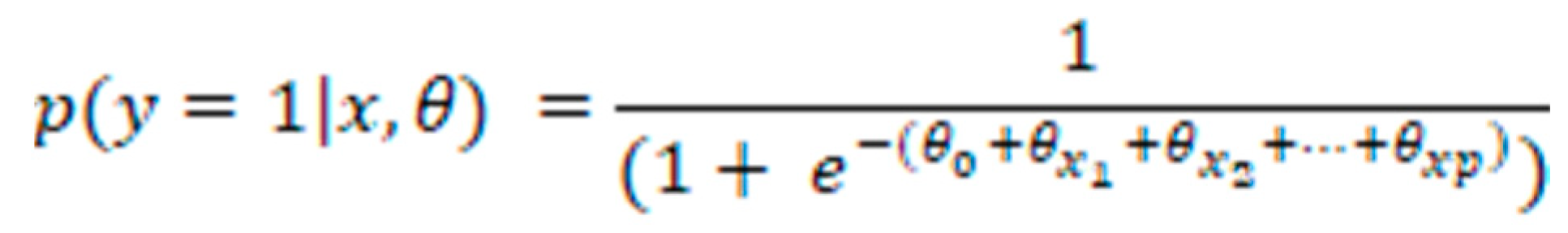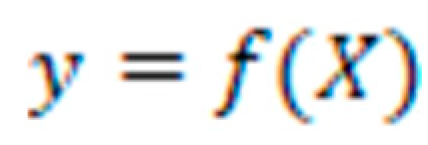1. Introduction
Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is a complex and debilitating disease. It may come by broad heterogeneity and common symptoms, including severe fatigue, post-exertional malaise (PEM), restless sleep, cognitive impairment, and orthostatic intolerance [
1]. The prevalence of ME/CFS is significant, with more than 65 million suffering individuals worldwide, indicating the significant impact of the disease on a global scale [
2]. In addition, the true prevalence of the disease is difficult to determine due to factors such as underdiagnoses and misdiagnoses [
3]. Although ME/CFS has been observed to be diagnosed more frequently in women, it is not a female-specific condition and approximately 35-40% of patients with ME/CFS are male [
4]. The reasons behind the higher prevalence in women are not fully understood [
4,
5] and may be influenced by a variety of factors such as hormonal differences, genetic predisposition, and social and cultural factors.
Dysfunctions of various systems, including the immune, neurological, gastrointestinal, and circulatory systems, have been reported in individuals with ME/CFS [
6,
7,
8,
9]. Studies focusing on the immune system have revealed abnormalities in various immune cell types among ME/CFS patients, suggesting that the disease is an immune disorder [
6,
7]. Increased levels of inflammatory cytokines were also observed in the plasma of ME/CFS patients compared with healthy controls, indicating an increased inflammatory response [
10].
Neuroimaging studies have identified abnormalities in the brains of ME/CFS patients, including changes in brain structure and function. These findings add to the understanding of cognitive impairment and other neurological symptoms experienced by individuals with ME/CFS [
11]. Digestive problems are common among ME/CFS patients, with a significant proportion reporting symptoms consistent with irritable bowel syndrome (IBS). This suggests that the gastrointestinal tract plays a potential role in the pathophysiology of the disease [
12,
13].
The circulatory system plays a very important role in providing essential compounds and removing metabolic wastes from various organs [
14]. Several studies have been conducted to characterize the blood metabolome of ME/CFS patients to gain insight into the underlying causes of the disease and to establish diagnostic strategies [
14]. These studies have highlighted differences in amino acids, lipids, and imbalances in energy and redox metabolisms. However, it is important to note that no consistently altered metabolites were identified in all studies, which poses a challenge to a full understanding of the disease.
The surprising nature of ME/CFS, in which multiple organ systems are affected [
6,
7,
8,
9,
10,
11,
12,
13,
14], underlines the complexity of the disease and the need for further research. ME/CFS is a heterogeneous condition, and individual variations in symptoms and underlying mechanisms may contribute to the difficulty in identifying consistent biomarkers or metabolic changes. More comprehensive and collaborative research efforts are required to uncover the underlying mechanisms for ME/CFS, identify reliable biomarkers, and develop targeted therapies. The involvement of multiple organ systems highlights the importance of a multidisciplinary approach in the diagnosis, treatment, and management of this complex disease. In this study, we comprehensively analyzed the metabolites of ME/CFS patients compared to normal controls to identify patterns of metabolites that could potentially serve as biomarkers for the disease. What makes our analysis comprehensive is that we examined metabolites belonging to nine different super pathways, aiming to address the heterogeneous nature of the disease and understand its mechanisms of development and progression. To achieve this, we employed a combination of explainable artificial intelligence (XAI) methodology combined with machine learning (ML). This methodology enabled us to identify discriminative metabolites for ME/CFS.
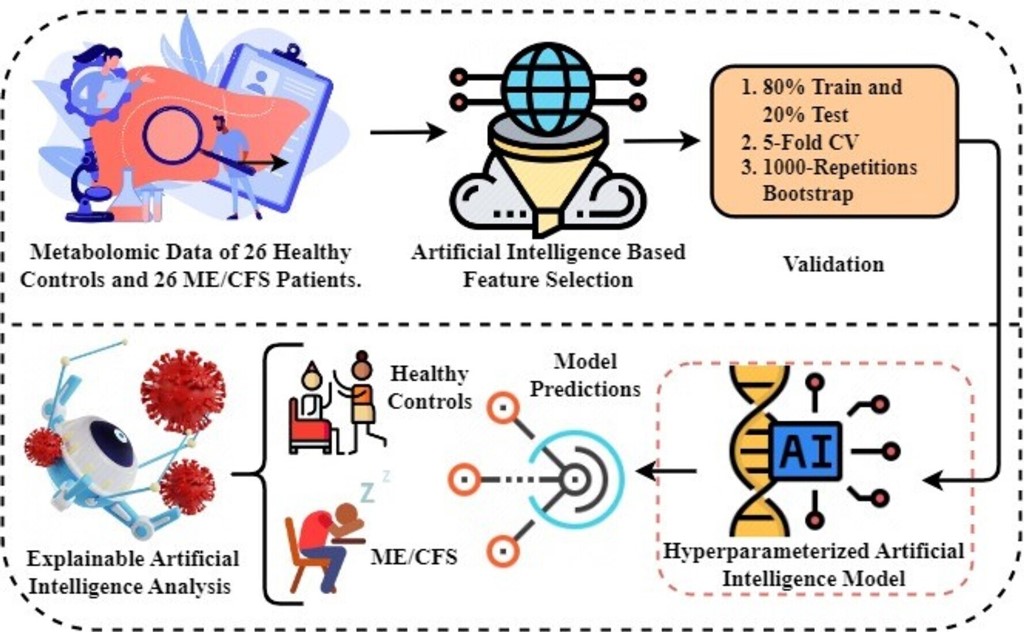
Experimental Setup and Proposed Framework
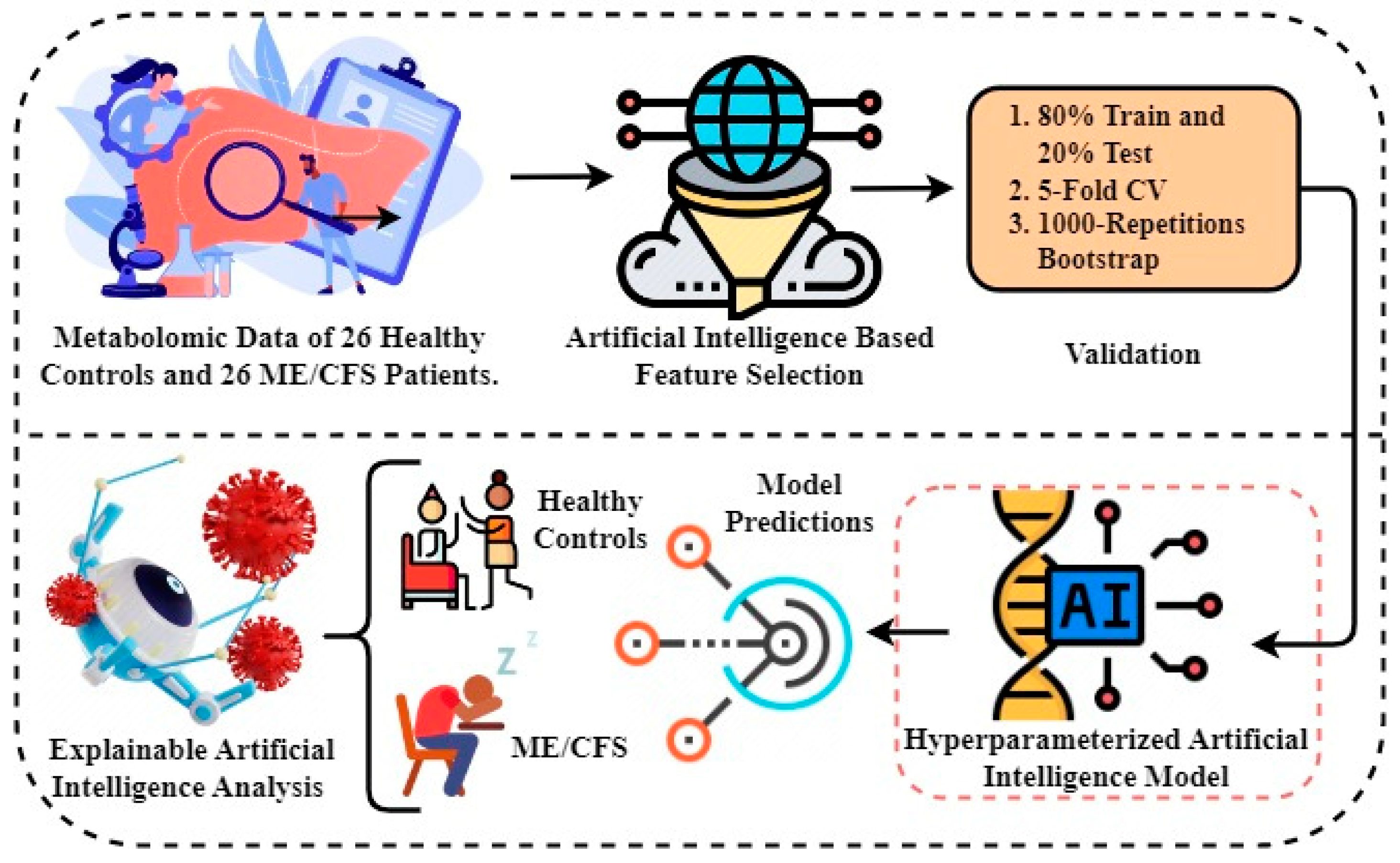
The Python programming language was used to perform the research experiments. The experiments were conducted in an environment containing a graphics processing unit (GPU) backend with 16GB of RAM and 90GB of disk space. An architectural representation of the proposed methodology is depicted in
Figure 1. Diagnosis and biomarker discovery of patients suffering from ME/CFS and healthy controls form the basis of the proposed study. Below is a step-by-step description of the proposed methodology:
The first step involves obtaining metabolomics data to be used in experiments. Metabolomics data are based on results from a study of 26 healthy controls and 26 ME/CFS patients aged 22 to 72 years with similar body mass index (BMI).
In the second step, artificial intelligence-based random forest (RF) feature selection is applied to identify biomarker candidate metabolites and to eliminate the high dimensionality problem in omic data. Because the metabolomics data has a large number of feature dimensions, the performance scores of the predicted models may be lower. Therefore, the twenty most important metabolites contributing to improved performance scores in ME/CFS prediction were identified.
In the third step, 80%-20% split, 5-fold cross-validation (CV), and 1000 replicates Bootstrap approaches were used to validate the prediction models to be generated using the selected biomarker candidate metabolites, and the results were compared.
In the fourth step, Bayesian hyper-parameter optimization was used to determine the optimal parameters.
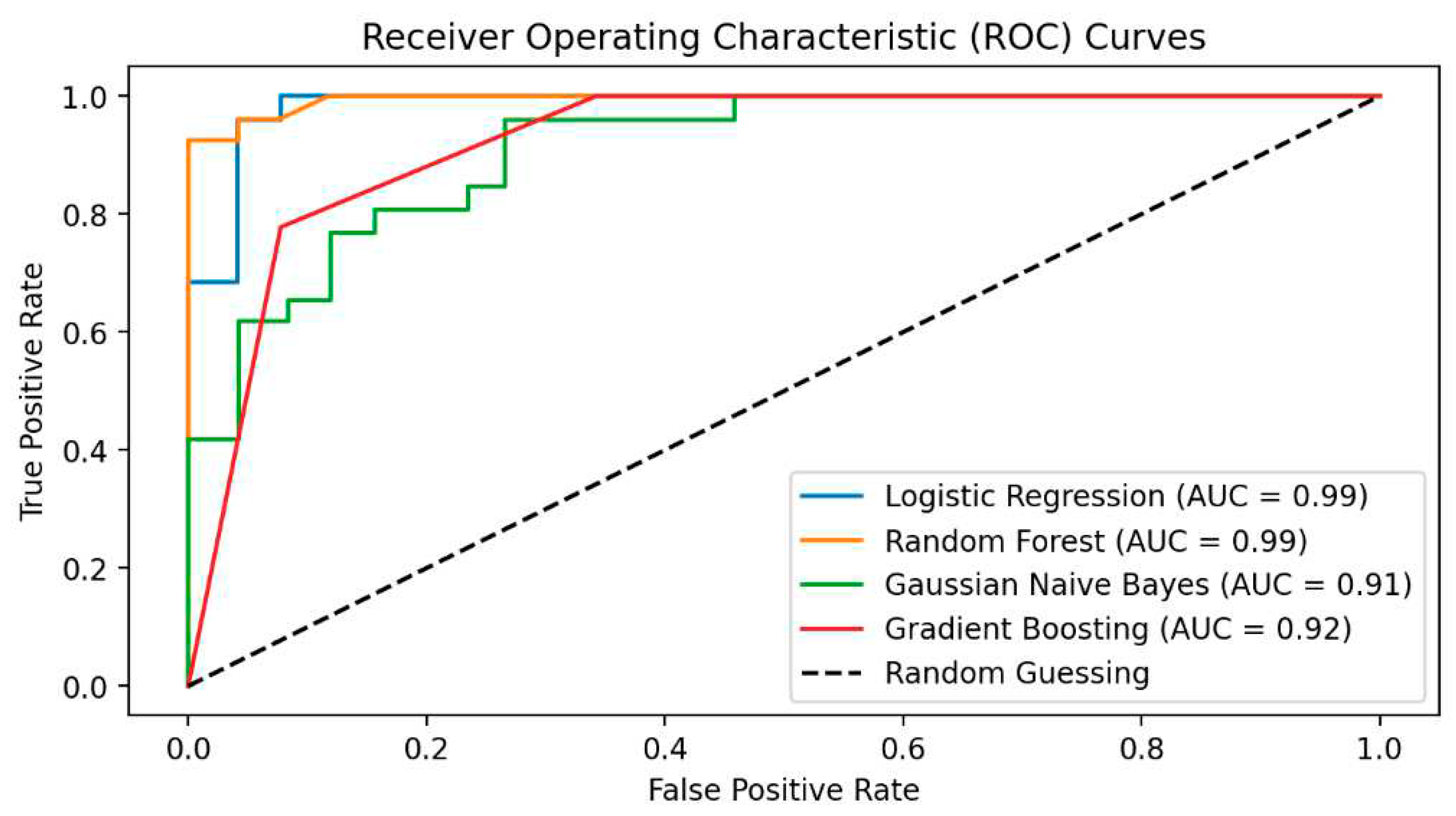
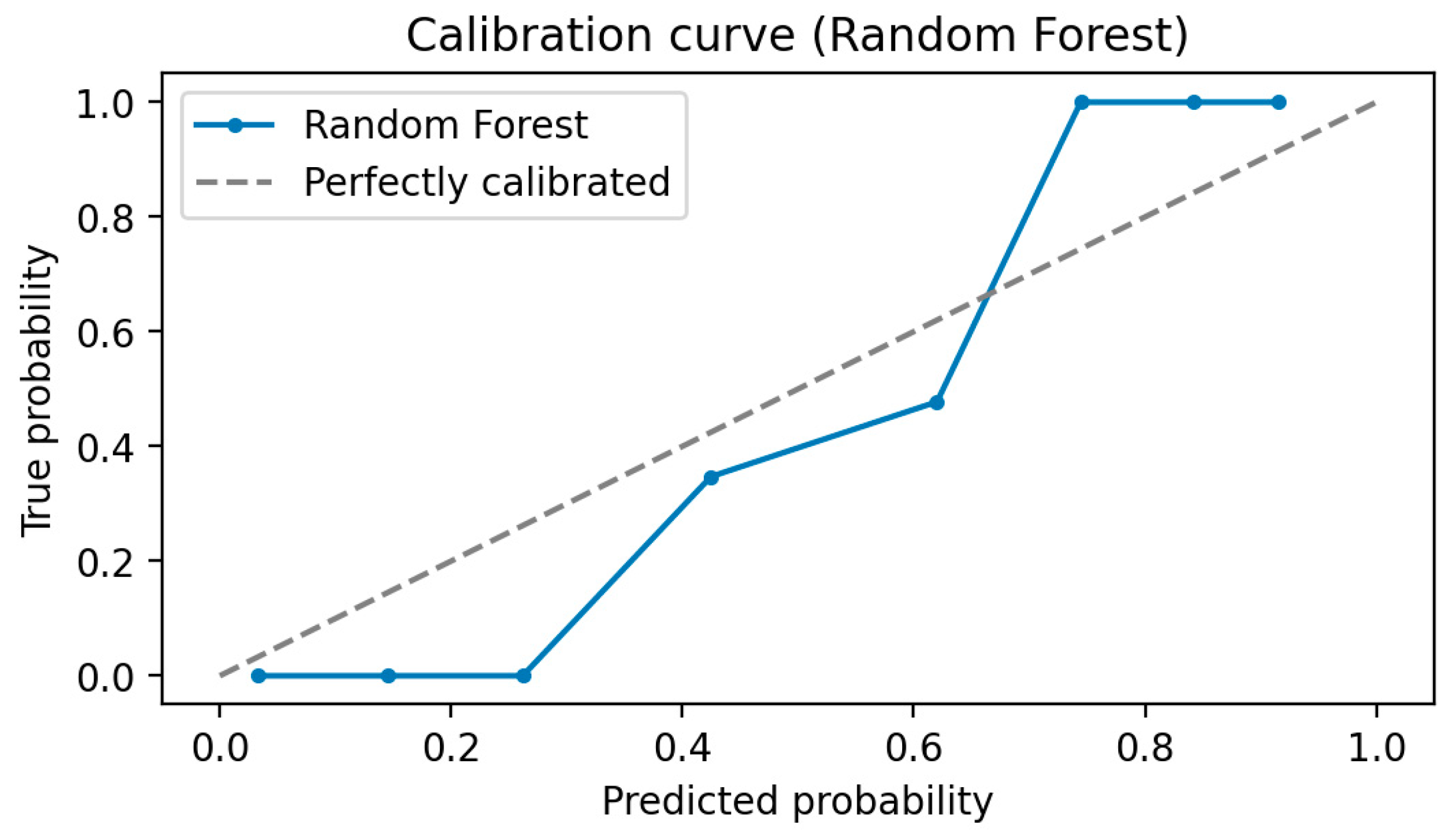
In the fifth step, predictive models were built to diagnose ME/CFS patients. For this purpose, Gaussian Naive Bayes (GNB), Gradient Boosting Classifier (GBC), Logistic regression (LR), and Random Forest Classifier (RFC) algorithms were constructed. Performance of the models was evaluated via area under (AUC) Receiver operating characteristic (ROC) Curve, Brier score, accuracy, precision, recall, and F1-score. While the primary purpose of the methodology is biomarker discovery and diagnosis of ME/CFS, an important secondary purpose is to provide users with indicative probability scores. Therefore, we evaluated the quality of the probabilities via a calibration curve and by calculating the Brier score.
Finally, XAI approaches were applied to the proposed model to provide transparency and interpretability to the model and to explain the decisions made by the model. Through the use of XAI, we can grasp both the rationale and the process behind a particular decision made by the proposed model.
Feature selection
For feature selection and dimensional reduction from the utilized metabolomics data, the RF method, which is based on artificial intelligence, has been applied in this study. The mean decrease impurity (MDI) method is commonly utilized to carry out the process of choosing features that are included in the random forest model. The impurity of the decision trees in the forest is used as a factor in the calculation of the important score for each feature. This score is based on the average amount that each feature reduces the impurity of the decision trees. The feature importance score has been normalized in such a way that the total of all features important values is equal to 1. After that, the most important features with the highest scores are chosen to be used for training the models that are being applied. The RF method of feature selection can be mathematically represented as follows:
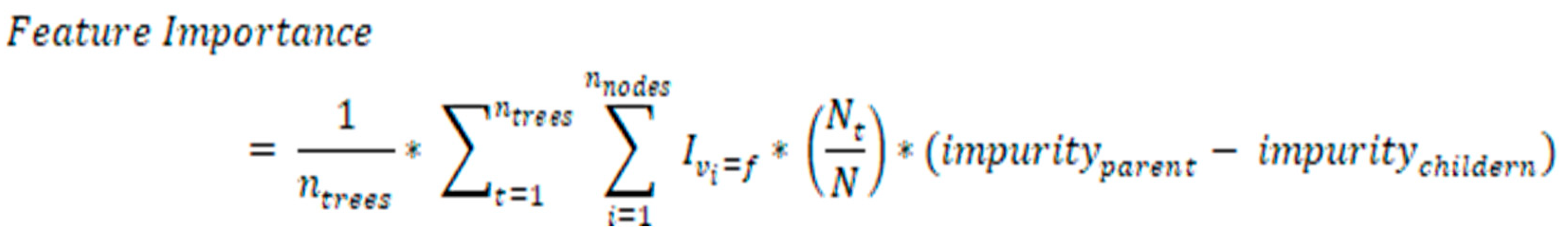
Where:
ntrees is the number of decision trees in the random forest.
vi is the feature used for the split at node i of the t-th tree.
f is the feature being evaluated for importance.
Ivi = f is an indicator function that equals 1 if vi = f and 0 otherwise.
Nt is the number of samples in the t-th tree that reaches node i.
N is the total number of samples in the training set.
impurityparent is the impurity of the set of samples at the parent node i.
impuritychildren is the weighted impurity of the two sets of samples after the split based on feature f.
Validation Methods
To evaluate the distinctive performance and calibration quality of our ML methodology on metabolomics ME/CFS data, in addition to the two classical approaches, hold-out, and k-fold cross-validation, we also used the bootstrap sampling described by Steyerberg et al [
15].
Hold-out Validation: It is the most basic validation method used for ML algorithms and is used to divide the dataset into two as training and test datasets. The training dataset, the training test dataset of the ML model, is used to evaluate the predictive performance of the model [
16].
k-fold Cross-validation: A statistical method for evaluating and comparing learning algorithms, in k-fold cross-validation, the data is first divided into k folds, each of which has a size that is equal to or very close to being equal to the others. Following this, k iterations of training and validation are carried out in such a way that, within each iteration, a different fold of the data is hold-out for validation while the remaining k minus one folds are used for learning [
17].
Bootstrap Validation: The Bootstrap resampling method is a way to predict the fit of a model to a hypothetical test set when an explicit test set is not available. It helps to avoid overfitting and improves the stability of ML algorithms. In this validation method, a set of artificial new datasets is "bootstrapped" by random sampling, replacing the original dataset. Each ML model was then trained on the sampled dataset and evaluated on the original dataset. This process was repeated 1000 times [
18].
The Bayesian Approach for hyper-parameter optimization
The effectiveness of every ML model is determined by the hyper-parameters associated with that model. They have influence over the learning process or the structure of the statistical model that lies beneath the surface. On the other hand, there is no standard approach to selecting hyper-parameters in real experiments. As a substitute, practitioners frequently set hyper-parameters through a process of trial and error or occasionally allow them to remain at their default settings, both of which result in inadequate generalization. By recasting it as an optimization problem, hyper-parameter optimization gives a methodical approach to solving this issue. According to this line of thinking, a good set of hyper-parameters should (at the very least) minimize a validation error. When compared to the vast majority of other optimization problems that can arise in machine learning, hyper-parameter optimization is a nested problem. This means that at each iteration, an ML model needs to be trained and validated. Many approaches have been developed to discover the optimal combination of ML model hyper-parameters. Grid search and random search are two optimization approaches that are often employed for this purpose. These strategies, however, have a few drawbacks. Grid searching is a time-consuming and inefficient strategy for the central processing unit (CPU) and graphics processing unit (GPU). The grid search strategy outperforms random search; nevertheless, the exact answer is more likely to be ignored. In comparison to these two strategies, Bayesian optimization is the best choice for searching for hyper-parameters. First, because the Gaussian process is involved, the Bayesian optimization technique may consider prior results. To put it another way, each step computation may be retrieved to assist in determining a better set of hyper-parameters. Second, compared to other methodologies (for example, grid search), Bayesian optimization takes fewer iterations and has a quicker processing time. Finally, even when working with non-convex issues, Bayesian optimization may be trusted [
19,
20,
21,
22].
Classification models
To identify patients into two categories, namely ME/CFS and healthy individuals, we made use of a variety of AI-based classification algorithms in this work. These included Gaussian Naive Bayes (GNB), Gradient Boosting Classifier (GBC), Logistic regression (LR), and Random Forest Classifier (RFC).
GNB: The GNB algorithm is a well-known classification method that is frequently utilized in the field of biomedical research to categorize various patient groups. GNB can be used to properly diagnose patients based on specific physiological traits or biomarkers in the case of healthy individuals as well as patients with ME/CFS. The Bayes theorem, which asserts that the probability of a hypothesis may be computed based on the probability of observing specific evidence, serves as the foundation for GNB's mathematical operation. This theorem underpins how GNB works. When using GNB, it is assumed that the conditional probability of each feature given the class is Gaussian, which indicates that the features are regularly distributed within each class. This is done to comply with the requirements of the GNB algorithm. This assumption makes the computation of the posterior probability much easier, which in turn enables classification that is both more efficient and more accurate. The GNB method works by first determining the posterior probability of each class for a certain set of features, and then designating the class that has the highest probability as the class that will be predicted [
23,
24]. The following mathematical notations can be used to express GNB:
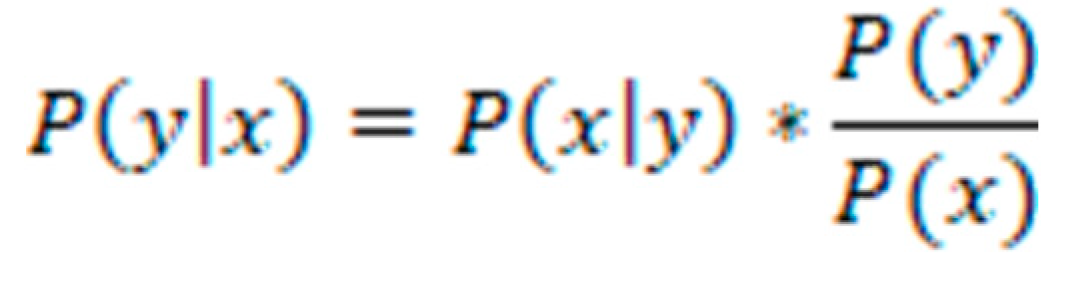
Where:
P(y|x) is the posterior probability of class y given input vector x.
P(x|y) is the likelihood of the input vector x given class y, modeled as a multivariate Gaussian distribution.
P(y) is the prior probability of class y, estimated as the relative frequency of y in the training set.
P(x) is the evidence or marginal likelihood of the input vector x, calculated as the sum of the joint probabilities of x and all possible classes y.
GBC: The GBC is a powerful ML algorithm that has shown great potential in the classification of healthy individuals and patients with ME/CFS. GBC operates by iteratively constructing an ensemble of weak prediction models, typically decision trees, and combining their outputs to make accurate predictions. During the training phase, GBC builds the ensemble by initially fitting a weak model to the training data. Subsequent models are then constructed in a way that each new model focuses on the instances that were previously misclassified by the ensemble [
25,
26]. The mathematical notations for the GBC model for classification are as follows:
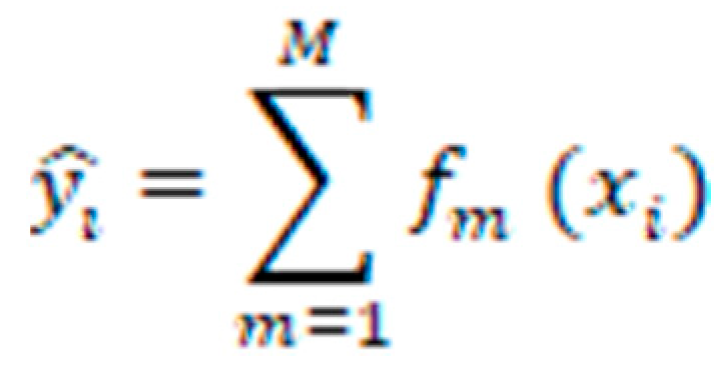
Where:
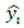
represents the predicted value for the i-th instance.
M denotes the number of weak classifiers (decision trees) used in the GBC.
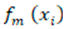
refers to the m-th weak classifier's prediction for the i-th instance.
LR: For binary classification problems such as disease categorization using patient data, LR is a common machine learning model. Given input data including patient demographics, symptoms, and laboratory test results, the LR model calculates the likelihood of a positive class. To maximize the likelihood of the positive class, the model learns the ideal set of weights or coefficients by minimizing the logistic loss function. To produce a probability between 0 and 1, the logistic function uses a linear combination of input features and their weights. Afterward, a threshold (such as 0.5) is used to the anticipated probability to determine the expected class; if the predicted probability is greater than the threshold, the positive class is predicted, and vice versa [
27,
28]. Here are some mathematical symbols for the LR model of binary classification:
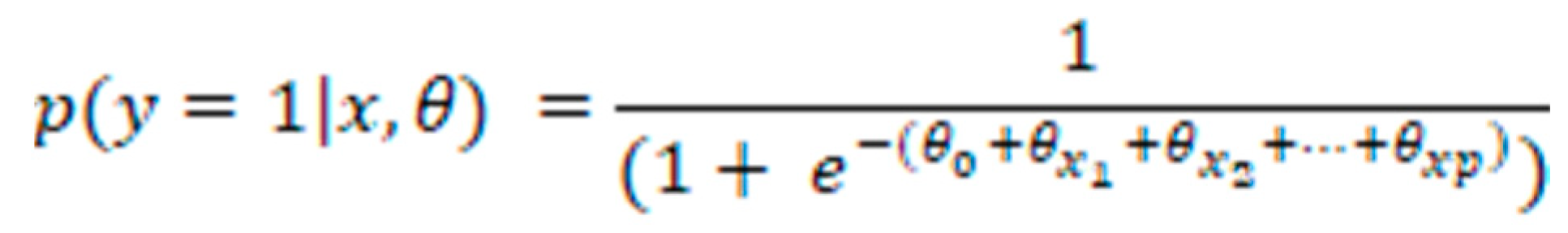
Where:
p(y=1|x,θ) is the predicted probability of the positive class given the input feature vector x and the model parameters θ.
e is the base of the natural logarithm (approximately 2.718).
θ0 is the intercept or bias term.
θx1, θx2, ..., θx0 are the coefficients or weights of the input features x1, x2, ..., xp
x = [x1, x2, ..., xp] is the input feature vector
RFC: The RFC, is a well-known technique for machine learning that is used for classification tasks, such as the classification of diseases based on patient data. Building an ensemble of decision trees that have been trained on random subsets of the input features and data samples is how RFC goes about doing its work. Each decision tree in the ensemble makes a prediction based on a subset of the input features, and the final prediction is generated by aggregating the predictions of all of the trees in the ensemble. RFC can handle high-dimensional data with a large number of features and can also capture nonlinear correlations between the input features and the output classes [
29,
30,
31]. The RFC equation can also be written as follows in its mathematical notation:
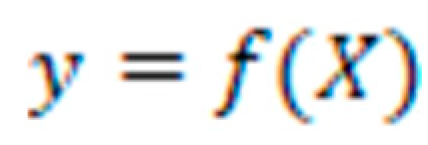
Where:
X is an input data matrix with n samples and p features, where X = [x1, x2, ..., xn], and each xi is a vector of p features.
y is a vector of predicted class labels, where y = [y_1, y_2, ..., y_n].
f(X) is the function that maps the input data X to the predicted class labels y using a random forest model.
Model Calibration
A well-calibrated model is one in which the estimated probability matches the true incidence of the outcome. For example, approximately 90% of patients with an estimated risk of ME/CFS of 0.9 would be classified as ME/CFS. This is critical for prediction models because clinical decision-makers need to know how confident the model is in making a particular prediction. Therefore, we calibrate the trained model to get the correctly predicted probability. In this article, we use the Brier score and Calibration curve for model calibration [
36,
37].
Brier Score: The Brier score is a metric used to evaluate the quality of probability estimates. It is especially used for probabilistic classification models. The Brier score provides a measure of the mean squared errors between the actual labels and the estimated probabilities. The lower the Brier score, the closer the predictions are to reality [
36,
37].
Calibration Curve: A calibration curve is a tool used to evaluate how close a classification model's estimates are to the true probabilities. This curve shows the accuracy of the probabilities predicted by the model. The calibration curve is important to determine the confidence level of the model and to evaluate the reliability of the predictions. A well-calibrated model means that high-probability predictions are more likely to happen, while low-probability predictions should be less likely to happen. It is verified that the probabilities predicted by a well-calibrated model are consistent with the realization rates [
36,
37].
XAI Approach
Interpretability is absolutely essential when using a complex ML model in a real-world environment such as the medical field. XAI is an emerging research area that aims to increase the interpretability and transparency of applied ML models. XAI ensures that decisions made by applied models are understood and trusted, especially in critical applications such as healthcare. XAI techniques can help users understand, validate, and trust the decisions made by these models in real-world applications [
38,
39]. In this research, Shapley values and the Treemap approach were used to interpret the estimation decision of the optimal ML model.
Shapley Additive Explanations (SHAP): It is an approach used to understand the contribution of each feature to the prediction to explain the predictions of SHAP ML models. This approach also takes into account the complexity of the ML model and the interactions between features that go into the model. It also measures the contribution of a feature to the prediction using Shapley values and thus produces graphical results for understanding the model's decisions [
39].
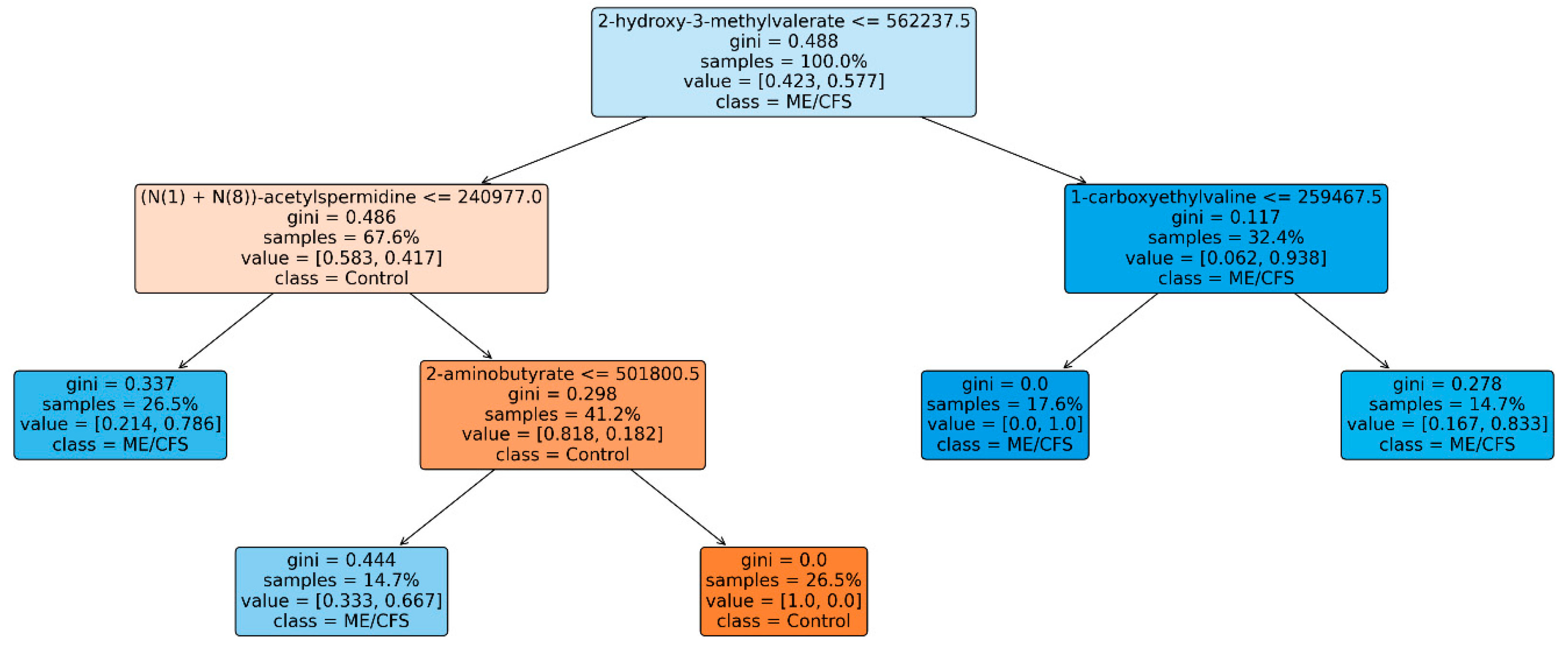
TreeMap: TreeMap provides an intuitive description for tree-based ML models, showing the name of the property used for each decision level and the split value for the condition. If an instance satisfies the condition, it goes to the left branch of the tree, otherwise, it goes to the right branch. When purity is high in TreeMap, the knuckle/leaf has a darker color. The samples row at each node shows the number of samples examined at that node [
40,
41].
3. Results
In this section, firstly, the results of biomarker candidate metabolites are given and the performance results of the applied predictive artificial intelligence algorithms are evaluated using various evaluation metrics. Predictive models were constructed based on both the original data and the metabolites identified as biomarker candidates, and the results were compared. The model showing the final performance was used for ME/CFS estimation and the decision-making function of the model was tried to be explained using XAI approaches.
Feature Selection Results
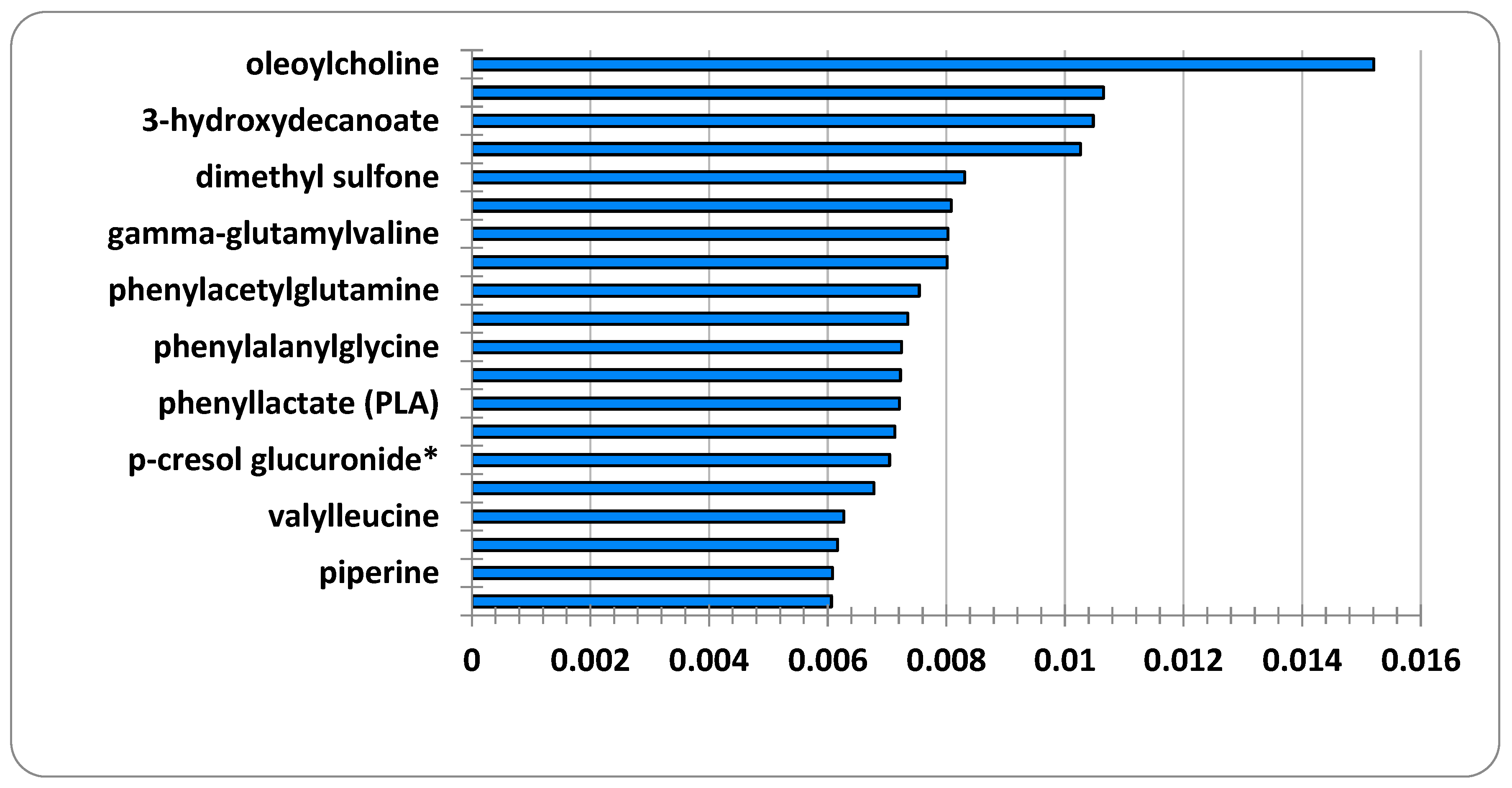
Figure 2 depicts the features that were selected together with their respective relevance ratings, which were determined using a random forest method based on machine learning. The results reveal that the metabolomics of C-glycosyltryptophan, oleoylcholine, cortisone, and 3-hydroxydecanoate are extremely relevant for the diagnosis of ME/CFS patients.
Hyper-parameters Optimization Results
Table 1, optimal hyper-parameters of ML models according to the Bayesian optimization are given.
XAI Results
The SHAP was used to identify metabolomics biomarkers according to their importance or contribution to the prediction of ME/CFS and to explain the prediction decisions of the model. The RFC-trained model was subjected to SHAP annotation and identified the most important trait metabolites responsible for the prediction of ME/CFS. The results pointed to a list of metabolites with importance scores. Metabolite biomarkers are arranged in decreasing order of importance. Oleoylcholine, phenylactate (PLA), octanoylcarnitine (CB), hydroxyasparagine**, and piperine are among the most prominent metabolite biomarkers important in the diagnosis of ME/CFS.
Figure 5 also visualizes the relationships between the relative value of biomarker candidate metabolites and the SHAP values for these metabolites. In each row of the graph, each patient is marked as a dot. The horizontal position of the dot reflects the SHAP values, and the color of the dot encodes the relative value of the metabolites and their mean in the dataset. For example, low values (relatively blue) of Oleoylcholine and phenylactate (PLA) metabolites contribute positively to ME/CFS, thus increasing disease risk (
Figure 5).
In addition to that, to gain an understanding of how the RFC model behaves, we have employed a method that is known as Treemap analysis.
Figure 6 is an illustration of the Treemap that is included in the RFC. The analysis explains how the proposed model came to its conclusion about the classification of patients as healthy controls and those with ME/CFS.
4. Discussion
Fatigue is a common occurrence in human beings and serves as an indicator of disrupted homeostasis within the body, resulting from either excessive physical and mental exertion or illness [
42]. In addition to being one of the most significant social concerns, chronic fatigue additionally constitutes the most significant economic losses [
43]. Pain, cognitive dysfunction, autonomic dysfunction, sleep disturbance, and neuroendocrine and immune symptoms are just some of the many symptoms associated with ME/CFS [
44]. A patient must have a symptom from neurological impairments, an immune/gastrointestinal/genitourinary impairment, and an energy metabolism/transport impairment to be diagnosed with ME/CFS and meet the criteria for post-exertional neuroimmune exhaustion. However, the strength and severity of such symptoms in a patient vary and are heterogeneous, from moderate to severe, with some patients even becoming bed-bound [
44]. Because it is challenging to identify the typical abnormal elements for this disorder utilizing general and conventional medical examination, artificially intelligent-based automated methods may aid in improving the diagnosis of ME/CFS. In recent years, a growing number of research have explained the pathology of ME/CFS and have established biomarkers for the same by employing a metabolome analysis technique [
45,
46,
47]. This has allowed for the development of a variety of diagnostic studies [
48].
The present investigation of the effectiveness of methodology combining ML and XAI techniques to investigate biomarkers of ME/CFS and develop an interpretable predictive model for disease diagnosis. Metabolomics data from patients diagnosed with ME/CFS and healthy controls were used. The classification algorithms employed include GNB, GBC, LR, and RFC. The classifiers' performance has been evaluated both with and without the implementation of the feature selection algorithm (RF). In addition to classical hold-out validation, cross-validation, and bootstrap approaches were also used to evaluate the performance of classification learning algorithms in the validation stage, and the effectiveness of these three validation approaches was also examined. Shapely values, an explainable AI system, have been utilized to interpret the classification models' predictions and decisions. After being trained and validated on the significant selected features using the Bootstrapping method, the RFC model is found to be superior to the other four models (GNB, GBC, and LR). Accuracy, precision, recall, F1 Score, and the AUC were all at or above 98% for the RFC model. The higher the values that attain for precision and sensitivity, the higher the proportion of correct diagnoses, also known as True Positives (TP), and the lower the value of false negatives (FN). Errors both positive and negative, known as false positives (FP) and FN, are widespread in comparative biology research. In addition, we demonstrated that our method was capable of demonstrating the main features as well as the interpretations of ML findings by utilizing SHAPley values and SHAP plots. The SHAP method's findings indicated that oleoylcholine, phenyllactate, octanoylcarnitine, hydrooxyasparagine, piperine, p-cresol glucuronide, and palmitoylcholine are all chemicals associated with ME/CFS and crucial to the model's final decision. The use of the SHAP technique revealed that the indolelactate, which has low Shapley values, is the least significant of all the features. On the other hand, the feature with the highest Shapley value is the oleoylcholine, which is also the one that contributes the most significant information for the diagnosis of ME/CFS. Oleoylcholine is a member of the class of chemical compounds known as acyl cholines. Germain et al. [
2] researched the metabolic pathways that influence the diagnosis of ME/CFS patients by performing statistical analysis in conjunction with pathway enrichment analysis. They found that acyl cholines, which are part of the sub-pathway of lipid metabolism known as fatty acid metabolism, are consistently reduced in two different patient cohorts that suffer from ME/CFS. Nagy-Szakal et al. [
49] have gained insights into ME/CFS phenotypes through comprehensive metabolomics. Biomarker identification and topological analysis of plasma metabolomics data were performed on a sample group consisting of fifty ME/CFS patients and fifty healthy controls. They have demonstrated that patients with ME/CFS have higher plasma levels of ceramide and observed that there is a variation in the level of carnitine, choline, and complex lipid metabolites. The study of plasma metabolomics data attained a more accurate prediction model of ME/CFS (AUC = 0.836).
A comprehensive metabolomics was conducted by Naviaux et al. [
49] to better understand the biology of CFS. They have investigated 612 plasma metabolites across 63 different metabolic pathways. Twenty metabolic pathways were revealed to be abnormal in patients with Chronic Fatigue Syndrome. Sphingolipid, phospholipid, purine, cholesterol, microbiota, pyrroline-5-carboxylate, riboflavin, branch chain amino acid, peroxisomal, and mitochondrial pathways were all disrupted. Diagnostic accuracies of 94% were found using AUC characteristic curve analysis. In our experiment utilizing the ML-based model, we were able to achieve a greater level of accuracy (98%) for our proposed prediction model, the RFC model. Petrick, and Shomron [
50] has been discussed how well the ML-based model performs. They highlighted how AI and ML have permitted important breakthroughs in untargeted metabolomics workflows and key findings in the fields of disease diagnosis. In conclusion, the proposed model (RFC) was successful in correctly diagnosing ME/FCS patients. The findings indicate that ML, when paired with the Shapely analysis, is able to explain the ME/FCS classification model and offer physicians a basic knowledge of the main metabolic chemicals that influence the model decision. Clinicians can get benefit from individual explanations of the important metabolic compounds in order to gain a better grasp of why the model yields certain diagnoses for individuals with ME/CFS.
5. Conclusions
Although research into the causes and mechanisms of ME/CFS continues, the exact underlying factors are not yet fully understood. It has been reported to result from a complex interaction of biological, genetic, environmental, and psychological factors. Advances in research are crucial to better understanding the disease, improving diagnosis and treatment options, and ultimately finding a cure. Based on this information, the RFC model proposed in this study correctly classified and evaluated ME/CFS patients through the selected biomarker candidate metabolites. The methodology combining ML and XAI can provide a clear interpretation of risk estimation for ME/CFS, helping physicians intuitively understand the impact of key metabolomics features in the model.
6. Limitations and future works
This study lacked a third-party verification by an independent biologist, which may have provided more explanation of the collected results, vital metabolic chemicals, and their significance to the diagnosis of patients with ME/FCS. It is vital to broaden the present investigation further by incorporating multicenter experiments in subsequent research or to make use of the associated data from multiple locations for external validation. The size of the metabolomics dataset might be increased by collecting additional samples from patients. This would be an improvement for this line of investigation. The performance of patient diagnosis will be improved by the development of advanced transfer learning-based methodologies.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Table S1: Supplementary file 1 and Table S2: Supplementary file 2.
Author Contributions
Conceptualization F.H.Y., A.A., C.C., and A.R.; methodology, F.Y.H.; software, F.H.Y., A.A., C.C., B.Y., and A.R.; validation F.H.Y. A.A., C.C., and A.R.; formal analysis, F.H.Y., B.Y., and A.R.; investigation, F.H.Y., A.A., C.C., B.Y., A.R., N.A.S., and N.F.M.; resources, F.H.Y., and B.Y.; data curation, F.H.Y.; writing—original draft preparation, F.H.Y., A.A., C.C., B.Y., A.R., N.A.S., and N.F.M.; writing—review and editing, F.H.Y., A.A., C.C., B.Y., A.R., N.A.S., and N.F.M; visualization, F.H.Y., and C.C.; supervision, C.C.. All authors have read and agreed to the published version of the manuscript.
Funding
Princess Nourah bint Abdulrahman University Researchers Supporting Project Number PNURSP2023R206, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Data Availability Statement
Acknowledgments
The authors express their gratitude to Princess Nourah bint Abdulrahman University Researchers Supporting Project Number PNURSP2023R206, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Toogood PL, Clauw DJ, Phadke S, Hoffman D. Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS): Where will the drugs come from? Pharmacological Research. 2021, 165, 105465. [CrossRef]
- Germain A, Barupal DK, Levine SM, Hanson MR. Comprehensive circulatory metabolomics in ME/CFS reveals disrupted metabolism of acyl lipids and steroids. Metabolites. 2020, 10, 34. [CrossRef] [PubMed]
- Malato J, Graça L, Sepúlveda N. Impact of imperfect diagnosis in ME/CFS association studies. medRxiv. 2022, 2022.06. 08.22276100.
- Valdez AR, Hancock EE, Adebayo S, Kiernicki DJ, Proskauer D, Attewell JR, et al. Estimating prevalence, demographics, and costs of ME/CFS using large scale medical claims data and machine learning. Frontiers in pediatrics. 2019, 6, 412. [CrossRef]
- Faro M, Sàez-Francás N, Castro-Marrero J, Aliste L, de Sevilla TF, Alegre J. Gender differences in chronic fatigue syndrome. Reumatología clínica (English edition). 2016, 12, 72–77.
- Marshall-Gradisnik S, Eaton-Fitch N. Understanding myalgic encephalomyelitis. Science. 2022, 377, 1150–1151. [CrossRef]
- Malkova A, Shoenfeld Y. Autoimmune autonomic nervous system imbalance and conditions: Chronic fatigue syndrome, fibromyalgia, silicone breast implants, COVID and post-COVID syndrome, sick building syndrome, post-orthostatic tachycardia syndrome, autoimmune diseases and autoimmune/inflammatory syndrome induced by adjuvants. Autoimmunity reviews. 2022, 103230.
- Dehhaghi M, Panahi HKS, Kavyani B, Heng B, Tan V, Braidy N, et al. The role of kynurenine pathway and NAD+ metabolism in myalgic encephalomyelitis/chronic fatigue syndrome. Aging and disease. 2022, 13, 698. [CrossRef] [PubMed]
- Nunes JM, Kell DB, Pretorius E. Cardiovascular and haematological pathology in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS): A role for viruses. Blood reviews. 2023, 101075.
- Hornig M, Montoya JG, Klimas NG, Levine S, Felsenstein D, Bateman L, et al. Distinct plasma immune signatures in ME/CFS are present early in the course of illness. Science advances. 2015, 1, e1400121. [CrossRef]
- Shan ZY, Barnden LR, Kwiatek RA, Bhuta S, Hermens DF, Lagopoulos J. Neuroimaging characteristics of myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS): a systematic review. Journal of translational medicine. 2020, 18, 1–11.
- Navaneetharaja N, Griffiths V, Wileman T, Carding SR. A role for the intestinal microbiota and virome in myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS)? Journal of clinical medicine. 2016, 5, 55. [CrossRef]
- Maes M, Leunis J-C, Geffard M, Berk M. Evidence for the existence of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) with and without abdominal discomfort (irritable bowel) syndrome. Neuroendocrinol Lett. 2014, 35, 445–453.
- Germain A, Giloteaux L, Moore GE, Levine SM, Chia JK, Keller BA, et al. Plasma metabolomics reveals disrupted response and recovery following maximal exercise in myalgic encephalomyelitis/chronic fatigue syndrome. JCI insight. 2022, 7.
- Steyerberg EW, Harrell Jr FE, Borsboom GJ, Eijkemans M, Vergouwe Y, Habbema JDF. Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. Journal of clinical epidemiology. 2001, 54, 774–781.
- Levman J, Ewenson B, Apaloo J, Berger D, Tyrrell PN. Error Consistency for Machine Learning Evaluation and Validation with Application to Biomedical Diagnostics. Diagnostics. 2023, 13, 1315. [CrossRef]
- Zhang X, Liu C-A. Model averaging prediction by K-fold cross-validation. Journal of Econometrics. 2023, 235, 280–301. [CrossRef]
- Diniz, MA. Statistical methods for validation of predictive models. Journal of Nuclear Cardiology. 2022, 29, 3248–3255. [Google Scholar] [CrossRef]
- Zhang J, Wang Q, Shen W. Hyper-parameter optimization of multiple machine learning algorithms for molecular property prediction using hyperopt library. Chinese Journal of Chemical Engineering. 2022, 52, 115–125. [CrossRef]
- Wu J, Chen X-Y, Zhang H, Xiong L-D, Lei H, Deng S-H. Hyperparameter optimization for machine learning models based on Bayesian optimization. Journal of Electronic Science and Technology. 2019, 17, 26–40.
- Jones, DR. A taxonomy of global optimization methods based on response surfaces. Journal of global optimization. 2001, 21, 345–383. [Google Scholar] [CrossRef]
- Yagin FH, Gülü M, Gormez Y, Castañeda-Babarro A, Colak C, Greco G, et al. Estimation of Obesity Levels with a Trained Neural Network Approach optimized by the Bayesian Technique. Applied Sciences. 2023, 13, 3875. [CrossRef]
- Mansourian P, Zhang N, Jaekel A, Zamanirafe M, Kneppers M. Anomaly Detection for Connected Autonomous Vehicles Using LSTM and Gaussian Naïve Bayes. International Conference on Wireless and Satellite Systems: Springer; 2023. p. 31-43.
- Maniruzzaman M, Rahman MJ, Ahammed B, Abedin MM, Suri HS, Biswas M, et al. Statistical characterization and classification of colon microarray gene expression data using multiple machine learning paradigms. Computer methods and programs in biomedicine. 2019, 176, 173–193. [CrossRef]
- Iqbal A, Barua K. A real-time emotion recognition from speech using gradient boosting. 2019 international conference on electrical, computer and communication engineering (ECCE): IEEE; 2019. p. 1-5.
- Alshboul O, Shehadeh A, Almasabha G, Almuflih AS. Extreme gradient boosting-based machine learning approach for green building cost prediction. Sustainability. 2022, 14, 6651. [CrossRef]
- Shah K, Patel H, Sanghvi D, Shah M. A comparative analysis of logistic regression, random forest and KNN models for the text classification. Augmented Human Research. 2020, 5, 1–16.
- Muharemi F, Logofătu D, Leon F. Machine learning approaches for anomaly detection of water quality on a real-world data set. Journal of Information and Telecommunication. 2019, 3, 294–307. [CrossRef]
- Ilyas H, Ali S, Ponum M, Hasan O, Mahmood MT, Iftikhar M, et al. Chronic kidney disease diagnosis using decision tree algorithms. BMC nephrology. 2021, 22, 1–11.
- Sattari MT, Apaydin H, Shamshirband S. Performance evaluation of deep learning-based gated recurrent units (GRUs) and tree-based models for estimating ETo by using limited meteorological variables. Mathematics. 2020, 8, 972. [CrossRef]
- Daneshvar D, Behnood A. Estimation of the dynamic modulus of asphalt concretes using random forests algorithm. International Journal of Pavement Engineering. 2022, 23, 250–260. [CrossRef]
- Yacouby R, Axman D. Probabilistic extension of precision, recall, and f1 score for more thorough evaluation of classification models. Proceedings of the first workshop on evaluation and comparison of NLP systems2020. p. 79-91.
- Bowers AJ, Zhou X. Receiver operating characteristic (ROC) area under the curve (AUC): A diagnostic measure for evaluating the accuracy of predictors of education outcomes. Journal of Education for Students Placed at Risk (JESPAR). 2019, 24, 20–46. [CrossRef]
- Nahm, FS. Receiver operating characteristic curve: overview and practical use for clinicians. Korean journal of anesthesiology. 2022, 75, 25–36. [Google Scholar] [CrossRef]
- Muschelli III, J. ROC and AUC with a binary predictor: a potentially misleading metric. Journal of classification. 2020, 37, 696–708. [Google Scholar] [CrossRef]
- Huang Y, Li W, Macheret F, Gabriel RA, Ohno-Machado L. A tutorial on calibration measurements and calibration models for clinical prediction models. Journal of the American Medical Informatics Association. 2020, 27, 621–633. [CrossRef]
- Liu J, Wang C, Yan R, Lu Y, Bai J, Wang H, et al. Machine learning-based prediction of postpartum hemorrhage after vaginal delivery: combining bleeding high risk factors and uterine contraction curve. Archives of Gynecology and Obstetrics. 2022, 306, 1015–1025. [CrossRef]
- Borys K, Schmitt YA, Nauta M, Seifert C, Krämer N, Friedrich CM, et al. Explainable AI in medical imaging: An overview for clinical practitioners–Beyond saliency-based XAI approaches. European journal of radiology. 2023, 110786.
- Yagin FH, Cicek İB, Alkhateeb A, Yagin B, Colak C, Azzeh M, et al. Explainable artificial intelligence model for identifying COVID-19 gene biomarkers. Computers in Biology and Medicine. 2023, 154, 106619. [CrossRef]
- Khanna VV, Chadaga K, Sampathila N, Prabhu S, Bhandage V, Hegde GK. A distinctive explainable machine learning framework for detection of polycystic ovary syndrome. Applied System Innovation. 2023, 6, 32. [CrossRef]
- Chatterjee J, Dethlefs N. Scientometric review of artificial intelligence for operations & maintenance of wind turbines: The past, present and future. Renewable and Sustainable Energy Reviews. 2021, 144, 111051.
- Tanaka M, Tajima S, Mizuno K, Ishii A, Konishi Y, Miike T, et al. Frontier studies on fatigue, autonomic nerve dysfunction, and sleep-rhythm disorder. The Journal of Physiological Sciences. 2015, 65, 483–498. [CrossRef]
- Yamano E, Watanabe Y, Kataoka Y. Insights into metabolite diagnostic biomarkers for myalgic encephalomyelitis/chronic fatigue syndrome. International Journal of Molecular Sciences. 2021, 22, 3423. [CrossRef] [PubMed]
- Group ICFSS. The chronic fatigue syndrome: A comprehensive approach to its definition and study. Annals of Internal Medicine. 1994, 121, 953–959. [CrossRef] [PubMed]
- Armstrong CW, McGregor NR, Lewis DP, Butt HL, Gooley PR. The association of fecal microbiota and fecal, blood serum and urine metabolites in myalgic encephalomyelitis/chronic fatigue syndrome. Metabolomics. 2017, 13, 1–13.
- Tomas C, Newton J. Metabolic abnormalities in chronic fatigue syndrome/myalgic encephalomyelitis: a mini-review. Biochemical Society Transactions. 2018, 46, 547–553. [CrossRef]
- Huth TK, Eaton-Fitch N, Staines D, Marshall-Gradisnik S. A systematic review of metabolomic dysregulation in chronic fatigue syndrome/myalgic encephalomyelitis/systemic exertion intolerance disease (CFS/ME/SEID). Journal of translational medicine. 2020, 18, 1–14.
- Jason LA, Boulton A, Porter NS, Jessen T, Njoku MG, Friedberg F. Classification of myalgic encephalomyelitis/chronic fatigue syndrome by types of fatigue. Behavioral Medicine. 2010, 36, 24–31. [CrossRef] [PubMed]
- Nagy-Szakal D, Barupal DK, Lee B, Che X, Williams BL, Kahn EJ, et al. Insights into myalgic encephalomyelitis/chronic fatigue syndrome phenotypes through comprehensive metabolomics. Scientific reports. 2018, 8, 10056. [CrossRef] [PubMed]
- Petrick LM, Shomron N. AI/ML-driven advances in untargeted metabolomics and exposomics for biomedical applications. Cell Reports Physical Science. 2022, 3.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
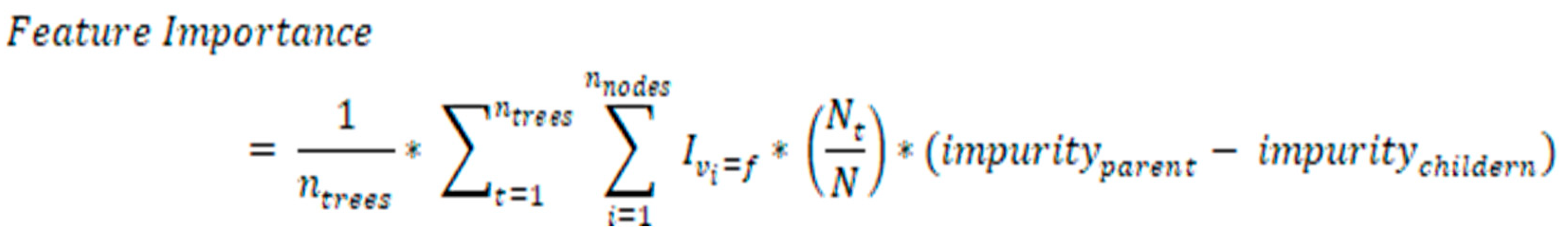
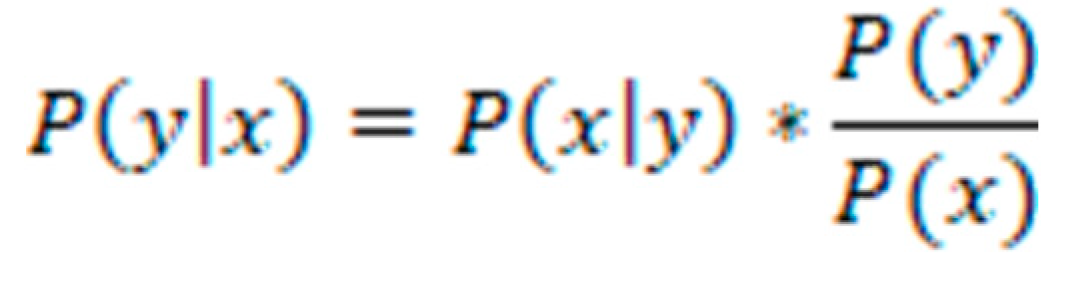
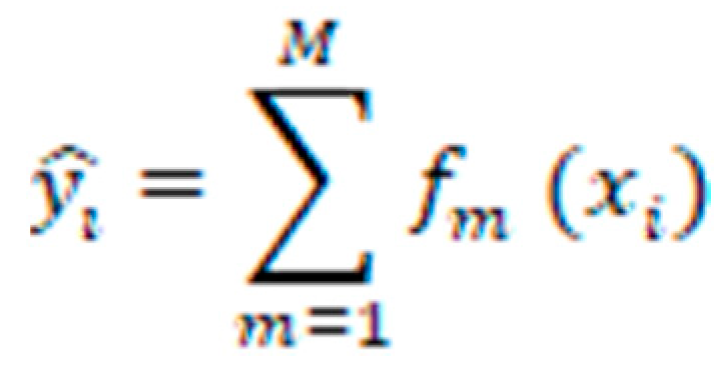
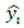 represents the predicted value for the i-th instance.
represents the predicted value for the i-th instance.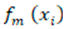 refers to the m-th weak classifier's prediction for the i-th instance.
refers to the m-th weak classifier's prediction for the i-th instance.