Submitted:
28 July 2023
Posted:
31 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Identification and Characterization of the SrbZIPs
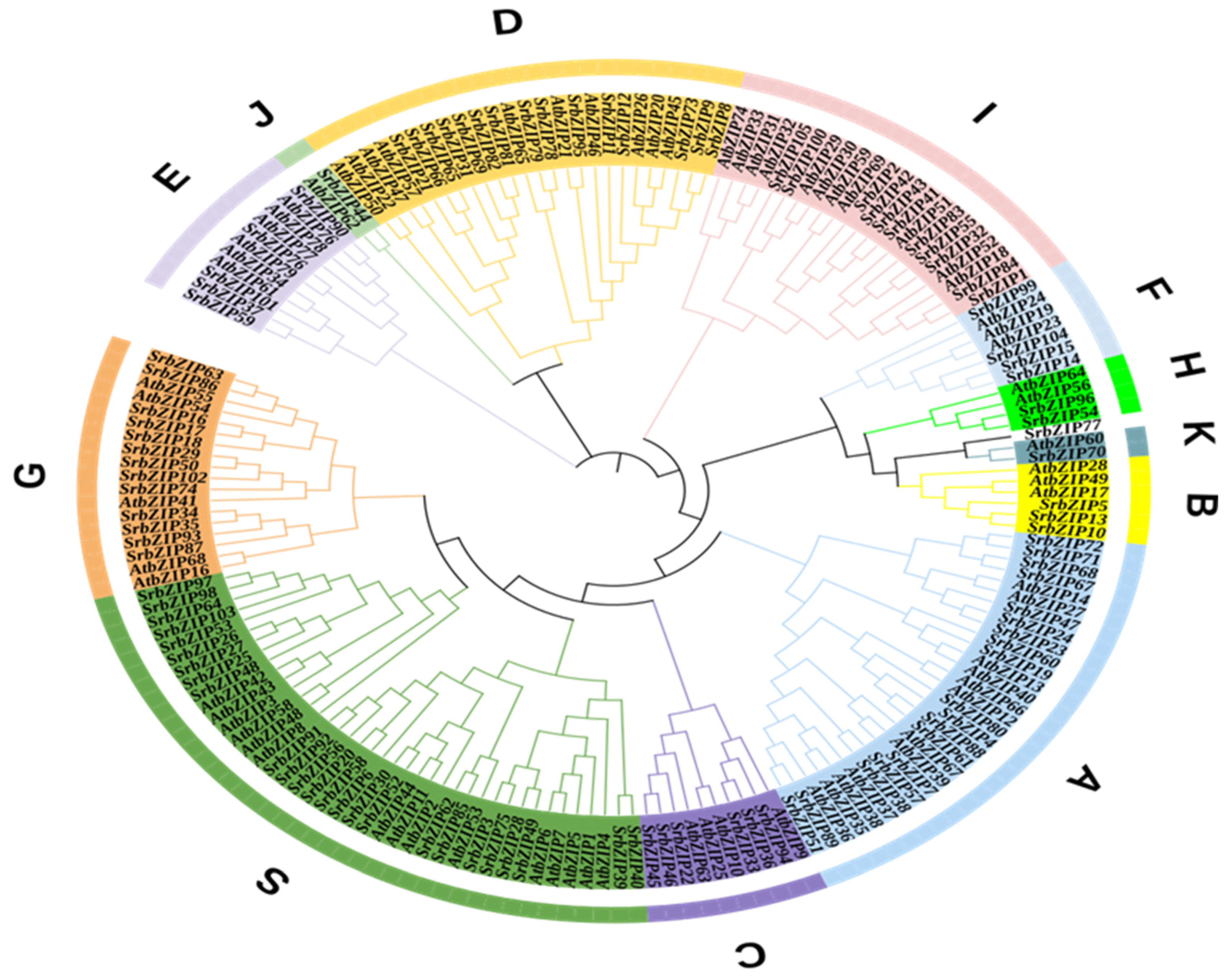
2.2. Classfication of SrbZIP Genes Based on Phylogram
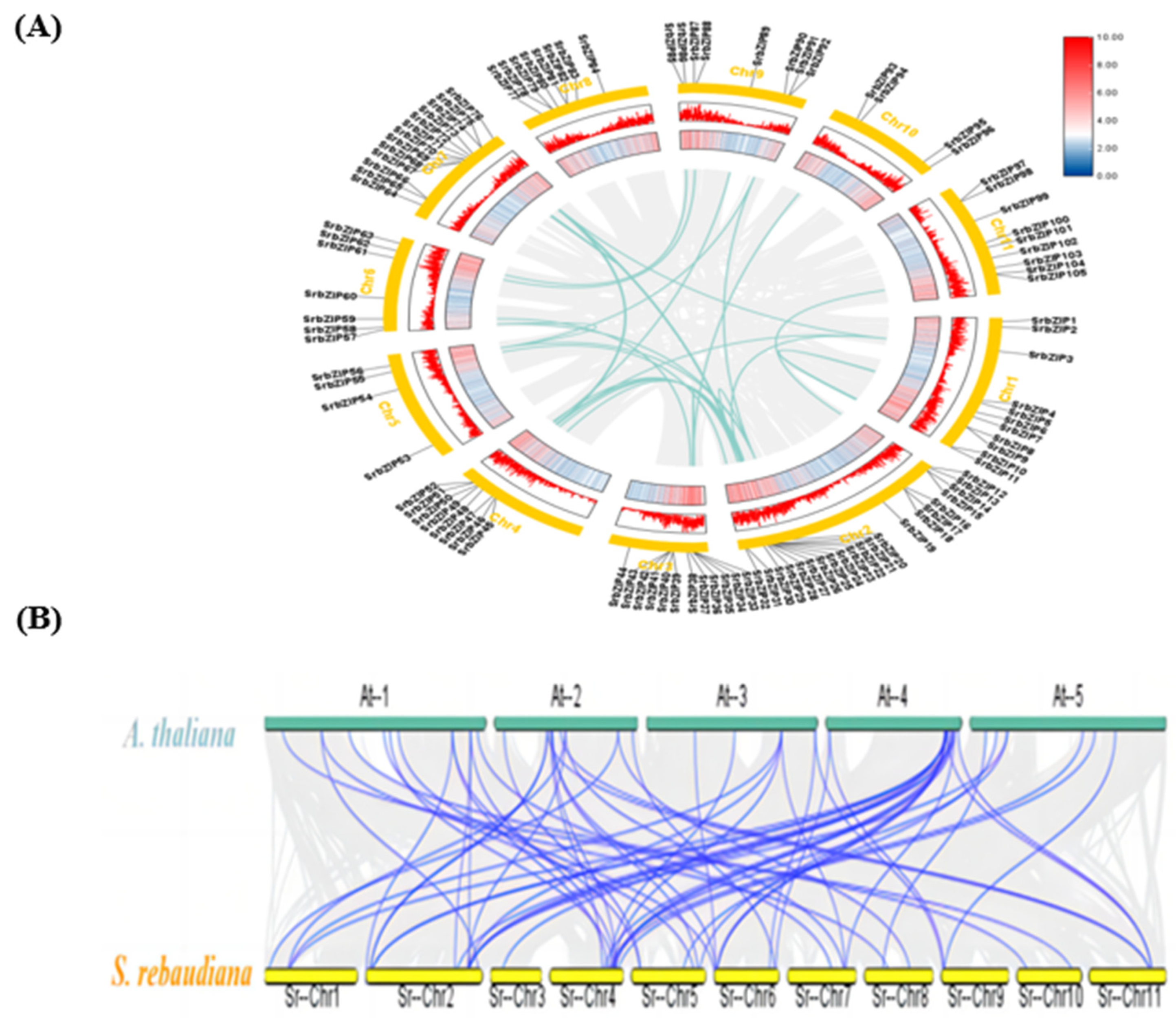
2.3. Chromosomal Location and Collinearity Analysis of SrbZIP Members
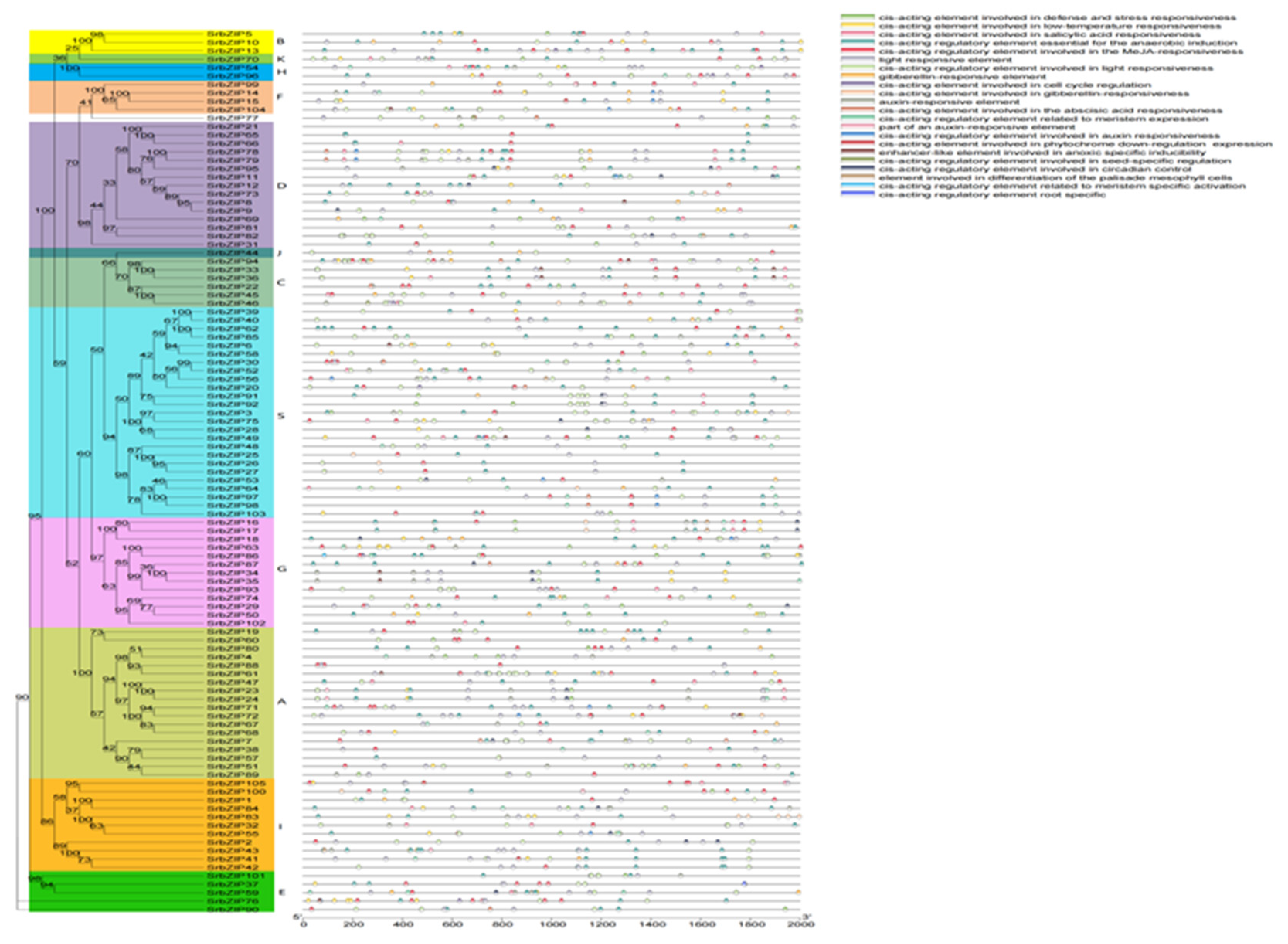
2.4. Gene Structure and Conserved Motif Analysis of SrbZIPs
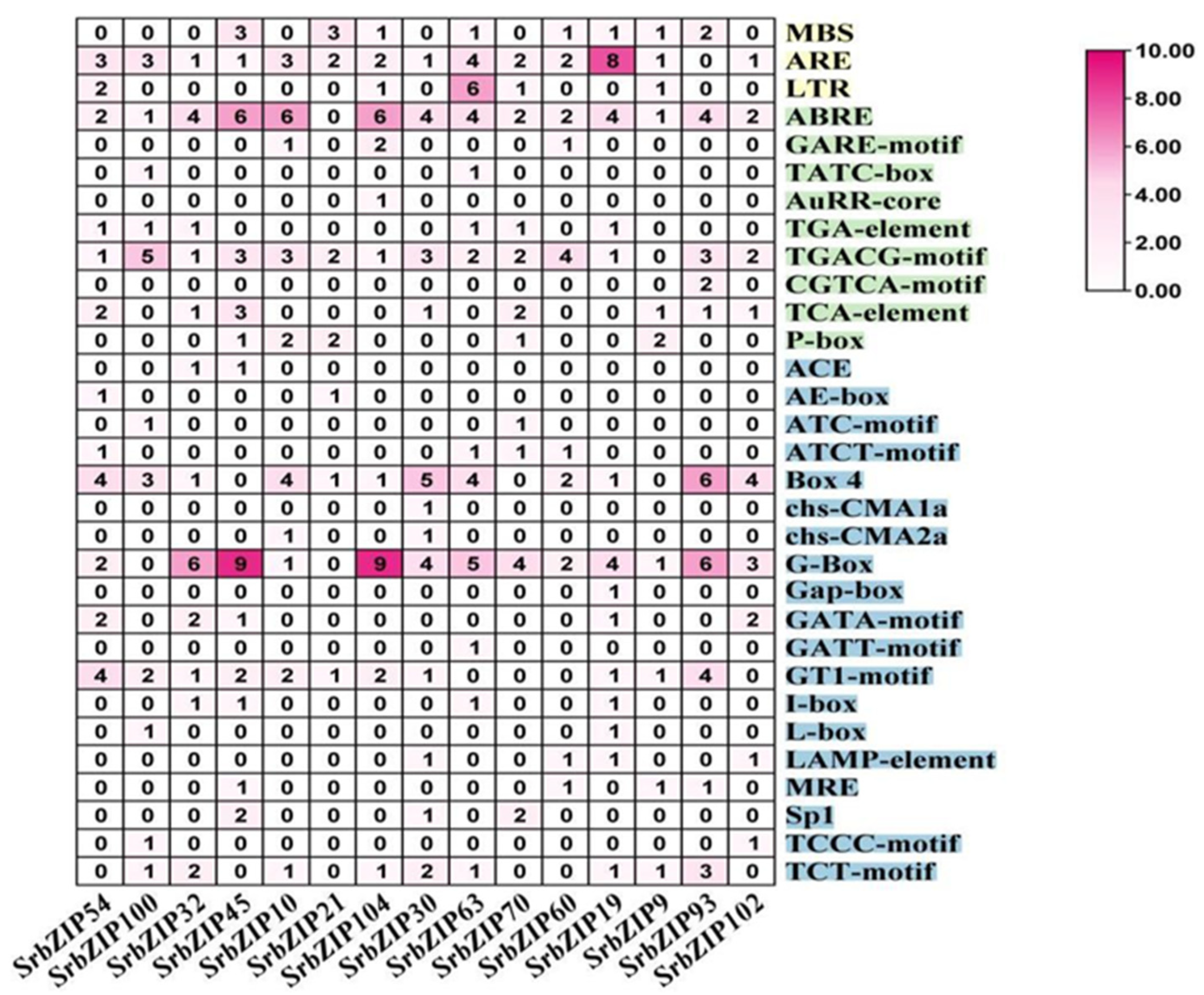
2.5. Cis-elements Analysis in SrbZIPs Promoter Regions
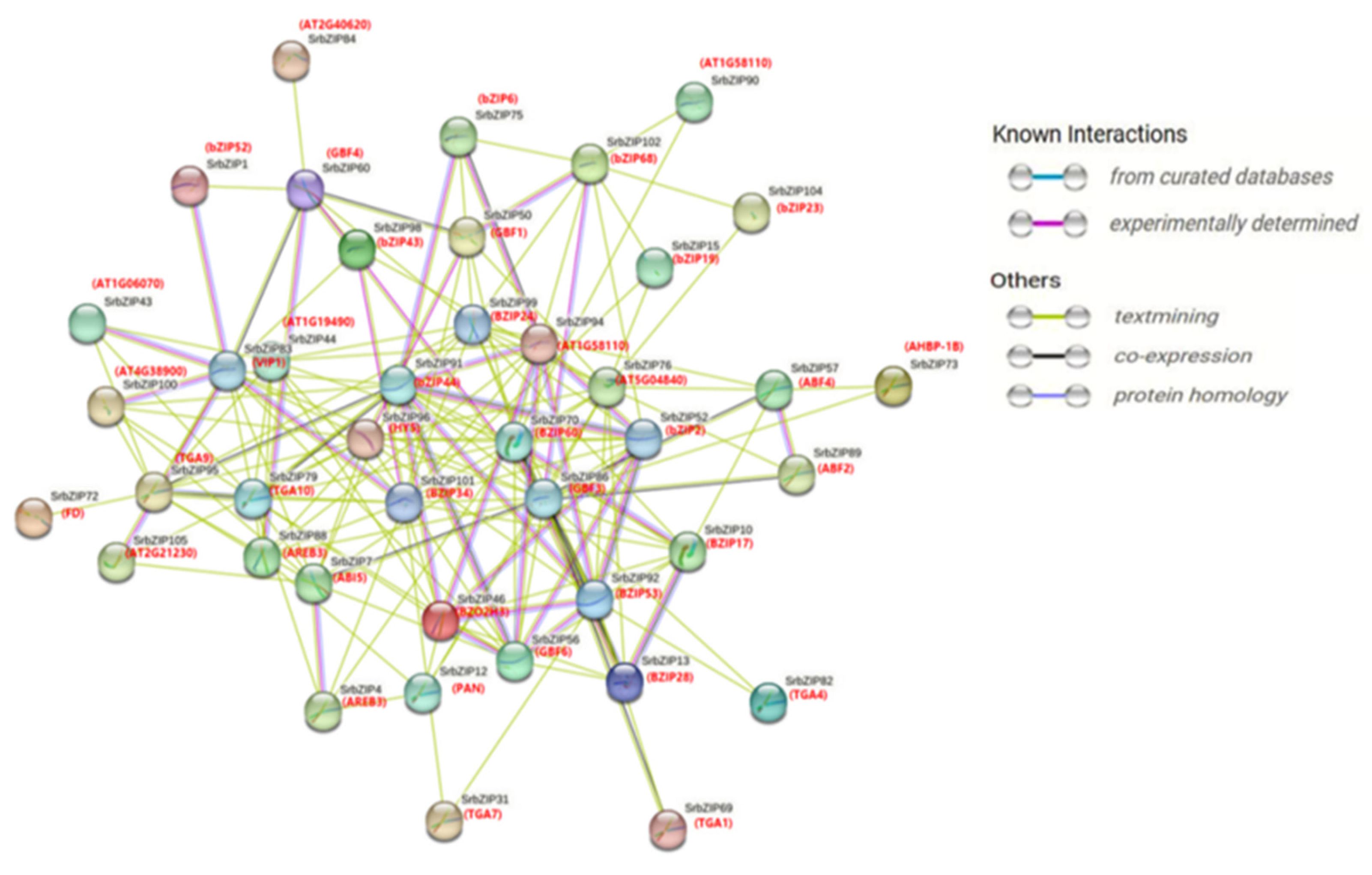
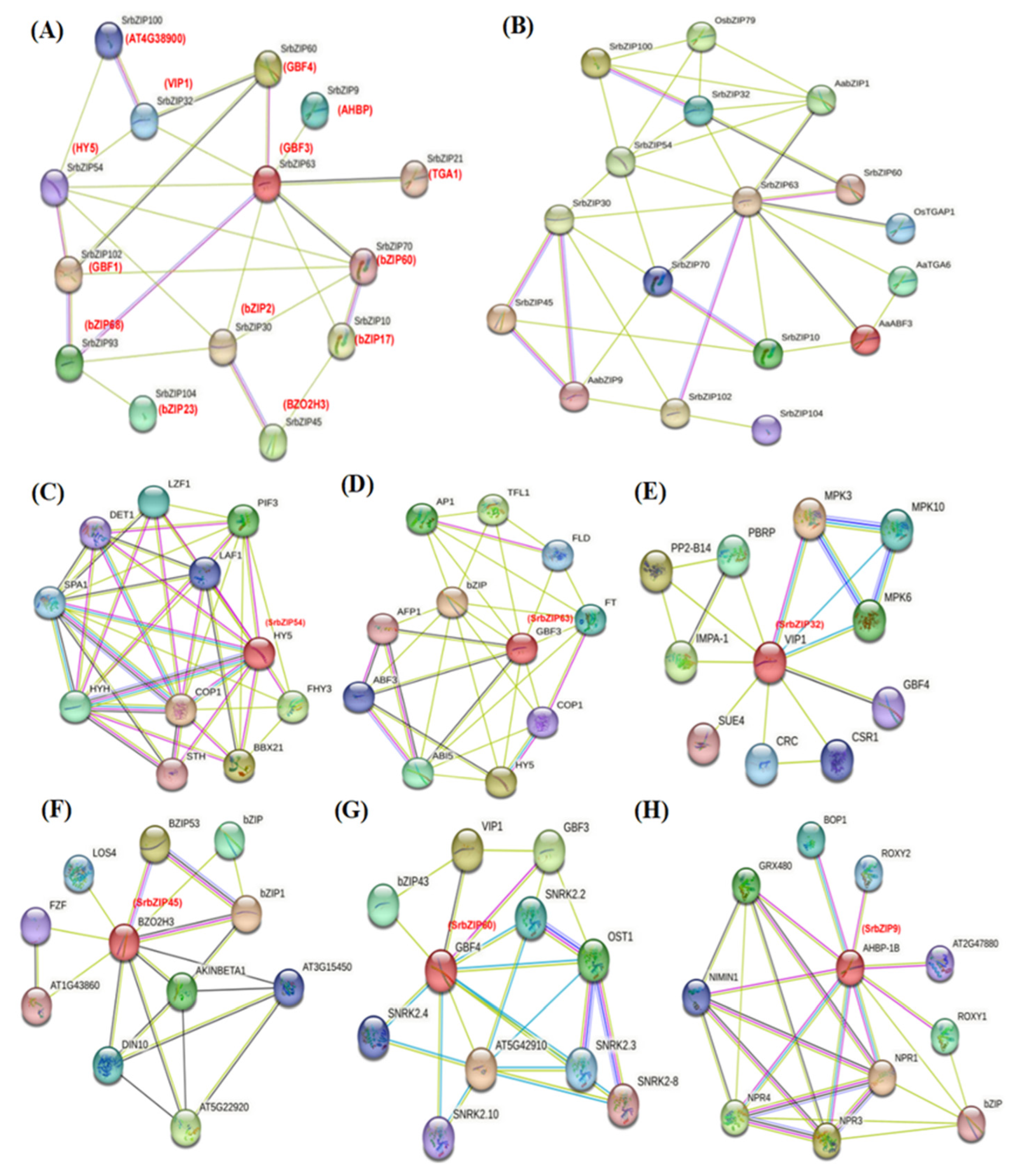
2.6. Protein Interaction Analysis Network of SrbZIP Proteins
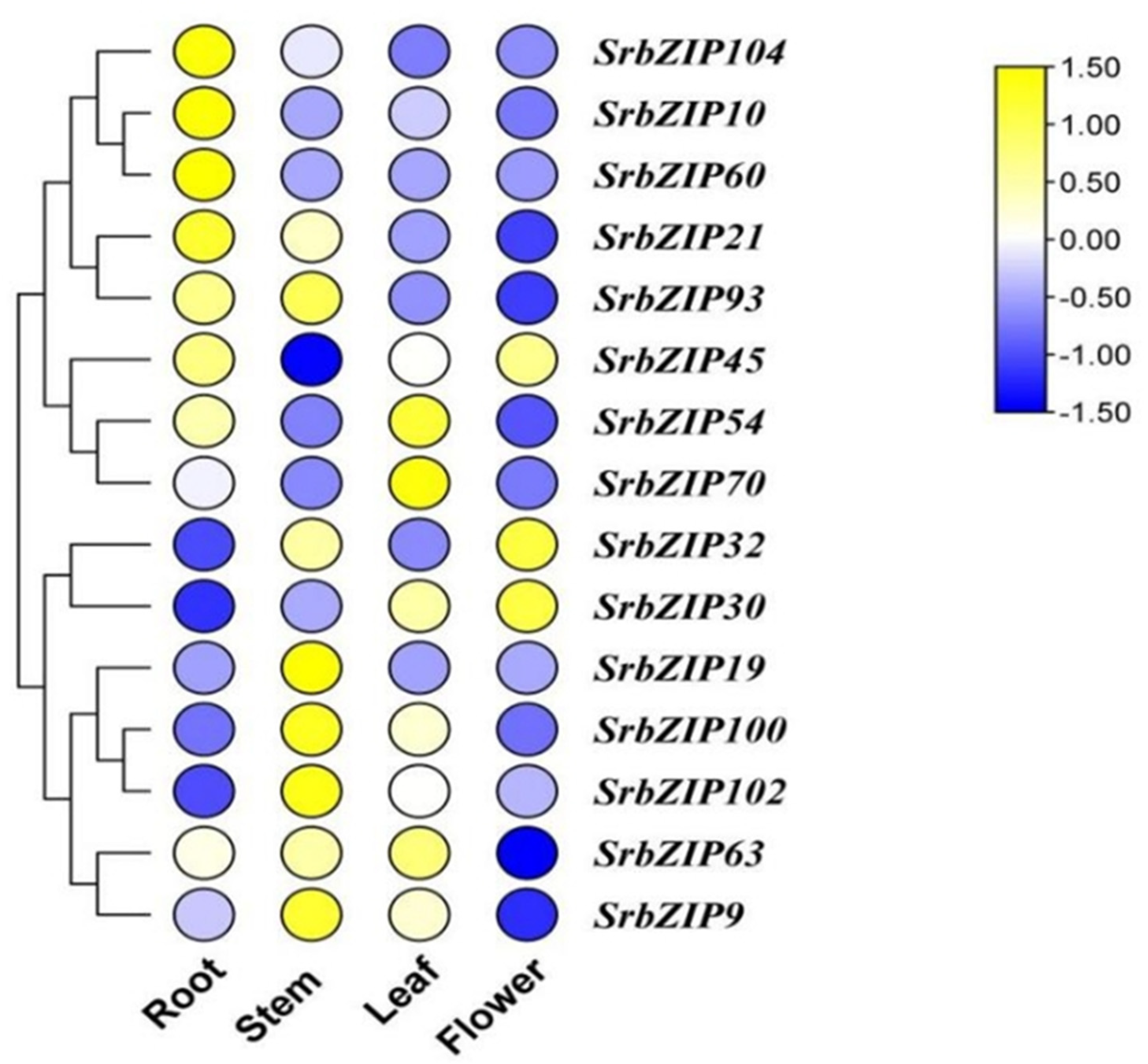
2.7. Expression Pattern and qRT-PCR Validation of SrbZIP Genes
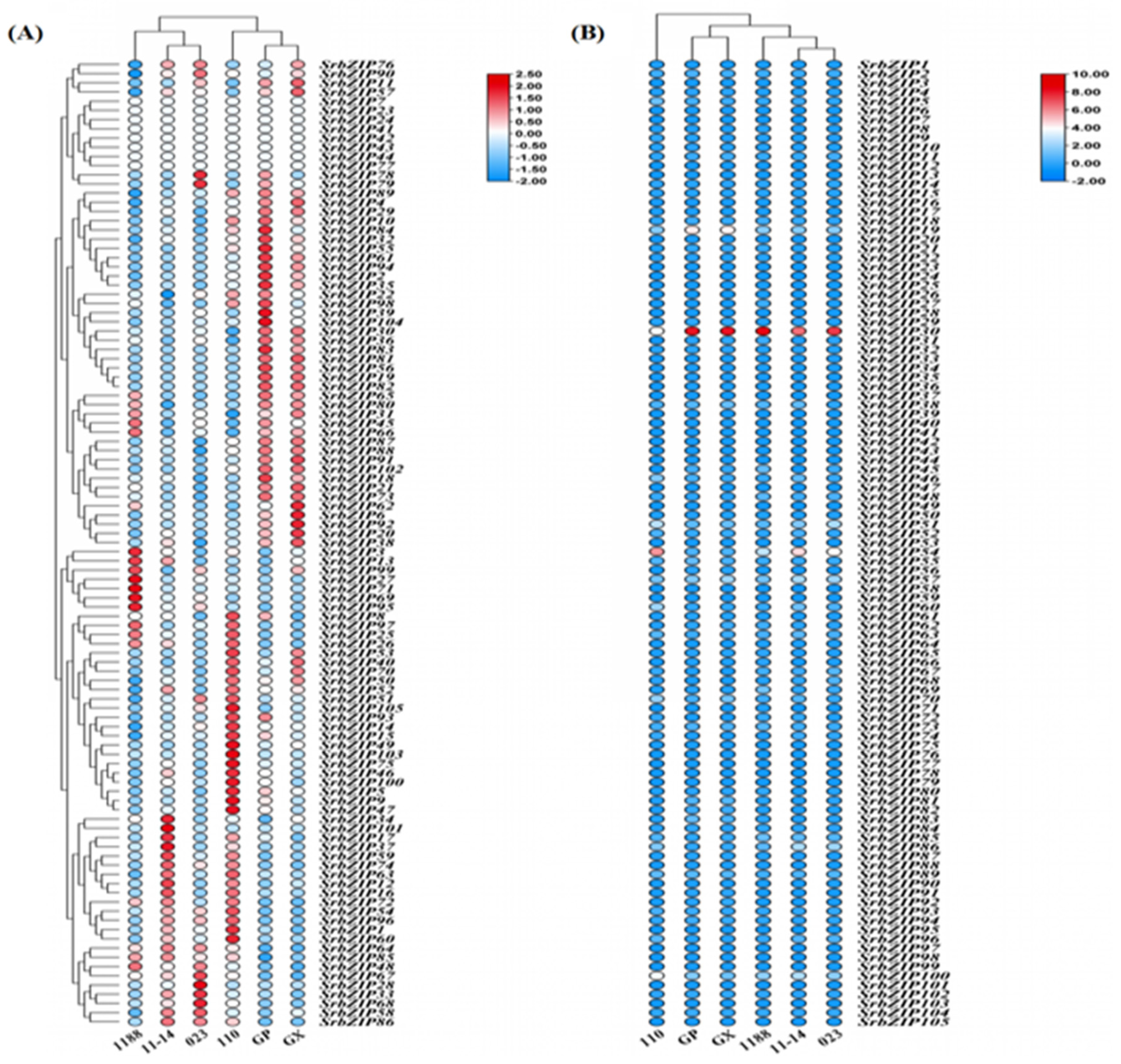
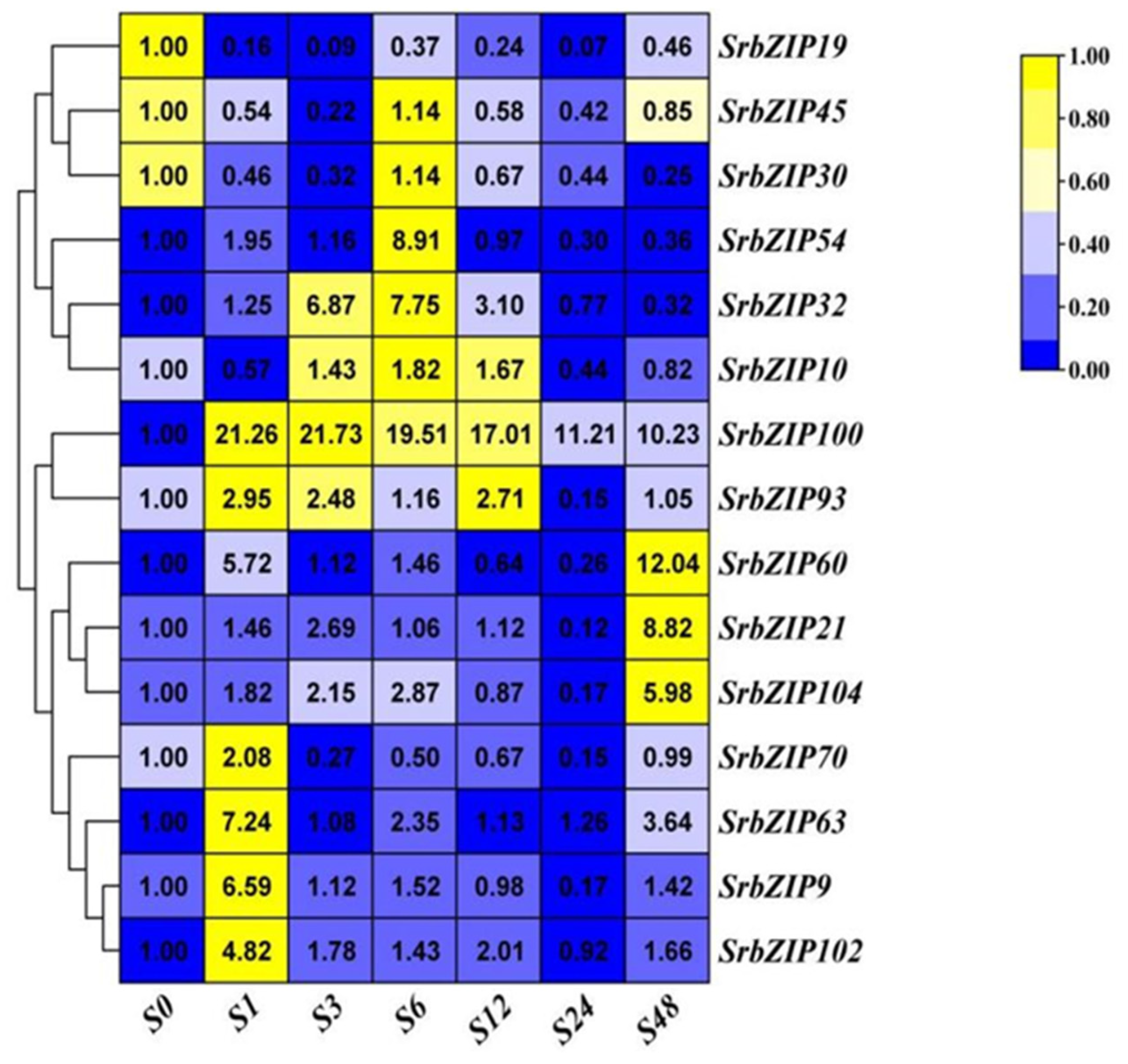
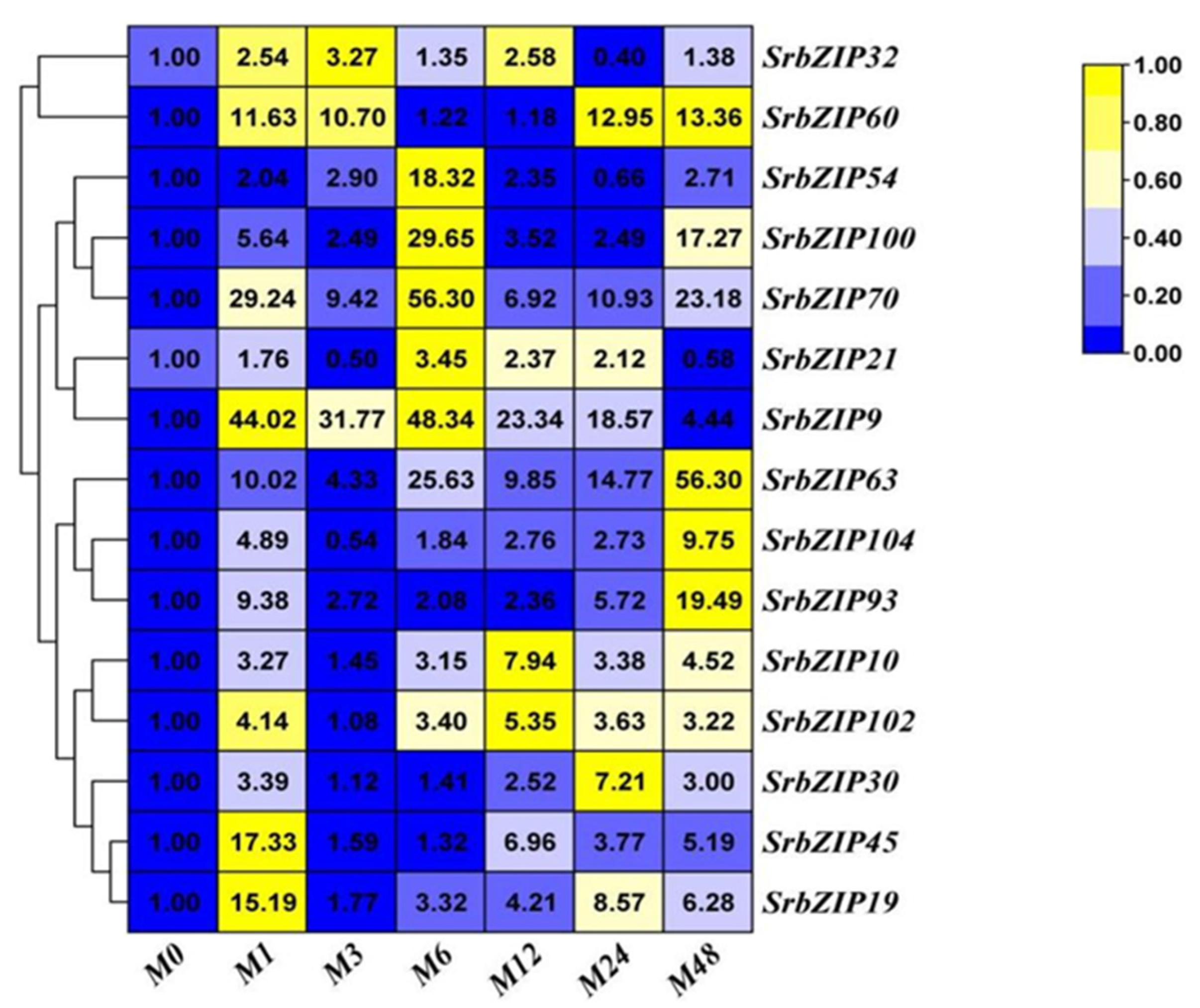
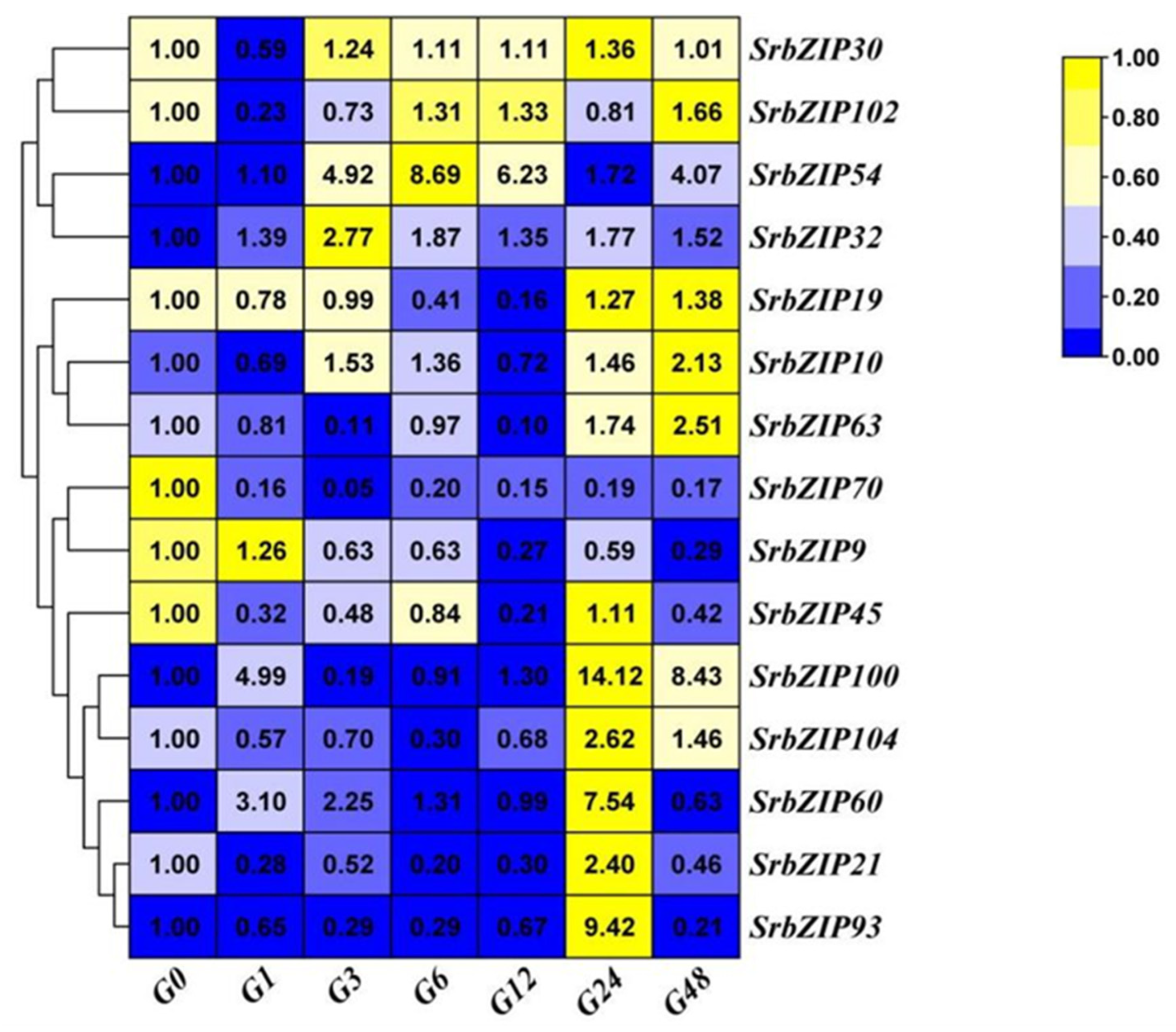
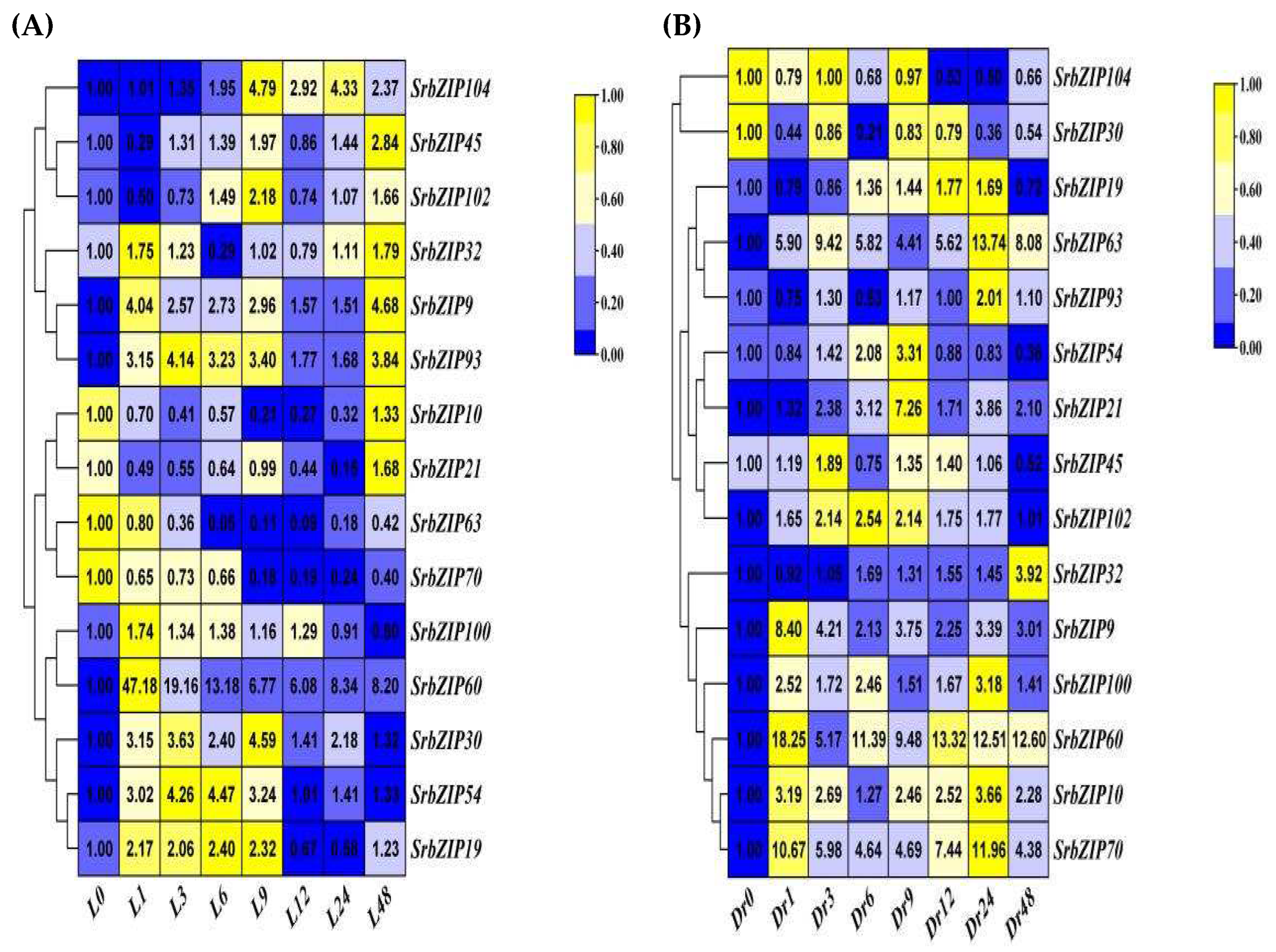
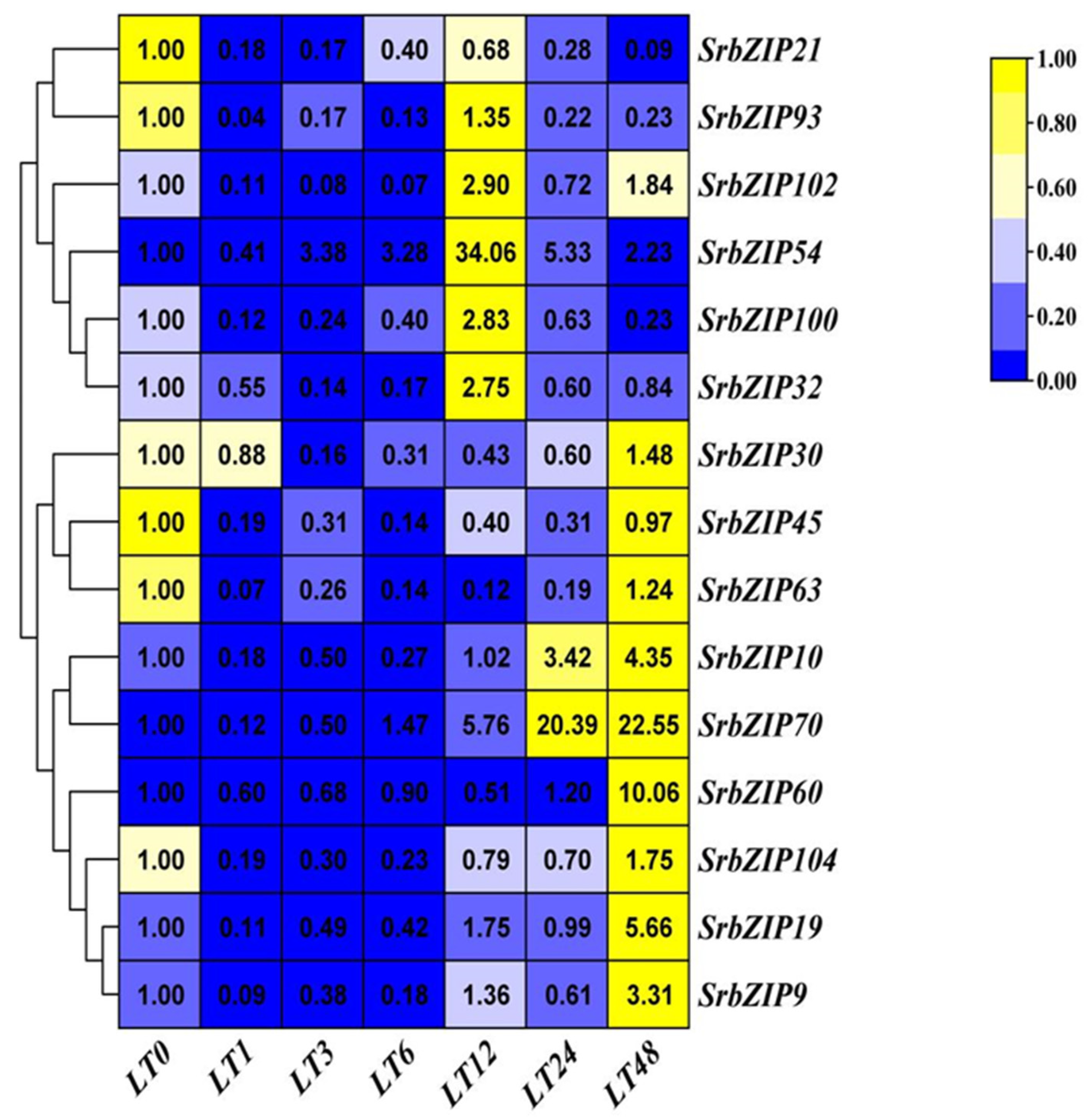
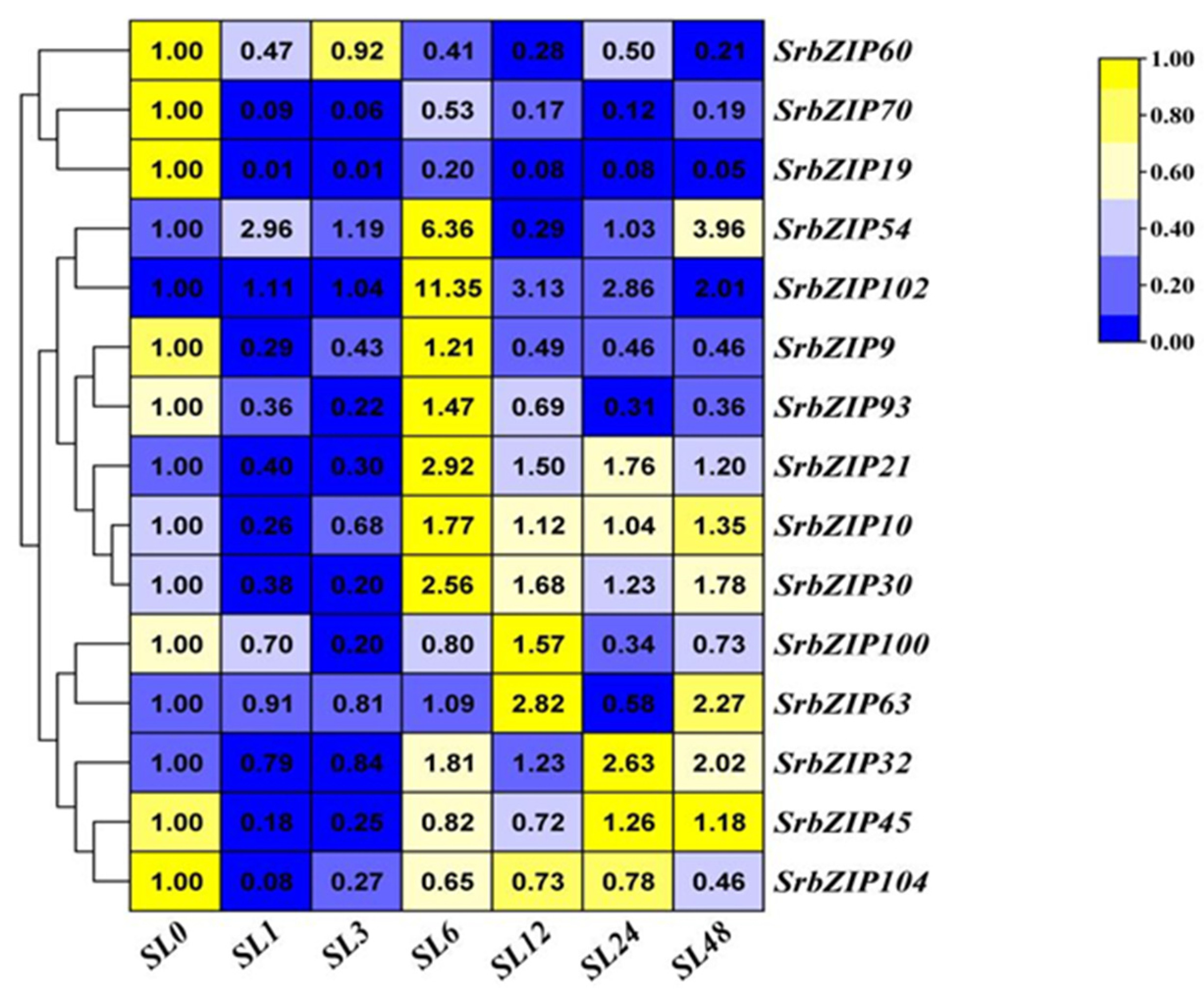
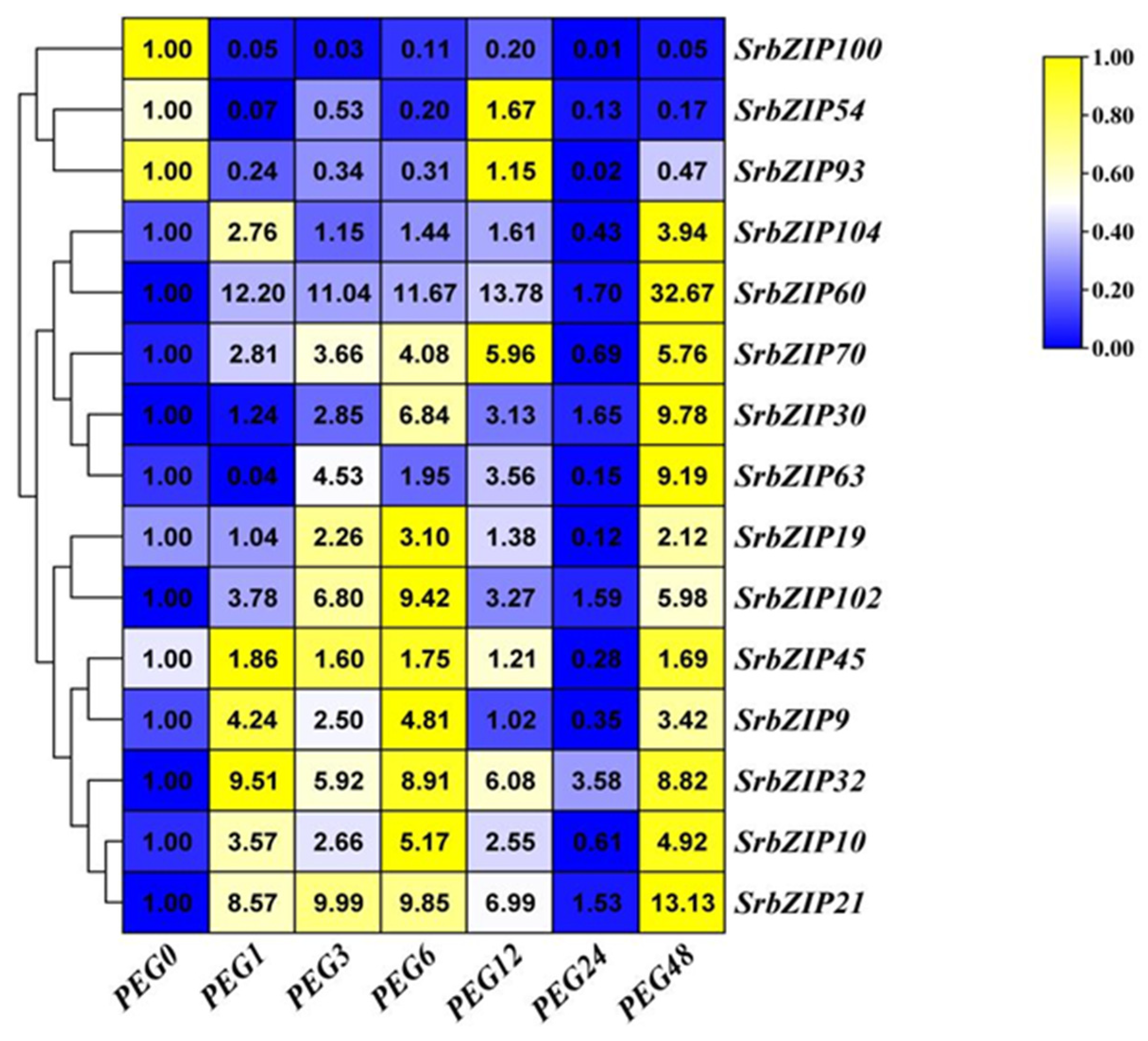
2.8. Expression Pattern of the SrbZIP Genes in Response to Phytohormones and Abiotic Stresses.
2.9. Analysis of Terpenoid Synthesis-Related SrbZIP Genes Which Responsed to Light-treatment, Phytohormones Treatment and Abiotic Stresses
3. Discussion
3.1. Classification and Gene Duplication of SrbZIPs
3.2. Structure Characteristic and Function Prediction of SrbZIPs
3.3. Expression Patterns of SrbZIP Genes and Light, Phytohormone and Abiotic Stress Response
4. Materials and methods
4.1. Plant Materials
4.2. Data Sources
4.3. Identification of S. rebaudiana bZIP Gene Family
4.4. Stevia rebaudiana bZIP Chromosomal Location and Collinearity Analysis
4.5. Phylogenetic Analysis of SrbZIP Genes
4.6. Gene Structure, Conserved Motif, and Cis-acting Element Analysis
4.7. Protein–Protein Interaction Analysis Network of SrbZIPs
4.8. Stevia rebaudiana bZIP Expression Pattern Analysis
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family-an update. Curr Opin Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021, 21, 122–138. [Google Scholar] [CrossRef]
- Jiang, M.; Wang, Z.; Ren, W.; Yan, S.; Xing, N.; Zhang, Z.; Li, H.; Ma, W. Identification of the bZIP gene family and regulation of metabolites under salt stress in isatis indigotica. Front Plant Sci. 2022, 13, 1011616. [Google Scholar] [CrossRef]
- Han, Y.; Hou, Z.; He, Q.; Zhang, X.; Yan, K.; Han, R.; Liang, Z. Genome-wide characterization and expression analysis of bZIP gene family under abiotic stress in Glycyrrhiza uralensis. Front Genet. 2021, 12, 754237. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, L.; ShangGuan, G.; Jia, C.; Deng, S.; Noman, M.; Liu, Y.; Guo, Y.; Han, L.; Zhang, X.; Dong, Y.; Ahmad, N.; Du, L.; Li, H.; Yang, J. Genome-wide identification and expression analysis of bZIP gene family in Carthamus tinctorius L. Sci Rep. 2020, 10, 15521. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Lara, P.; Oñate-Sánchez, L.; Abraham, Z.; et al. Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J Biol Chem. 2003, 278, 21003–21011. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.C. The ULTRAPETALA gene controls shoot and floral meristem size in Arabidopsis. Development. 2001, 128, 1323–1333. [Google Scholar] [CrossRef]
- Lee, J. et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007, 19, 731–749. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.F.; Sun, L.; Valdés, A.E.; Engström, P.; Song, Z.T.; Lu, S.J.; et al. Membrane-associated transcription factor peptidase, site-2 protease, antagonizes ABA signaling in Arabidopsis. New Phytol. 2015, 208, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.S.; Sharma, E.; Jain, N.; Singh, B.; Burman, N.; and Khurana, J.P. A rice bZIP transcription factor, OsbZIP16, regulates abiotic stress tolerancewhen over-expressed in Arabidopsis. J. Plant Biochem. 2018, 27, 393–400. [Google Scholar] [CrossRef]
- Hao, X.; Zhong, Y.; Nï Tzmann, H.W.; Fu, X.; Yan, T.; Shen, Q.; Chen, M.; Ma, Y.; Zhao, J.; Osbourn, A.; Li, L.; Tang, K. Light-induced artemisinin biosynthesis is regulated by the bZIP transcription factor AaHY5 in Artemisia annua. Plant Cell Physiol. 2019, 60, 1747–1760. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Fang, J.; Zhao, R.; Liu, X.; Yin, W.; Wang, Y.; Wang, X.; Wang, X.; Fang, Y. CRISPR/Cas9-mediated mutagenesis of VvbZIP36 promotes anthocyanin accumulation in grapevine (Vitis vinifera). Hortic Res. 2022, 9, uhac022. [Google Scholar] [CrossRef]
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic Res. 2017, 4, 17023. [Google Scholar] [CrossRef]
- Xu, X.; Yuan, H.; Yu, X.; Huang, S.; Sun, Y.; Zhang, T.; Liu, Q.; Tong, H.; Zhang, Y.; Wang, Y.; Liu, C.; Wu, L.; Hou, M.; Yang, Y. The chromosome-level stevia genome provides insights into steviol glycoside biosynthesis. Hortic Res. 2021, 8, 129–139. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; et al. “Protein identification and analysis tools on the ExPASy server,” in the proteomics protocols handbook Springer Protocols Handbooks. Editor J. M. Walker (Totowa, NJ: Humana Press), 2005, 571-607. [CrossRef]
- Liu, Y.; Guan, X.; Liu, S.; Yang, M.; Ren, J.; Guo, M.; Huang, Z.; Zhang, Y. Genomewide identification and analysis of TCP transcription factors involved in the formation of leafy head in Chinese Cabbage. Int. J. Mol. Sci. 2018, 19, 847–862. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, Q.; Lyu, C.; Chen, J.; Xiao, R.; Chen, J.; Yang, Y.; Zhang, H.; Hou, K.; Wu, W. Characterizing glycosyltransferases by a combination of sequencing platforms applied to the leaf tissues of Stevia rebaudiana. BMC Genomics. 2020, 21:794-810. PMID: 33187479; PMCID: PMC7664074. [CrossRef]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of HY5 in plant growth and development. Mol Plant. 2016, 9, 1353–1365. [Google Scholar] [CrossRef]
- Yang, Y.; Liang, T.; Zhang, L.; Shao, K.; Gu, X.; Shang, R.; Shi, N.; Li, X.; Zhang, P.; Liu, H. UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat Plants. 2018, 4, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Miladinova-Georgieva, K.; Geneva, M.; Stancheva, I.; Petrova, M.; Sichanova, M.; Kirova, E. Effects of different elicitors on micropropagation, biomass and secondary metabolite production of Stevia rebaudiana Bertoni-A Review. Plants (Basel). 2022, 12, 153–170. [Google Scholar] [CrossRef]
- Tahmasi, S.; Garoosi, G.; Ahmadi, J.; Farjaminezhad, R. Effect of salicylic acid on stevioside and rebaudioside A production and transcription of biosynthetic genes in in vitro culture of Stevia rebaudiana. Iran. J. Genet. Plant Breed. 2017, 6, 1–8. [Google Scholar]
- Bayraktar, M.; Naziri, E.; Karabey, F.; Akgun, I.H.; Bedir, E.; Bärbel, R.O.; Gürel, A. Enhancement of stevioside production by using biotechnological approach in in vitro culture of Stevia rebaudiana. Int. J. Second. Metab. 2018, 5, 362–374. [Google Scholar] [CrossRef]
- Ahmad, A.; Ali, H.; Khan, H.; Begam, A.; Khan, S.; Ali, S.S.; Abbasi, B.H. Effect of gibberellic acid on production of biomass, polyphenolics and steviol glycosides in adventitious root cultures of Stevia rebaudiana (Bert.). Plants. 2020, 9, 420-436. doi: 10.3390/plants9040420. PMID: 32235525; PMCID: PMC7238111. [CrossRef]
- Ceunen, S.; Geuns, J.M. Influence of photoperiodism on the spatio-temporal accumulation of steviol glycosides in Stevia rebaudiana (Bertoni). Plant Sci. 2013, 198, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Hernández, K.V.; Moreno-Romero, J.; Hernández de la Torre, M.; Manríquez, C.P.; Leal, D.R.; Martínez-Garcia, J.F. Effect of light intensity on steviol glycosides production in leaves of Stevia rebaudiana plants. Phytochemistry. 2022, 194, 113027–113050. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Huang, S.; Han, Y.; Yuan, H.; Gu, C.; Wang, Z. Environmental cues induce changes of steviol glycosides contents and transcription of corresponding biosynthetic genes in Stevia rebaudiana. Plant Physiol. Biochem. 2015, 86, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Sharma, S.; Saxena, S. Effect of salts (NaCl and Na2CO3) on callus and suspension culture of Stevia rebaudiana for Steviol glycoside production. Appl. Biochem. Biotechnol. 2014, 172, 2894–2906. [Google Scholar] [CrossRef] [PubMed]
- Lucho, S.R.; do Amaral, M.N.; Auler, P.A.; Bianchi, V.J.; Ferrer, M.A.; Calderón, A.A.; Braga, E.J.B. Salt stress-induced changes in in vitro cultured Stevia rebaudiana Bertoni: Effect on metabolite contents, antioxidant capacity and expression of steviol glycosides-related biosynthetic genes. J. Plant Growth Regul. 2019, 38, 1341–1353. [Google Scholar] [CrossRef]
- Gupta, P.; Sharma, S.; Saxena, S. Biomass yield and steviol glycoside production in callus and suspension culture of Stevia rebaudiana treated with proline and polyethylene glycol. Appl. Biochem. Biotechnol. 2015, 176, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Fu, X.; Lv, Z.; Lu, X.; Shen, Q.; Zhang, L.; Zhu, M.; Wang, G.; Sun, X.; Liao, Z.; Tang, K. A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Mol Plant. 2015, 8, 163–75. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Huang, H.; Zhao, Y.; Xie, L.; He, Q.; Zhong, Y.; Wang, Y.; Wang, Y.; Tang, K. The Transcription factor AabZIP9 positively regulates the biosynthesis of artemisinin in Artemisia annua. Front Plant Sci. 2019, 10, 1294–1307. [Google Scholar] [CrossRef]
- Lv, Z.; Guo, Z.; Zhang, L.; Zhang, F.; Jiang, W.; Shen, Q.; Fu, X.; Yan, T.; Shi, P.; Hao, X.; Ma, Y.; Chen, M.; Li, L.; Zhang, L.; Chen, W.; Tang, K. Interaction of bZIP transcription factor TGA6 with salicylic acid signaling modulates artemisinin biosynthesis in Artemisia annua. J Exp Bot. 2019, 70, 3969–3979. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, L.; Hao, X.; Fu, X.; Ma, Y.; Xie, L.; Shen, Q.; Kayani, S.; Pan, Q.; Sun, X.; Tang, K. AaABF3, an abscisic acid-responsive transcription factor, positively regulates artemisinin biosynthesis in Artemisia annua. Front Plant Sci. 2018, 9, 1777–1786. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Nishizawa, Y.; Minami, E.; Nojiri, H.; Yamane, H.; Okada, K. Overexpression of the bZIP transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells. J Plant Physiol. 2015, 173, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Okada, A.; Okada, K.; Miyamoto, K.; Koga, J.; Shibuya, N.; Nojiri, H.; Yamane, H. OsTGAP1, a bZIP transcription factor, coordinately regulates the inductive production of diterpenoid phytoalexins in rice. J Biol Chem. 2009, 284, 26510–26518. [Google Scholar] [CrossRef]
- Zhou, J.; Wu, Y.; Zhang, X.; Zhao, L.; Feng, Z.; Wei, F.; Zhang, Y.; Feng, H.; Zhou, Y.; Zhu, H. MPK homolog GhNTF6 was involved in cotton against Verticillium wilt by interacted with VdEPG1. Int J Biol Macromol. 2022, 195, 456–465. [Google Scholar] [CrossRef] [PubMed]
- 38. Hartmann, L,; Pedrotti, L.; Weiste, C.; Fekete, A.; Schierstaedt, J.; Göttler, J.; Kempa, S.; Krischke, M.; Dietrich, K.; Mueller, M.J.; Vicente-Carbajosa, J.; Hanson, J.; Dröge-Laser, W. Crosstalk between two bZIP signaling pathways orchestrates salt-induced metabolic reprogramming in Arabidopsis roots. Plant Cell. 2015, 27, 2244-60. doi: 10.1105/tpc.15.00163. PMID: 26276836; PMCID: PMC4568499. [CrossRef]
- Li, X.F.; Li, Y.J.; An, Y.H.; Xiong, L.J.; Shao, X.H.; Wang, Y.; Sun, Y. AKINbeta1 is involved in the regulation of nitrogen metabolism and sugar signaling in Arabidopsis. J Integr Plant Biol. 2009, 51, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Wang, Y.; Gai, R.; Xi, D.; Mao, C.; Ming, F. Rice SnRK protein kinase OsSAPK8 acts as a positive regulator in abiotic stress responses. Plant Sci. 2020, 292, 110373–110381. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Lv, J.; Shi, Y.; Gao, J.; Hua, J.; Song, C.; Gong, Z.; Yang, S. EGR2 phosphatase regulates OST1 kinase activity and freezing tolerance in Arabidopsis. EMBO J. 2019, 38, e99819. [Google Scholar] [CrossRef]
- Ding, Y.; Sun, T.; Ao, K.; Peng, Y.; Zhang, Y.; Li, X.; Zhang, Y. Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell. 2018, 173, 1454–1467. [Google Scholar] [CrossRef] [PubMed]
- Rayapuram, C.; Baldwin, I.T. Increased SA in NPR1-silenced plants antagonizes JA and JA-dependent direct and indirect defenses in herbivore-attacked Nicotiana attenuata in nature. Plant J. 2007, 52, 700–715. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, J.; Li, H.; Yang, Y.; Guang, Y.; Zhou, Y. The bZIP gene family in watermelon: genome-wide identifcation and expression analysis under cold stress and root-knot nematode infection. PeerJ. 2019, 7, e7878. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, J.; Zhang, B.; Vanitha, J.; Ramachandran, S.; Jiang, S.Y. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J. Integr. Plant Biol. 2011, 53, 212–231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, Y.; Shi, H.; Guo, M.; Chai, M.; He, Q.; Yan, M.; Cao, D.; Zhao, L.; Cai, H.; et al. Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genomics. 2018, 19, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Meng, X.X.; Zhang, Y.M.; Zhu, X.W.; Li, J.; Chen, W.Q.; Wan, H.H.; Wang, S.F.; Cao, X.; Sun, W.; Mi, Y.L.; Zhai, J.W. Genome-wide identification and expression profiles of bZIP Genes in Cannabis sativa L. Cannabis Cannabinoid Res. 2022, 7, 882–895. [Google Scholar] [CrossRef] [PubMed]
- Song, X.M.; Liu, T.K.; Duan, W.K.; Ma, Q.H.; Ren, J.; Wang, Z.; Li, Y.; Hou, X.L. Genome-wide analysis of the GRAS gene family in Chinese cabbage (Brassica rapa ssp pekinensis). Genomics. 2014, 103, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10–31. [Google Scholar] [CrossRef] [PubMed]
- Patthy, L. Intron-dependent evolution: preferred types of exons and introns. FEBS Lett. 1987, 214, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Fu, F.; Zhang, H.; Song, F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genomics. 2015, 16, 771-789. doi: 10.1186/s12864-015-1990-6. PMID: 26459863; PMCID: PMC4603586. [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Oyama, T.; Shimura, Y.; Okada, K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997, 11, 2983–2995. [Google Scholar] [CrossRef]
- Jing, Y.; Zhang, D.; Wang, X.; Tang, W.; Wang, W.; Huai, J.; Xu, G.; Chen, D.; Li, Y.; Lin, R. Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell. 2013, 25, 242–56. [Google Scholar] [CrossRef]
- Oravecz, A.; Baumann, A.; Máté, Z.; Brzezinska, A.; Molinier, J.; Oakeley, E.J.; Adám, E.; Schäfer, E.; Nagy, F.; Ulm, R. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 2006, 18, 1975–1990. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.A.; Jenkins, G.I. UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 2008, 146, 576–588. [Google Scholar] [CrossRef]
- Michael, R.; Ranjan, A.; Kumar, R.S.; Pathak, P.K.; Trivedi, P.K. Light-regulated expression of terpene synthase gene, AtTPS03, is controlled by the bZIP transcription factor, HY5, in Arabidopsis thaliana. Biochem Biophys Res Commun. 2020, 529, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Ramegowda, V.; Gill, U.S.; Sivalingam, P.N.; Gupta, A.; Gupta, C.; Govind, G.; Nataraja, K.N.; Pereira, A.; Udayakumar, M.; Mysore, K.S.; Senthil-Kumar, M. GBF3 transcription factor imparts drought tolerance in Arabidopsis thaliana. Sci Rep. 2017, 7, 9148–9161. [Google Scholar] [CrossRef] [PubMed]
- Schindler, U.; Menkens, A.E.; Beckmann, H.; Ecker, J.R.; Cashmore, A.R. Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 1992, 11, 1261–1273. [Google Scholar] [CrossRef] [PubMed]
- Djamei, A.; Pitzschke, A.; Nakagami, H.; Rajh, I.; Hirt, H. Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science. 2007, 318, 453–456. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: anintegrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; et al. Mcscanx: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Li, M.; Christina, K.; Koichiro, T. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol.Evol. 2018, 35, 1547–1549, PMID: 29722887; PMCID: PMC5967553. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.Y.; Li, W.W.; Noble, W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. Correction to 'The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets'. Nucleic Acids Res. 2021, 49, 10800. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




