Submitted:
27 July 2023
Posted:
28 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. DNA extraction and genome sequencing
2.2. Genome assembly and annotation
2.3. The selection and analysis of MFS-domain proteins
3. Results
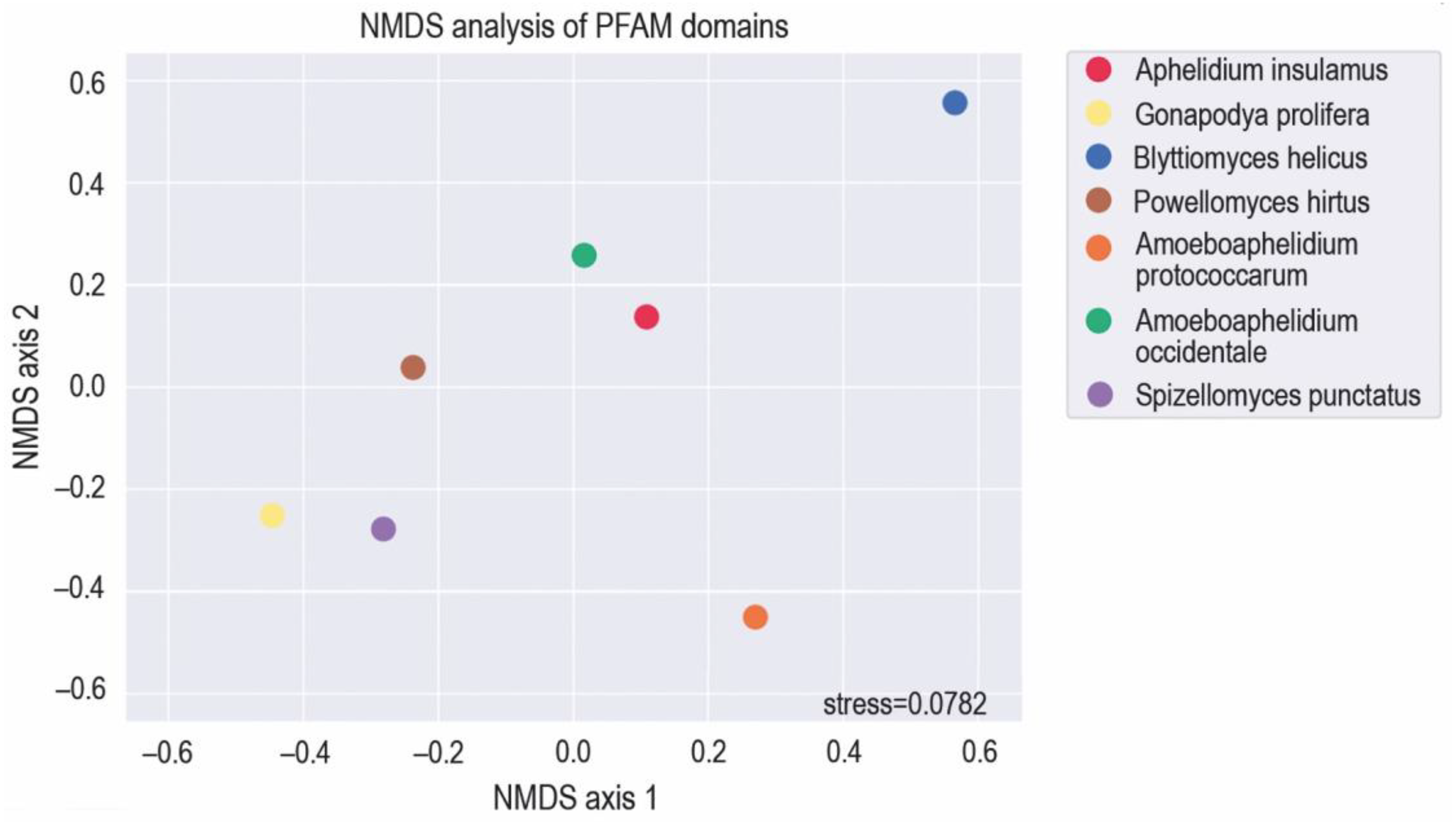
3.1. Assembly of Aphelidium insulamus genome
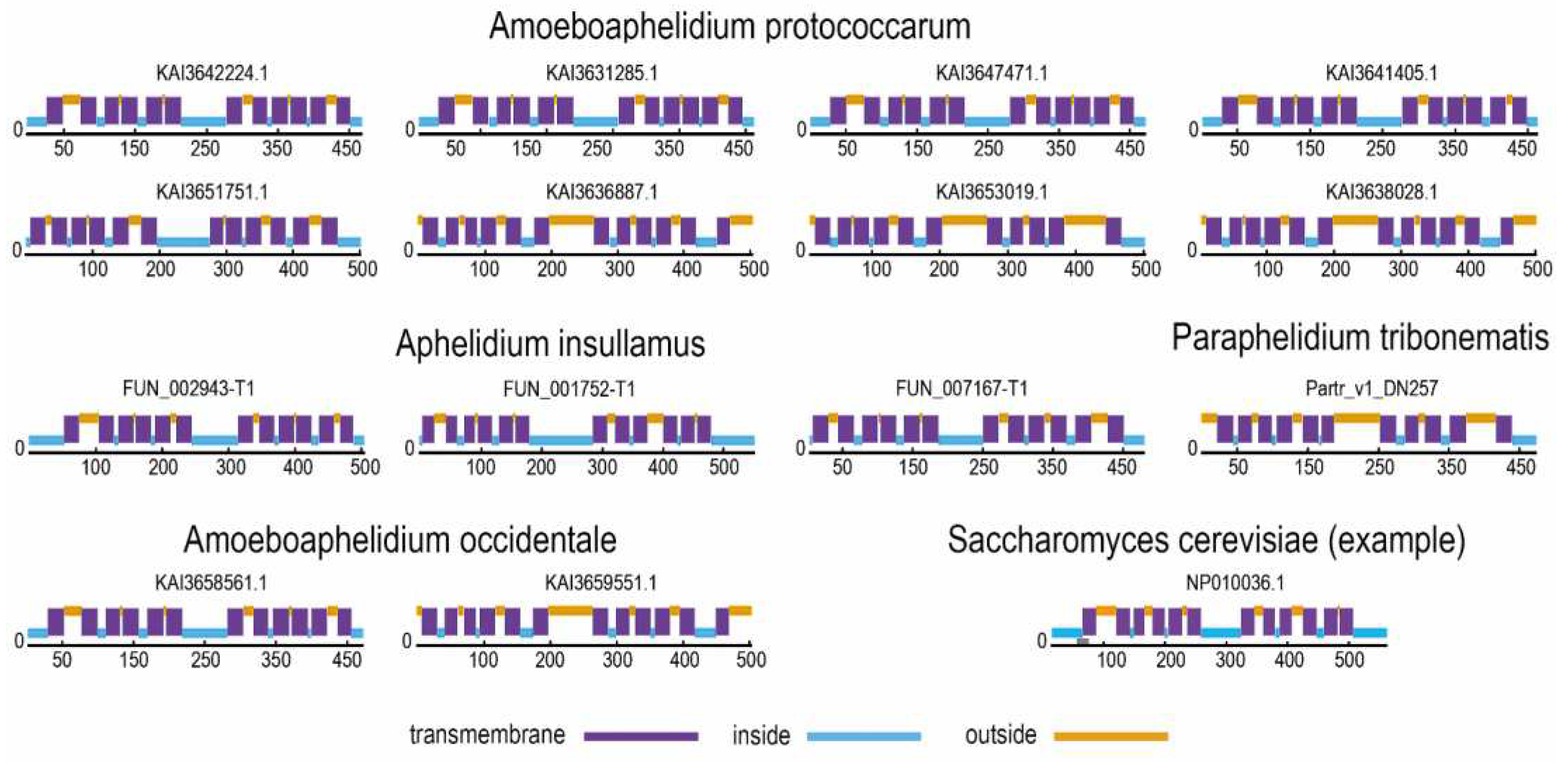
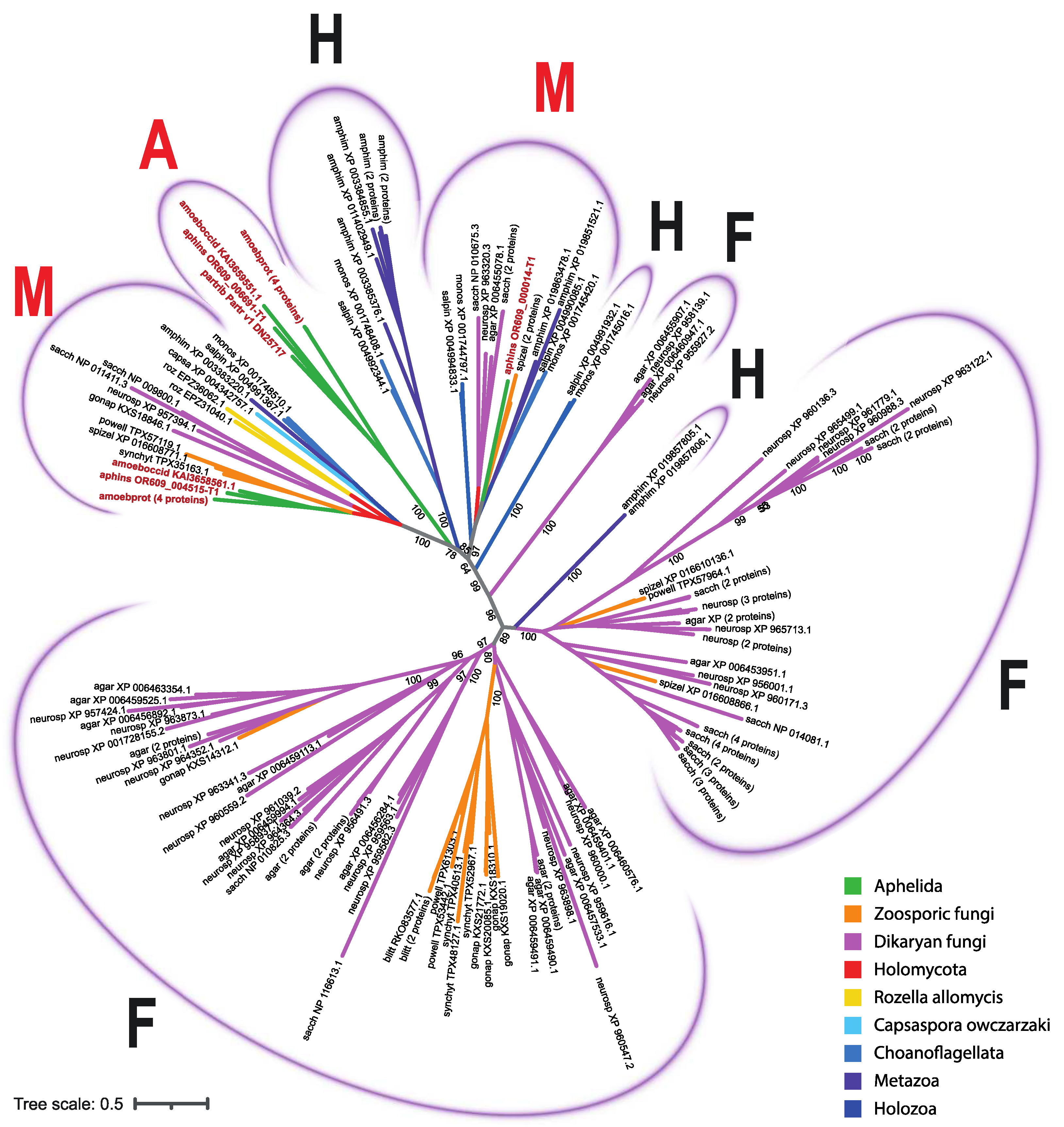
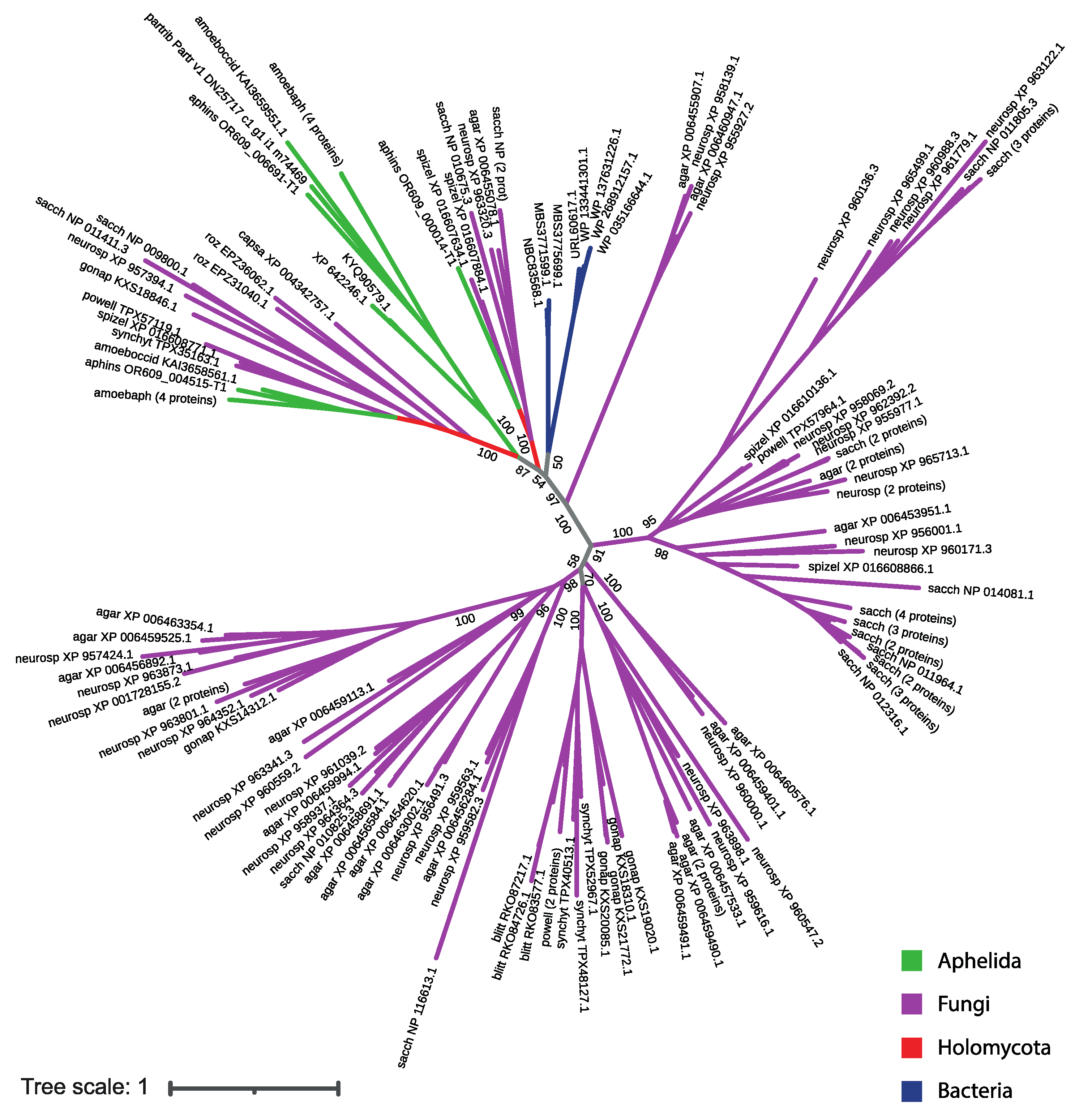
3.2. MFS Protein Analysis
4. Discussion
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Gromov, B.V. Algal parasites of the genera Aphelidium, Amoeboaphelidium and Pseudoaphelidium from the Cienkovski’s “Monadea” group as representatives of new class. Zool. Z. 2000, 79, 517–525.
- Karpov, S.A.; Mamkaeva, M.A.; Aleoshin, V.V.; Nassonova, E.; Lilje, O.; Gleason, F.H. Morphology, phylogeny, and ecology of the aphelids (Aphelidea, Opisthokonta) and proposal for the new superphylum Opisthosporidia. Front. Microbiol. 2014, 5;. https://doi.org/10.3389/fmicb.2014.00112. [CrossRef]
- Letcher, P.M.; Lopez, S.; Schmieder, R.; Lee, P.A.; Behnke, C.; Powell, M.J.; McBride, R.C. Characterization of Amoeboaphelidium protococcarum, an algal parasite new to the cryptomycota isolated from an outdoor algal pond used for the production of biofuel. PLoS One 2013; 8(2);. https://doi.org/10.1371/journal.pone.0056232. [CrossRef]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukes, J.; Schoch, C.L.; Smirnov, A. et al. Revisions to the classification, nomenclature, and diversity of eukaryotes., J. Eukaryot. Microbiol. 2019, 66, 4–119;. https://doi.org/10.1111/jeu.12691. [CrossRef]
- Letcher, P.M.; Powell, M.J. A taxonomic summary of Aphelidiaceae. IMA Fungus 2019, 10(4);. https://doi.org/10.1186/s43008-019-0005-7. [CrossRef]
- Torruella, G.; Grau-Bové, X.; Moreira, D.; Karpov, S.A.; Burns, J.A.; Sebé-Pedrós, A.; Völcker, E.; López-García, P. Global transcriptome analysis of the aphelid Paraphelidium tribonemae supports the phagotrophic origin of fungi. Commun. Biol. 2018, 1(231);. https://doi.org/10.1038/s42003-018-0235-z. [CrossRef]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018, 90, 135–159;. https://doi.org/10.1007/s13225-018-0401-0. [CrossRef]
- Galindo, L.J.; Torruella, G.; López-García, P.; Ciobanu, M.; Gutiérrez-Preciado, A.; Karpov, S.A.; Moreira, D. Phylogenomics supports the monophyly of aphelids and fungi and identifies new molecular synapomorphies. Syst. Biol. 2022, syac054;. https://doi.org/10.1093/sysbio/syac054. [CrossRef]
- Mikhailov, K.V.; Karpov, S.A.; Letcher, P.M.; Lee, Ph.A.; Logacheva, M.D.; Penin, A.A.; Nesterenko, M.A.; Pozdnyakov, I.R.; Potapenko, E.V.; Sherbakov, D.Y.; Panchin, Y.V.; Aleoshin, V.V. Genomic analysis reveals cryptic diversity in aphelids and sheds light on the emergence of Fungi. Curr. Biol. 2022, 32, 1–13;. https://doi.org/10.1016/j.cub.2022.08.071. [CrossRef]
- Guarro, J.; Gené, J.; Stchigel, A.M. Developments in fungal taxonomy. Clin Microbiol Rev. 1999, 12(3), 454–500. https://doi.org/10.1128/CMR.12.3.454. [CrossRef]
- Margulis, L.; Chapman, M.J. Chapter Four - Kingdom Fungi. In Kingdoms and Domains, 4th ed; Margulis, L.; Chapman, M.J.; Eds.; Academic Press, 2009, pp. 379–409;. https://doi.org/10.1016/B978-0-12-373621-5.00004-0. [CrossRef]
- Naranjo-Ortiz, M.A.; Gabaldón, T. Fungal evolution: diversity, taxonomy and phylogeny of the Fungi. Biol. Rev. 2019, 94, 2101–2137;. https://doi.org/10.1111/brv.12550. [CrossRef]
- Richards, T.A.; Leonard, G.; Wideman, J.G. What Defines the “Kingdom” Fungi? Microbiol. Spectr. 2017, 5(3). https://doi.org/10.1128/microbiolspec.FUNK-0044-2017. [CrossRef]
- Henderson, P.J. The homologous glucose transport proteins of prokaryotes and eukaryotes. Res. Microbiol. 1990, 141(3), 316–28;. https://doi.org/10.1016/0923-2508(90)90005-b. [CrossRef]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H.; jr. Major Facilitator Superfamily. Microbiol. Mol. Biol. R. 1998, 62(1), 1092–2172;. https://doi.org/10.1128/MMBR.62.1.1-34.1998. [CrossRef]
- Yan, N. Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 2013, 38(3), 151–159;. https://doi.org/10.1016/j.tibs.2013.01.003; PMID 23403214. [CrossRef]
- InterPro. Classification of protein families. Major facilitator superfamily domain. Available online: https://www.ebi.ac.uk/interpro/entry/InterPro/IPR020846/ (accessed on 18.07.2023).
- Reddy, V.S.; Shlykov, M.A.; Castillo, R.; Sun, E.I.; Saier, M.H. The major facilitator superfamily (MFS) revisited. FEBS J. 2012, 279(11), 2022-2035;. https://doi.org/10.1111/j.1742-4658.2012.08588.x. [CrossRef]
- Gonçalves, C.; Coelho, M.A.; Salema-Oom, M.; Gonçalves, P. Stepwise Functional Evolution in a Fungal Sugar Transporter Family. Mol. Biol. Evol. 2016, 33(2), 352–366;. https://doi.org/10.1093/molbev/msv220. [CrossRef]
- Merényi, Z.; Krizsán, K.; Sahu, N.; Liu, X.; Bálint, B.; Stajich, J.; Spatafora, J.W.; Nagy, L.G. Taxonomic vs genomic fungi: contrasting evolutionary loss of protistan genomic heritage and emergence of fungal novelties. bioRxiv 2022, 11.15.516418;. https://doi.org/10.1101/2022.11.15.516418. [CrossRef]
- Van Dijck, P.; Brown, N.A.; Goldman, G.H.; Rutherford, J.; Xue, C.; Van Zeebroeck, G. Nutrient sensing at the plasma membrane of fungal cells. Microbiol. Spectrum 2017, 5(2);. https://doi.org/10.1128/microbiolspec.FUNK-0031-2016. [CrossRef]
- Bisson, L.F.; Coons, D.M.; Kruckeberg, A.L.; Lewis, D.A. Yeast sugar transporters. Crit. Rev. Biochem. Mol Biol. 1993, 28(4):, 259–308;. https://doi.org/10.3109/10409239309078437. [CrossRef]
- Goffeau, A.; Park, J.; Paulsen, I.T.; Jonniaux, J.L.; Dinh, T.; Mordant, P.; Saier, M.H. Multidrug-resistant transport proteins in yeast: complete inventory and phylogenetic characterization of yeast open reading frames with the major facilitator superfamily. Yeast 1997, 13(1):43–54;. https://doi.org/10.1002/(SICI)1097-0061(199701)13:1<43:AID-YEA56>3.0.CO;2-J. [CrossRef]
- Dias, P.J, Sá-Correia, I. The drug:H+ antiporters of family 2 (DHA2), siderophore transporters (ARN) and glutathione:H+antiporters (GEX) have a common evolutionary origin in hemiascomycete yeasts. BMC Genomics 2013, 14, 901;. https://doi.org/10.1186/1471-2164-14-901. [CrossRef]
- Karpov, S.A.; Vishnyakov, A.E.; López-García, P.; Zorina, N.A.; Ciobanu, M.; Tcvetkova, V.S.; Moreira, D. Morphology and molecular phylogeny of Aphelidium insulamus sp. nov. (Aphelida, Opisthosporidia). Protistology 2020, 14(4), 191–203;. https://doi.org/10.21685/1680-0826-2020-14-4-3. [CrossRef]
- Fahrni, J.F.; Bolivar, I.; Berney, C.; Nassonova, E.; Smirnov A & Pawlowski, J. Phylogeny of lobose amoebae based on actin and small-subunit ribosomal RNA genes. Mol. Biol. Evol. 2003, 20(11),1881–1886;. https://doi.org/10.1093/molbev/msg201. [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Yu; Pevzner, P. Assembly of Long Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37(5), 540–546;. https://doi.org/10.1038/s41587-019-0072-8. [CrossRef]
- Challis, R.; Richards, E.; Rajan, J.; Cochrane, G.; & Blaxter, M. BlobToolKit–interactive quality assessment of genome assemblies. G3-Genes Genom. Genet. 2020, 10(4), 1361–1374;. https://doi.org/10.1534/g3.119.400908. [CrossRef]
- Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinform. 2018, 34(18), 3094–3100;. https://doi.org/10.1093/bioinformatics/bty191. [CrossRef]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017, 27(5), 737–746;. https://doi.org/10.1101/gr.214270.116. [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; Earl, A.M. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 19;9(11):e112963;. https://doi.org/10.1371/journal.pone.0112963. [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinform. 2015, 31(19), 3210–3212;. https://doi.org/10.1093/bioinformatics/btv351. [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinform. 2013, 29(8), 1072–1075;. https://doi.org/10.1093/bioinformatics/btt086. [CrossRef]
- Palmer, J.; Stajich, J. Nextgenusfs/funannotate: funannotate v1.8.1 (Version 1.8.1). Zenodo 2020;. https://doi.org/10.5281/zenodo.4054262. [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, R7;. https://doi.org/10.1186/gb-2008-9-1-r7. [CrossRef]
- Käll, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007, 35 (Web Server issue), W429–432;. https://doi.org/10.1093/nar/gkm256. [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. In: Gene Prediction. Methods in Molecular Biology; Kollmar, M; Ed.; 2019; Humana: New York, NY; Volume 1962;. https://doi.org/10.1007/978-1-4939-9173-0_1. [CrossRef]
- Jones, P.; Binns, D.; Chang H-Yu, Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; Pesseat, S.; Quinn, A.F.; Sangrador-Vegas, A.; Scheremetjew, M.; Yong S-Y, Lopez, R.; Hunter, S. InterProScan 5: genome-scale protein function classification. Bioinform. 2014, 30(9), 1236–1240;. https://doi.org/10.1093/bioinformatics/btu031. [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer K & Madden, T.L. BLAST+: architecture and applications. BMC Bioinform. 2009, 10, 421;. https://doi.org/10.1186/1471-2105-10-421. [CrossRef]
- Wheeler, T.J.; Eddy, S.R. nhmmer: DNA homology search with profile HMMs. Bioinform. 2013, 29(19), 2487–2489;. https://doi.org/10.1093/bioinformatics/btt403. [CrossRef]
- Di Tommaso, P.; Moretti, S.; Xenarios, I.; Orobitg, M.; Montanyola, A.; Chang, J.M.; Taly, J.F.; Notredame, C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011, 39 (Web Server issue), W13-7;. https://doi.org/10.1093/nar/gkr245. [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinform. 2009, 25, 1972–1973;. https://doi.org/10.1093/bioinformatics/btp348. [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32(1), 268–274;. https://doi.org/10.1093/molbev/msu300. [CrossRef]
- Ocaña-Pallarès, E.; Williams, T.A.; López-Escardó, D.; Arroyo, A.S.; Pathmanathan, J.S.; Bapteste, E.; Tikhonenkov, D.V.;.Keeling, P.J.;.Szöllősi, G.J.; Ruiz-Trillo, I. Divergent genomic trajectories predate the origin of animals and fungi. Nature 2022, 609, 747–753;. https://doi.org/10.1038/s41586-022-05110-4. [CrossRef]
- Chang, Y.; Wang, S.; Sekimoto, S.; Aerts, A.L.; Choi, C.; Clum, A.; LaButti, K.M.; Lindquist, E.A.; Ngan CYe, Ohm, R.A.; Salamov, A.A.; Grigoriev, I.V.; Spatafora, J.W.; Berbee, M.L. Phylogenomic analyses indicate that early fungi evolved digesting cell walls of algal ancestors of land plants. Genome Biol. Evol. 2015, 7(6), 1590–1601;. https://doi.org/10.1093/gbe/evv090. [CrossRef]
- Tikhonenkov, D.V.; Hehenberger, E.; Esaulov, A.S.; Belyakova, O.I.; Mazei, Y.A.; Mylnikov, A.P.; Keeling, P.J. Insights into the origin of metazoan multicellularity from predatory unicellular relatives of animals. BMC Biol. 2020, 18(39);. https://doi.org/10.1186/s12915-020-0762-1. [CrossRef]
- Brun, S; Silar, P. Convergent evolution of morphogenetic processes in Fungi. In: Evolutionary biology – Concepts, molecular and morphological evolution; Pontarotti, P.; Ed.; Springer: Berlin, Heidelberg, 2010; pp. 317–328;. https://doi.org/10.1007/ 978-3-642-12340-5_19. [CrossRef]
- Kożyczkowska, A.;. Najle, S.R.; Ocaña-Pallarès, E.; Aresté, C.; Shabardina, V.; Ara, P.S.; Ruiz-Trillo, I.; Casacuberta, E. Stable transfection in protist Corallochytrium limacisporum identifies novel cellular features among unicellular animals relatives. Curr. Biol. 2021, 31(18), 4104-4110.e5;. https://doi.org/10.1016/j.cub.2021.06.061. [CrossRef]
- Torruella, G.; de Mendoza, A.; Grau-Bové, X.; Antó, M.; Chaplin, M.A.; del Campo, J.; Eme, L.; Pérez-Cordón, G.; Whipps, C.M.; Nichols, K.M.; Paley, R.; Roger, A.J.; Sitjà-Bobadilla, A.; Donachie, S.; Ruiz-Trillo, I. Phylogenomics Reveals Convergent Evolution of Lifestyles in Close Relatives of Animals and Fungi. Curr. Biol. 2015, 25(18), 2404–2410;. https://doi.org/10.1016/j.cub.2015.07.053. [CrossRef]
- Pozdnyakov, I.R.; Zolotarev, A.V.; Karpov, S.A. Comparative analysis of zoosporogenesis’ genes of the bastoclad Blastocladiella emersonii and the aphelid Paraphelidium tribonematis reveals the new directions of evolutionary research. Protistology 2021, 15(1), 10–23;. https://doi.org/10.21685/1680-0826-2021-15-1-2. [CrossRef]
- Nagy, L.G.; Kovács GM and Krizsán, K. Complex multicellularity in fungi: evolutionary convergence, single origin, or both? Biol. Rev. Camb. Philos. Soc. 2018, 93(4), 1778–1794;. https://doi.org/10.1111/brv. 12418. [CrossRef]

| Species | Assembly size (bp) | Num scaffolds | Scaffold N50 (bp) | Average scaffold (bp) | Largest scaffold (bp) | GC, % | Num genes | Num proteins | Unique BUSCOs |
|---|---|---|---|---|---|---|---|---|---|
| Aphelidium insulamus | 18,927,283 | 274 | 252,907 | 69,078 | 1,020,338 | 52.05 | 7,925 | 7,820 | 277 |
| Amoeboaphelidium protococcarum | 24,734,778 | 258 | 2,170,272 | 95,871 | 3,250,117 | 40.50 | 13,180 | 13,180 | 281 |
| Amoeboaphelidium occidentale | 13,559,732 | 951 | 73,507 | 14,258 | 366,412 | 39.93 | 7,568 | 7,495 | 277 |
| Gonapodya prolifera | 48,794,828 | 352 | 347,324 | 138,622 | 1,572,201 | 51.75 | 13,911 | 13,831 | 283 |
| Blyttiomyces helicus | 46,468,912 | 8,398 | 6,675 | 5,533 | 73,981 | 53.75 | 12,446 | 12,167 | 168 |
| Powellomyces hirtus | 26,238,698 | 482 | 157,542 | 54,437 | 764,225 | 51.37 | 6,536 | 6,536 | 292 |
| Spizellomyces punctatus | 24,131,112 | 38 | 1,465,700 | 635,029 | 2,242,449 | 47.16 | 9,164 | 9,422 | 295 |
 | |||
| Species | Designation in the phylogenetic tree | Number of SP proteins | Accession numbers |
| Aphelidium insulamus | aphins | 3 | OR609_004515-T1 (FUN_002943-T1), OR609_000014-T1 (FUN_001752-T1), OR609_006691-T1 (FUN_007167-T1) |
| Paraphelidium tribonematis | partrib | 1 | Partr_v1_DN257 |
| Amoeboaphelidium protococcarum | amoebprot | 8 | KAI3642224.1, KAI3631285.1, KAI3647471.1, KAI3641405.1, KAI3636887.1, KAI3651751.1, KAI3653019.1, KAI3638028.1 |
| Amoeboaphelidium occidentale | amoeboccid | 2 | KAI3658561.1, KAI3659551.1 |
| Blyttiomyces. helicus | blitt | 3 | RKO83577.1, RKO87217.1, RKO84726.1 |
| Gonapodya prolifera | gonap | 6 | KXS20085.1, KXS21772.1, KXS18310.1, KXS19020.1, KXS14312.1, KXS18846.1 |
| Powellomyces hirtus | powell | 4 | TPX57964, TPX61303, TPX53442, TPX57119 |
| Spizellomyces punctatus | spizel | 5 | XP_016610136.1, XP_016608866.1, XP_016607884.1, XP_016608771.1, XP_016607634.1 |
| Synchytrium endobioticum | synchyt | 4 | TPX52967.1, TPX40513.1, TPX48127.1, TPX35163.1 |
| Agaricus bisporus | agar | 26 | XP_006454719.1, XP_006461850.1, XP_006453951.1, XP_006456284.1, XP_006459401.1, XP_006454620.1, XP_006457533.1, XP_006460576.1, XP_006459994.1, XP_006459113.1, XP_006463002.1, XP_006459487.1, XP_006459489.1, XP_006459491.1, XP_006458691.1, XP_006459490.1, XP_006456584.1, XP_006460947.1, XP_006455907.1, XP_006455078.1, XP_006456754.1, XP_006456796.1, XP_006456892.1, XP_006463354.1, XP_006459525.1, XP_006458285.1 |
| Neurospora crassa | neurosp | 34 | XP_959573.2, XP_959411.2, XP_965713.1, XP_955977.1, XP_962392.2, XP_958069.2, XP_960171.3, XP_956001.1, XP_959563.1, XP_960000.1, XP_964364.3, XP_958937.1, XP_956491.3, XP_959616.1, XP_959582.3, XP_961039.2, XP_963898.1, XP_963341.3, XP_963320.3, XP_960136.3, XP_960988.3, XP_961779.1, XP_960559.2, XP_958139.1, XP_955927.2, XP_964352.1, XP_963873.1, XP_965499.1, XP_960547.2, XP_963122.1, XP_957424.1, XP_001728155.2, XP_963801.1, XP_957394.1 |
| Saccharomyces cerevisiae | sacch | 30 | NP_010087.1, NP_010143.1, NP_011960.2, NP_116644.1, NP_013724.1, NP_013182.1, NP_011962.1, NP_010632.1, NP_012316.1, NP_010629.3, NP_010630.1, NP_011964.1, NP_014486.1, NP_012321.1, NP_014470.1, NP_010845.1, NP_010036.1, NP_012692.3, NP_014081.1, NP_010825.3, NP_010785.1, NP_014538.2, NP_012694.1, NP_010034.1, NP_011805.3, NP_116613.1, NP_009857.1, NP_010675.3, NP_009800.1, NP_011411.3 |
| Rozella allomycis | roz | 2 | EPZ36062, EPZ31040 |
| Capsaspora owczarzaki | capsa | 1 | XP_004342757.1 |
| Monosiga brevicollis | monos | 5 | XP_001748408.1, XP_001745016.1, XP_001745420.1, XP_001748510.1, XP_001744797.1 |
| Salpingoeca rosetta | salpin | 5 | XP_004992344.1, XP_004991932.1, XP_004990085.1, XP_004994833.1, XP_004991367.1 |
| Amphimedon queenslandica | amphim | 13 | XP_019857806.1, XP_019857805.1, XP_019851521.1, XP_011406421.1, XP_003389392.2, XP_003383220.1, XP_019863478.1, XP_011402949.1, XP_019856859.1, XP_003384062.3, XP_003385376.1, XP_011408594.2, XP_003384855.1 |
| Description | Max Score | Total Score | Query Cover | E value | Per. Ident | Acc. Len | Accession |
|---|---|---|---|---|---|---|---|
| hypothetical protein MP228_003054 [Amoeboaphelidium protococcarum] | 1009 | 1009 | 100% | 0.0 | 100.00% | 501 | KAI3651751.1 |
| hypothetical protein MIR68_003639 [Amoeboaphelidium protococcarum] | 882 | 882 | 100% | 0.0 | 92.66% | 503 | KAI3638028.1 |
| hypothetical protein MP228_002444 [Amoeboaphelidium protococcarum] | 881 | 881 | 100% | 0.0 | 92.64% | 502 | KAI3653019.1 |
| hypothetical protein MIR68_005154 [Amoeboaphelidium protococcarum] | 863 | 863 | 100% | 0.0 | 90.89% | 504 | KAI3636887.1 |
| hypothetical protein MP638_005237 [Amoeboaphelidium occidentale] | 196 | 196 | 89% | 2e-52 | 30.95% | 483 | KAI3659551.1 |
| sugar transporter family protein [Tieghemostelium lacteum] | 197 | 197 | 93% | 7e-52 | 30.42% | 631 | KYQ90579.1 |
| sugar transporter family protein [Dictyostelium discoideum AX4] | 190 | 190 | 92% | 4e-49 | 30.29% | 630 | XP_642246.1 |
| sugar porter family MFS transporter [Bacteroidales bacterium] | 186 | 186 | 92% | 1e-48 | 28.48% | 495 | MBS3771599.1 |
| sugar porter family MFS transporter [Bacteroidales bacterium] | 185 | 185 | 92% | 2e-48 | 28.14% | 495 | MBS3775699.1 |
| sugar porter family MFS transporter [Acetilactobacillus jinshanensis] | 181 | 181 | 90% | 6e-47 | 31.30% | 467 | WP_133441301.1 |
| sugar porter family MFS transporter [Bacteroidota bacterium] | 181 | 181 | 92% | 6e-47 | 27.35% | 486 | NBC83568.1 |
| sugar porter family MFS transporter [uncultured bacterium] | 181 | 181 | 90% | 7e-47 | 31.30% | 467 | URL60617.1 |
| sugar porter family MFS transporter [Lentilactobacillus sp. SPB1-3] | 178 | 178 | 90% | 5e-46 | 29.00% | 467 | WP_268912157.1 |
| sugar porter family MFS transporter [Lentilactobacillus curieae] | 177 | 177 | 90% | 6e-46 | 29.93% | 460 | WP_035166644.1 |
| sugar porter family MFS transporter [Secundilactobacillus hailunensis] | 176 | 176 | 90% | 4e-45 | 30.79% | 463 | WP_137631226.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





