of Sciences, 100(10):5591–5596, 2003. [CrossRef]
- Laurens Van der Maaten and Geoffrey Hinton. Visualizing data using t-sne. Journal of machine learning research, 9(11), 2008.
- Leland McInnes, John Healy, and James Melville. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv preprint arXiv:1802.03426, 2018.
- Bryan Perozzi, Rami Al-Rfou, and Steven Skiena. Deepwalk: Online learning of social representations. In Proceedings of the 20th ACM SIGKDD international conference on Knowledge discovery and data mining, pages 701–710, 2014.
- Aditya Grover and Jure Leskovec. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD international conference on Knowledge discovery and data mining, pages 855–864, 2016.
- Jian Tang, Meng Qu, Mingzhe Wang, Ming Zhang, Jun Yan, and Qiaozhu Mei. Line: Large-scale information network embedding. In Proceedings of the 24th international conference on world wide web, pages 1067–1077, 2015.
- Bryan Perozzi, Vivek Kulkarni, Haochen Chen, and Steven Skiena. Don’t walk, skip! online learning of multi-scale network embeddings. In Proceedings of the 2017 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining 2017, pages 258–265, 2017.
- Leonardo FR Ribeiro, Pedro HP Saverese, and Daniel R Figueiredo. struc2vec: Learning node representations from structural identity. In Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining, pages 385–394, 2017.
- Yuxiao Dong, Nitesh V Chawla, and Ananthram Swami. metapath2vec: Scalable representation learning for heterogeneous networks. In Proceedings of the 23rd ACM SIGKDD international conference on knowledge discovery and data mining, pages 135–144, 2017.
- Tao-yang Fu, Wang-Chien Lee, and Zhen Lei. Hin2vec: Explore meta-paths in heterogeneous information networks for representation learning. In Proceedings of the 2017 ACM on Conference on Information and Knowledge Management, pages 1797–1806, 2017.
- Yukuo Cen, Xu Zou, Jianwei Zhang, Hongxia Yang, Jingren Zhou, and Jie Tang. Representation learning for attributed multiplex heterogeneous network. In Proceedings of the 25th ACM SIGKDD international conference on knowledge discovery & data mining, pages 1358–1368, 2019.
- Daixin Wang, Peng Cui, and Wenwu Zhu. Structural deep network embedding. In Proceedings of the 22nd ACM SIGKDD international conference on Knowledge discovery and data mining, pages 1225–1234, 2016.
- Shaosheng Cao, Wei Lu, and Qiongkai Xu. Deep neural networks for learning graph representations. In Proceedings of the AAAI conference on artificial intelligence, volume 30, 2016.
- Shiyu Chang, Wei Han, Jiliang Tang, Guo-Jun Qi, Charu C Aggarwal, and Thomas S Huang. Heterogeneous network embedding via deep architectures. In Proceedings of the 21th ACM SIGKDD international conference on knowledge discovery and data mining, pages 119–128, 2015.
- Jiawei Zhang, Congying Xia, Chenwei Zhang, Limeng Cui, Yanjie Fu, and S Yu Philip. Bl-mne: emerging heterogeneous social network embedding through broad learning with aligned autoencoder. In 2017 IEEE International Conference on Data Mining (ICDM), pages 605–614. IEEE, 2017. [CrossRef]
- Cheng Yang, Zhiyuan Liu, Deli Zhao, Maosong Sun, and Edward Y Chang. Network representation learning with rich text information. In IJCAI, volume 2015, pages 2111–2117, 2015.
- Xiao Huang, Jundong Li, and Xia Hu. Label informed attributed network embedding. In Proceedings of the tenth ACM international conference on web search and data mining, pages 731–739, 2017.
- Lizi Liao, Xiangnan He, Hanwang Zhang, and Tat-Seng Chua. Attributed social network embedding. IEEE Transactions on Knowledge and Data Engineering, 30(12):2257–2270, 2018.
- Hongchang Gao and Heng Huang. Deep attributed network embedding. In Twenty-Seventh International Joint Conference on Artificial Intelligence (IJCAI)), 2018.
- Zhen Zhang, Hongxia Yang, Jiajun Bu, Sheng Zhou, Pinggang Yu, Jianwei Zhang, Martin Ester, and Can Wang. Anrl: attributed network representation learning via deep neural networks. In Ijcai, volume 18, pages 3155–3161, 2018. [CrossRef]
- Hitoshi Iuchi, Taro Matsutani, Keisuke Yamada, Natsuki Iwano, Shunsuke Sumi, Shion Hosoda, Shitao Zhao, Tsukasa Fukunaga, and Michiaki Hamada. Representation learning applications in biological sequence analysis. Computational and Structural Biotechnology Journal, 19:3198–3208, 2021. [CrossRef]
- Hai-Cheng Yi, Zhu-Hong You, De-Shuang Huang, and Chee Keong Kwoh. Graph representation learning in bioinformatics: trends, methods and applications. Briefings in Bioinformatics, 23(1):bbab340, 2022. [CrossRef]
- Thomas N Kipf and Max Welling. Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:1609.02907, 2016.
- Chenyi Zhuang and Qiang Ma. Dual graph convolutional networks for graph-based semi-supervised classification. In Proceedings of the 2018 world wide web conference, pages 499–508, 2018. [CrossRef]
- Ruoyu Li, Sheng Wang, Feiyun Zhu, and Junzhou Huang. Adaptive graph convolutional neural networks. In Proceedings of the AAAI conference on artificial intelligence, volume 32, 2018.
- Hongyang Gao, Zhengyang Wang, and Shuiwang Ji. Large-scale learnable graph convolutional networks. In Proceedings of the 24th ACM SIGKDD international conference on knowledge discovery & data mining, pages 1416–1424, 2018.
- Jie Chen, Tengfei Ma, and Cao Xiao. Fastgcn: fast learning with graph convolutional networks via importance sampling. arXiv preprint arXiv:1801.10247, 2018.
- Will Hamilton, Zhitao Ying, and Jure Leskovec. Inductive representation learning on large graphs. Advances in neural information processing systems, 30, 2017.
- Keyulu Xu, Weihua Hu, Jure Leskovec, and Stefanie Jegelka. How powerful are graph neural networks? arXiv preprint arXiv:1810.00826, 2018.
- Johannes Gasteiger, Aleksandar Bojchevski, and Stephan Günnemann. Predict then propagate: Graph neural networks meet personalized pagerank. arXiv preprint arXiv:1810.05997, 2018.
- Petar Velickovic, Guillem Cucurull, Arantxa Casanova, Adriana Romero, Pietro Lio, Yoshua Bengio, et al. Graph attention networks. stat, 1050(20):10–48550, 2017.
- Kiran K Thekumparampil, Chong Wang, Sewoong Oh, and Li-Jia Li. Attention-based graph neural network for semi-supervised learning. arXiv preprint arXiv:1803.03735, 2018.
- Aravind Sankar, Yanhong Wu, Liang Gou, Wei Zhang, and Hao Yang. Dynamic graph representation learning via self-attention networks. arXiv preprint arXiv:1812.09430, 2018.
- Jiani Zhang, Xingjian Shi, Junyuan Xie, Hao Ma, Irwin King, and Dit-Yan Yeung. Gaan: Gated attention networks for learning on large and spatiotemporal graphs. arXiv preprint arXiv:1803.07294, 2018.
- Xiao Wang, Houye Ji, Chuan Shi, Bai Wang, Yanfang Ye, Peng Cui, and Philip S Yu. Heterogeneous graph attention network. In The world wide web conference, pages 2022–2032, 2019. [CrossRef]
- Guangtao Wang, Rex Ying, Jing Huang, and Jure Leskovec. Multi-hop attention graph neural network. arXiv preprint arXiv:2009.14332, 2020. [CrossRef]
- Haiyun Xu, Shaojie Zhang, Bo Jiang, and Jin Tang. Graph context-attention network via low and high order aggregation. Neurocomputing, 2023. [CrossRef]
- Thomas N Kipf and Max Welling. Variational graph auto-encoders. arXiv preprint arXiv:1611.07308, 2016.
- Martin Simonovsky and Nikos Komodakis. Graphvae: Towards generation of small graphs using variational autoencoders. In Artificial Neural Networks and Machine Learning–ICANN 2018: 27th International Conference on Artificial Neural Networks, Rhodes, Greece, October 4-7, 2018, Proceedings, Part I 27, pages 412–422. Springer, 2018.
- Aditya Grover, Aaron Zweig, and Stefano Ermon. Graphite: Iterative generative modeling of graphs. In International conference on machine learning, pages 2434–2444. PMLR, 2019.
- Aleksandar Bojchevski and Stephan Günnemann. Deep gaussian embedding of graphs: Unsupervised inductive learning via ranking. arXiv preprint arXiv:1707.03815, 2017.
- Dingyuan Zhu, Peng Cui, Daixin Wang, and Wenwu Zhu. Deep variational network embedding in wasserstein space. In Proceedings of the 24th ACM SIGKDD international conference on knowledge discovery & data mining, pages 2827–2836, 2018.
- Petar Velickovic, William Fedus, William L Hamilton, Pietro Liò, Yoshua Bengio, and R Devon Hjelm. Deep graph infomax. ICLR (Poster), 2(3):4, 2019.
- Fan-Yun Sun, Jordan Hoffmann, Vikas Verma, and Jian Tang. Infograph: Unsupervised and semi-supervised graph-level representation learning via mutual information maximization. arXiv preprint arXiv:1908.01000, 2019.
- Jintang Li, Ruofan Wu, Wangbin Sun, Liang Chen, Sheng Tian, Liang Zhu, Changhua Meng, Zibin Zheng, and Weiqiang Wang. Maskgae: masked graph modeling meets graph autoencoders. arXiv preprint arXiv:2205.10053, 2022.
- Ian Goodfellow, Jean Pouget-Abadie, Mehdi Mirza, Bing Xu, David Warde-Farley, Sherjil Ozair, Aaron Courville, and Yoshua Bengio. Generative adversarial networks. Communications of the ACM, 63(11):139–144, 2020.
- Hongwei Wang, Jia Wang, Jialin Wang, Miao Zhao, Weinan Zhang, Fuzheng Zhang, Xing Xie, and Minyi Guo. Graphgan: Graph representation learning with generative adversarial nets. In Proceedings of the AAAI conference on artificial intelligence, volume 32, 2018. [CrossRef]
- Shirui Pan, Ruiqi Hu, Sai-fu Fung, Guodong Long, Jing Jiang, and Chengqi Zhang. Learning graph embedding with adversarial training methods. IEEE transactions on cybernetics, 50(6):2475–2487, 2019. [CrossRef]
- Quanyu Dai, Qiang Li, Jian Tang, and Dan Wang. Adversarial network embedding. In Proceedings of the AAAI Conference on Artificial Intelligence, volume 32, 2018.
- Wenchao Yu, Cheng Zheng, Wei Cheng, Charu C Aggarwal, Dongjin Song, Bo Zong, Haifeng Chen, and Wei Wang. Learning deep network representations with adversarially regularized autoencoders. In Proceedings of the 24th ACM SIGKDD international conference on knowledge discovery & data mining, pages 2663–2671, 2018.
- Aleksandar Bojchevski, Oleksandr Shchur, Daniel Zügner, and Stephan Günnemann. Netgan: Generating graphs via random walks. In International conference on machine learning, pages 610–619. PMLR, 2018.
- Nicola De Cao and Thomas Kipf. Molgan: An implicit generative model for small molecular graphs. arXiv preprint arXiv:1805.11973, 2018.
- Zhitao Ying, Jiaxuan You, Christopher Morris, Xiang Ren, Will Hamilton, and Jure Leskovec. Hierarchical graph representation learning with differentiable pooling. Advances in neural information processing systems, 31, 2018.
- Muhan Zhang, Zhicheng Cui, Marion Neumann, and Yixin Chen. An end-to-end deep learning architecture for graph classification. In Proceedings of the AAAI conference on artificial intelligence, volume 32, 2018. [CrossRef]
- Junhyun Lee, Inyeop Lee, and Jaewoo Kang. Self-attention graph pooling. In International conference on machine learning, pages 3734–3743. PMLR, 2019.
- Frederik Diehl, Thomas Brunner, Michael Truong Le, and Alois Knoll. Towards graph pooling by edge contraction. In ICML 2019 workshop on learning and reasoning with graph-structured data, 2019.
- Gabriele Corso, Zhitao Ying, Michal Pándy, Petar Veličković, Jure Leskovec, and Pietro Liò. Neural distance embeddings for biological sequences. Advances in Neural Information Processing Systems, 34:18539–18551, 2021.
- Jun Xia, Lirong Wu, Jintao Chen, Bozhen Hu, and Stan Z Li. Simgrace: A simple framework for graph contrastive learning without data augmentation. In Proceedings of the ACM Web Conference 2022, pages 1070–1079, 2022. [CrossRef]
- Chang Tang, Xiao Zheng, Wei Zhang, Xinwang Liu, Xinzhong Zhu, and En Zhu. Unsupervised feature selection via multiple graph fusion and feature weight learning. Science China Information Sciences, 66(5):1–17, 2023. [CrossRef]
- Jiaqi Sun, Lin Zhang, Guangyi Chen, Kun Zhang, Peng XU, and Yujiu Yang. Feature expansion for graph neural networks. arXiv preprint arXiv:2305.06142, 2023.
- Anh-Tien Ton, Francesco Gentile, Michael Hsing, Fuqiang Ban, and Artem Cherkasov. Rapid identification of potential inhibitors of sars-cov-2 main protease by deep docking of 1.3 billion compounds. Molecular informatics, 39(8):2000028, 2020.
- Konda Mani Saravanan, Haiping Zhang, Md Tofazzal Hossain, Md Selim Reza, and Yanjie Wei. Deep learning-based drug screening for covid-19 and case studies. In Silico Modeling of Drugs Against Coronaviruses: Computational Tools and Protocols, pages 631–660, 2021.
- Deshan Zhou, Shaoliang Peng, Dong-Qing Wei, Wu Zhong, Yutao Dou, and Xiaolan Xie. Lunar: drug screening for novel coronavirus based on representation learning graph convolutional network. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 18(4):1290–1298, 2021. [CrossRef]
- Zhengyang Wang, Meng Liu, Youzhi Luo, Zhao Xu, Yaochen Xie, Limei Wang, Lei Cai, Qi Qi, Zhuoning Yuan, Tianbao Yang, et al. Advanced graph and sequence neural networks for molecular property prediction and drug discovery. Bioinformatics, 38(9):2579–2586, 2022. [CrossRef]
- Xiao-Shuang Li, Xiang Liu, Le Lu, Xian-Sheng Hua, Ying Chi, and Kelin Xia. Multiphysical graph neural network (mp-gnn) for covid-19 drug design. Briefings in Bioinformatics, 23(4), 2022. [CrossRef]
- Jiangsheng Pi, Peishun Jiao, Yang Zhang, and Junyi Li. Mdgnn: Microbial drug prediction based on heterogeneous multi-attention graph neural network. Frontiers in Microbiology, 13, 2022. [CrossRef]
- Yiyue Ge, Tingzhong Tian, Suling Huang, Fangping Wan, Jingxin Li, Shuya Li, Hui Yang, Lixiang Hong, Nian Wu, Enming Yuan, et al. A data-driven drug repositioning framework discovered a potential therapeutic agent targeting covid-19. BioRxiv, pages 2020–03, 2020.
- Raghvendra Mall, Abdurrahman Elbasir, Hossam Al Meer, Sanjay Chawla, and Ehsan Ullah. Data-driven drug repurposing for covid-19. 2020.
- Seyed Aghil Hooshmand, Mohadeseh Zarei Ghobadi, Seyyed Emad Hooshmand, Sadegh Azimzadeh Jamalkandi, Seyed Mehdi Alavi, and Ali Masoudi-Nejad. A multimodal deep learning-based drug repurposing approach for treatment of covid-19. Molecular diversity, 25:1717–1730, 2021. [CrossRef]
- Rosa Aghdam, Mahnaz Habibi, and Golnaz Taheri. Using informative features in machine learning based method for covid-19 drug repurposing. Journal of cheminformatics, 13:1–14, 2021. [CrossRef]
- Kanglin Hsieh, Yinyin Wang, Luyao Chen, Zhongming Zhao, Sean Savitz, Xiaoqian Jiang, Jing Tang, and Yejin Kim. Drug repurposing for covid-19 using graph neural network with genetic, mechanistic, and epidemiological validation. Research Square, 2020.
- Thai-Hoang Pham, Yue Qiu, Jucheng Zeng, Lei Xie, and Ping Zhang. A deep learning framework for high-throughput mechanism-driven phenotype compound screening and its application to covid-19 drug repurposing. Nature machine intelligence, 3(3):247–257, 2021. [CrossRef]
- Kanglin Hsieh, Yinyin Wang, Luyao Chen, Zhongming Zhao, Sean Savitz, Xiaoqian Jiang, Jing Tang, and Yejin Kim. Drug repurposing for covid-19 using graph neural network and harmonizing multiple evidence. Scientific reports, 11(1):23179, 2021. [CrossRef]
- Siddhant Doshi and Sundeep Prabhakar Chepuri. A computational approach to drug repurposing using graph neural networks. Computers in Biology and Medicine, 150:105992, 2022. [CrossRef]
- Bo Ram Beck, Bonggun Shin, Yoonjung Choi, Sungsoo Park, and Keunsoo Kang. Predicting commercially available antiviral drugs that may act on the novel coronavirus (sars-cov-2) through a drug-target interaction deep learning model. Computational and structural biotechnology journal, 18:784–790, 2020.
- Sovan Saha, Piyali Chatterjee, Anup Kumar Halder, Mita Nasipuri, Subhadip Basu, and Dariusz Plewczynski. Ml-dtd: Machine learning-based drug target discovery for the potential treatment of covid-19. Vaccines, 10(10):1643, 2022. [CrossRef]
- Xianfang Wang, Qimeng Li, Yifeng Liu, Zhiyong Du, and Ruixia Jin. Drug repositioning of covid-19 based on mixed graph network and ion channel. Mathematical Biosciences and Engineering, 19(4):3269–3284, 2022. [CrossRef]
- Peiliang Zhang, Ziqi Wei, Chao Che, and Bo Jin. Deepmgt-dti: Transformer network incorporating multilayer graph information for drug–target interaction prediction. Computers in biology and medicine, 142:105214, 2022.
- Guodong Li, Weicheng Sun, Jinsheng Xu, Lun Hu, Weihan Zhang, and Ping Zhang. Ga-ens: A novel drug–target interactions prediction method by incorporating prior knowledge graph into dual wasserstein generative adversarial network with gradient penalty. Applied Soft Computing, 139:110151, 2023. [CrossRef]
- Zhenchao Tang, Guanxing Chen, Hualin Yang, Weihe Zhong, and Calvin Yu-Chian Chen. Dsil-ddi: A domain-invariant substructure interaction learning for generalizable drug–drug interaction prediction. IEEE Transactions on Neural Networks and Learning Systems, 2023. [CrossRef]
- Sarina Sefidgarhoseini, Leila Safari, and Zanyar Mohammady. Drug-drug interaction extraction using transformer-based ensemble model. 2023.
- Zhong-Hao Ren, Zhu-Hong You, Chang-Qing Yu, Li-Ping Li, Yong-Jian Guan, Lu-Xiang Guo, and Jie Pan. A biomedical knowledge graph-based method for drug–drug interactions prediction through combining local and global features with deep neural networks. Briefings in Bioinformatics, 23(5), 2022. [CrossRef]
- Yujie Chen, Tengfei Ma, Xixi Yang, Jianmin Wang, Bosheng Song, and Xiangxiang Zeng. Muffin: multi-scale feature fusion for drug–drug interaction prediction. Bioinformatics, 37(17):2651–2658, 2021. [CrossRef]
- Deng Pan, Lijun Quan, Zhi Jin, Taoning Chen, Xuejiao Wang, Jingxin Xie, Tingfang Wu, and Qiang Lyu. Multisource attention-mechanism-based encoder–decoder model for predicting drug–drug interaction events. Journal of Chemical Information and Modeling, 62(23):6258–6270, 2022. [CrossRef]
- Zimeng Li, Shichao Zhu, Bin Shao, Tie-Yan Liu, Xiangxiang Zeng, and Tong Wang. Multi-view substructure learning for drug-drug interaction prediction. arXiv preprint arXiv:2203.14513, 2022.
- Mei Ma and Xiujuan Lei. A dual graph neural network for drug–drug interactions prediction based on molecular structure and interactions. PLOS Computational Biology, 19(1):e1010812, 2023. [CrossRef]
- Lopamudra Dey, Sanjay Chakraborty, and Anirban Mukhopadhyay. Machine learning techniques for sequence-based prediction of viral–host interactions between sars-cov-2 and human proteins. Biomedical journal, 43(5):438–450, 2020.
- Hangyu Du, Feng Chen, Hongfu Liu, and Pengyu Hong. Network-based virus-host interaction prediction with application to sars-cov-2. Patterns, 2(5):100242, 2021.
- Hongpeng Yang, Yijie Ding, Jijun Tang, and Fei Guo. Inferring human microbe–drug associations via multiple kernel fusion on graph neural network. Knowledge-Based Systems, 238:107888, 2022. [CrossRef]
- Bihter Das, Mucahit Kutsal, and Resul Das. A geometric deep learning model for display and prediction of potential drug-virus interactions against sars-cov-2. Chemometrics and Intelligent Laboratory Systems, 229:104640, 2022.
- Farah Shahid, Aneela Zameer, and Muhammad Muneeb. Predictions for covid-19 with deep learning models of lstm, gru and bi-lstm. Chaos, Solitons & Fractals, 140:110212, 2020. [CrossRef]
- Hossein Abbasimehr and Reza Paki. Prediction of covid-19 confirmed cases combining deep learning methods and bayesian optimization. Chaos, Solitons & Fractals, 142:110511, 2021. [CrossRef]
- Trisha Sinha, Titash Chowdhury, Rabindra Nath Shaw, and Ankush Ghosh. Analysis and prediction of covid-19 confirmed cases using deep learning models: a comparative study. In Advanced Computing and Intelligent Technologies: Proceedings of ICACIT 2021, pages 207–218. Springer, 2022. [CrossRef]
- Junyi Gao, Rakshith Sharma, Cheng Qian, Lucas M Glass, Jeffrey Spaeder, Justin Romberg, Jimeng Sun, and Cao Xiao. Stan: spatio-temporal attention network for pandemic prediction using real-world evidence. Journal of the American Medical Informatics Association, 28(4):733–743, 2021. [CrossRef]
- Myrsini Ntemi, Ioannis Sarridis, and Constantine Kotropoulos. An autoregressive graph convolutional long short-term memory hybrid neural network for accurate prediction of covid-19 cases. IEEE Transactions on Computational Social Systems, 2022. [CrossRef]
- Dong Li, Xiaofei Ren, and Yunze Su. Predicting covid-19 using lioness optimization algorithm and graph convolution network. Soft Computing, pages 1–65, 2023. [CrossRef]
- Konstantinos Skianis, Giannis Nikolentzos, Benoit Gallix, Rodolphe Thiebaut, and Georgios Exarchakis. Predicting covid-19 positivity and hospitalization with multi-scale graph neural networks. Scientific Reports, 13(1):5235, 2023. [CrossRef]
- Zohair Malki, El-Sayed Atlam, Ashraf Ewis, Guesh Dagnew, Osama A Ghoneim, Abdallah A Mohamed, Mohamed M Abdel-Daim, and Ibrahim Gad. The covid-19 pandemic: prediction study based on machine learning models. Environmental science and pollution research, 28:40496–40506, 2021. [CrossRef]
- Dixizi Liu, Weiping Ding, Zhijie Sasha Dong, and Witold Pedrycz. Optimizing deep neural networks to predict the effect of social distancing on covid-19 spread. Computers & Industrial Engineering, 166:107970, 2022. [CrossRef]
- Devante Ayris, Maleeha Imtiaz, Kye Horbury, Blake Williams, Mitchell Blackney, Celine Shi Hui See, and Syed Afaq Ali Shah. Novel deep learning approach to model and predict the spread of covid-19. Intelligent Systems with Applications, 14:200068, 2022. [CrossRef]
- Valerio La Gatta, Vincenzo Moscato, Marco Postiglione, and Giancarlo Sperli. An epidemiological neural network exploiting dynamic graph structured data applied to the covid-19 outbreak. IEEE Transactions on Big Data, 7(1):45–55, 2020. [CrossRef]
- Truong Son Hy, Viet Bach Nguyen, Long Tran-Thanh, and Risi Kondor. Temporal multiresolution graph neural networks for epidemic prediction. In Workshop on Healthcare AI and COVID-19, pages 21–32. PMLR, 2022.
- Ru Geng, Yixian Gao, Hongkun Zhang, and Jian Zu. Analysis of the spatio-temporal dynamics of covid-19 in massachusetts via spectral graph wavelet theory. IEEE Transactions on Signal and Information Processing over Networks, 8:670–683, 2022. [CrossRef]
- Baoling Shan, Xin Yuan, Wei Ni, Xin Wang, and Ren Ping Liu. Novel graph topology learning for spatio-temporal analysis of covid-19 spread. IEEE Journal of Biomedical and Health Informatics, 2023. [CrossRef]
- Jose Luis Izquierdo, Julio Ancochea, Savana COVID-19 Research Group, and Joan B Soriano. Clinical characteristics and prognostic factors for intensive care unit admission of patients with covid-19: retrospective study using machine learning and natural language processing. Journal of medical Internet research, 22(10):e21801, 2020. [CrossRef]
- Isotta Landi, Benjamin S Glicksberg, Hao-Chih Lee, Sarah Cherng, Giulia Landi, Matteo Danieletto, Joel T Dudley, Cesare Furlanello, and Riccardo Miotto. Deep representation learning of electronic health records to unlock patient stratification at scale. NPJ digital medicine, 3(1):96, 2020. [CrossRef]
- Tyler Wagner, FNU Shweta, Karthik Murugadoss, Samir Awasthi, AJ Venkatakrishnan, Sairam Bade, Arjun Puranik, Martin Kang, Brian W Pickering, John C O’Horo, et al. Augmented curation of clinical notes from a massive ehr system reveals symptoms of impending covid-19 diagnosis. Elife, 9:e58227, 2020.
- Tingyi Wanyan, Hossein Honarvar, Suraj K Jaladanki, Chengxi Zang, Nidhi Naik, Sulaiman Somani, Jessica K De Freitas, Ishan Paranjpe, Akhil Vaid, Jing Zhang, et al. Contrastive learning improves critical event prediction in covid-19 patients. Patterns, 2(12):100389, 2021. [CrossRef]
- Tingyi Wanyan, Mingquan Lin, Eyal Klang, Kartikeya M Menon, Faris F Gulamali, Ariful Azad, Yiye Zhang, Ying Ding, Zhangyang Wang, Fei Wang, et al. Supervised pretraining through contrastive categorical positive samplings to improve covid-19 mortality prediction. In Proceedings of the 13th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, pages 1–9, 2022. [CrossRef]
- Liantao Ma, Xinyu Ma, Junyi Gao, Xianfeng Jiao, Zhihao Yu, Chaohe Zhang, Wenjie Ruan, Yasha Wang, Wen Tang, and Jiangtao Wang. Distilling knowledge from publicly available online emr data to emerging epidemic for prognosis. In Proceedings of the Web Conference 2021, pages 3558–3568, 2021. [CrossRef]
- Tingyi Wanyan, Akhil Vaid, Jessica K De Freitas, Sulaiman Somani, Riccardo Miotto, Girish N Nadkarni, Ariful Azad, Ying Ding, and Benjamin S Glicksberg. Relational learning improves prediction of mortality in covid-19 in the intensive care unit. IEEE transactions on big data, 7(1):38–44, 2020. [CrossRef]
- Doudou Zhou, Ziming Gan, Xu Shi, Alina Patwari, Everett Rush, Clara-Lea Bonzel, Vidul A Panickan, Chuan Hong, Yuk-Lam Ho, Tianrun Cai, et al. Multiview incomplete knowledge graph integration with application to cross-institutional ehr data harmonization. Journal of Biomedical Informatics, 133:104147, 2022. [CrossRef]
- Junyi Gao, Chaoqi Yang, Joerg Heintz, Scott Barrows, Elise Albers, Mary Stapel, Sara Warfield, Adam Cross, Jimeng Sun, et al. Medml: Fusing medical knowledge and machine learning models for early pediatric covid-19 hospitalization and severity prediction. Iscience, 25(9):104970, 2022. [CrossRef]
- Kaize Ding, Zhe Xu, Hanghang Tong, and Huan Liu. Data augmentation for deep graph learning: A survey. ACM SIGKDD Explorations Newsletter, 24(2):61–77, 2022. [CrossRef]
- Travers Ching, Daniel S Himmelstein, Brett K Beaulieu-Jones, Alexandr A Kalinin, Brian T Do, Gregory P Way, Enrico Ferrero, Paul-Michael Agapow, Michael Zietz, Michael M Hoffman, et al. Opportunities and obstacles for deep learning in biology and medicine. Journal of The Royal Society Interface, 15(141):20170387, 2018.
- Riccardo Miotto, Fei Wang, Shuang Wang, Xiaoqian Jiang, and Joel T Dudley. Deep learning for healthcare: review, opportunities and challenges. Briefings in bioinformatics, 19(6):1236–1246, 2018. [CrossRef]
- Guido Zampieri, Supreeta Vijayakumar, Elisabeth Yaneske, and Claudio Angione. Machine and deep learning meet genome-scale metabolic modeling. PLoS computational biology, 15(7):e1007084, 2019. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





