1. Introduction
Heart failure (HF) is a significant global health concern, leading to substantial rates of illness and death. The prevalence of heart failure (HF) is on the rise in both developed and developing nations, primarily attributed to the extension of life expectancy. The leading factors contributing to this phenomenon are the escalating incidence of chronic ischemic heart diseases and hypertension. Therefore, healthcare costs are increasing day by day [
1] and this situation negatively affects the quality of life of individuals. In the European Society of Cardiology (ESC) 2021 HF guidelines, HF is classified as preserved ejection fraction HF (HFpEF) (Left Ventricular Ejection Fraction (LVEF) > 50%), mildly reduced ejection fraction HF (HFmrEF) (LVEF < 41%-49%) and reduced ejection fraction HF (HFrEF) (LVEF < 40%), respectively [
2].
Acute heart failure (AHF) is characterized by the sudden or gradual sign of symptoms and indications associated with heart failure. The prevailing etiology of this condition is the abrupt deterioration of chronic heart failure. This clinical condition holds significant importance in the medical field as it necessitates hospitalization and has the potential to result in mortality. Hence, prompt initiation of diagnosis and treatment for patients admitted to the emergency department is imperative [
3]. In the diagnosis of AHF, brain natriuretic peptide (BNP) and N-terminal proBNP (NT-proBNP) are employed alongside electrocardiography (ECG), echocardiography (ECO), blood tests, as well as clinical signs and complaints. When the B-type natriuretic peptide (BNP) is not available, a chest X-ray of the pulmonary artery (PA) can be utilized. A BNP level below 100 pg/ml, and NT-proBNP level below 300 pg/ml are indication of the absence of acute heart failure [
4,
5,
6]. However, BNP and NT-proBNP values may be low in end-stage heart failure, acute pulmonary edema, and acute right heart failure. On the contrary, elevated levels of certain markers may not necessarily be indicative of the presence of acute heart failure. The prevalence of this condition is notably elevated in individuals diagnosed with chronic renal failure and atrial fibrillation [
7].
Anemia is the most significant and independent driver of HF mortality, according to numerous studies that have used hematological markers to predict HF prognosis[
8,
9]. Furthermore, it has been observed that red-cell distribution width (RDW) is correlated with mortality, regardless of the presence of anemia [
10,
11,
12]. Previous research has indicated that elevated leukocyte levels and decreased lymphocyte counts are indicative of increased mortality risk in AHF [
13]. At present, there is a lack of a prompt and conclusive laboratory assay for the diagnosis of acute heart failure. The existing literature primarily focuses on the prognostic implications of hematological parameters.
Artificial intelligence (AI) plays a crucial role in numerous clinical decision support systems, facilitating the use of computational methods to make inferences that are comparable to human reasoning processes [
14]. The strategies presented in this context are founded upon medical information that has been either explicitly encoded or automatically generated from medical data using machine learning techniques. Explainable AI has the potential to facilitate the prioritisation of patients' well-being and enable them to make independent and well-informed choices regarding their healthcare in conjunction with medical professionals [
15]. There is considerable heterogeneity in the quality of currently available prognostic models for acute heart failure. Conversely, the predictive models for post-AHF disability and mortality are constrained by limited sample sizes, restricted clinical and risk factor ranges, and limited generalizability in clinical practise. Based on the projected life expectancy and clinical background, an accurate prognosis can enhance the process of post-acute heart failure discharge planning and aid healthcare professionals in tailoring intensive treatment or palliative care to the individual patient. The present study seeks to examine hematological indicators as predictors of AHF by analyzing the explanations provided by XAI, drawing from significant findings in various medical studies.
2. Materials and Methods
2.1. Study design and dataset
The present study was designed an observational type of research. Patients admitted to our hospital due to acute heart failure between January 2020 and April 2023 were retrospectively included in the study. Patients with AHF were divided into three groups according to their left ventricular ejection fraction: HFpEF (LVEF > 50%), HFmrEF (LVEF < 41%-49%), and HFrEF (LVEF < 40%) as specified by transthoracic echocardiography. The inclusion criteria in the current study were as follows: 425 patients older than 18 years hospitalized for worsening HF with two or more signs or symptoms of fluid retention (eg, dyspnea, paroxysmal nocturnal dyspnea, orthopnea, ankle edema, or jugular venous distension) and 430 healthy control group, individuals without HF who applied to the cardiology department. With G*Power software (University of Dusseldorf, Dusseldorf, Germany, version 3.0.1), the independent sample t-test was used to calculate sample size and actual power (α = 0.05, power = 0.80, effect size = 0.35). The results revealed that with a sample size of 260 participants, the actual power was 80.2% [
16].
The exclusion criteria from the study were as follows: Patients with hematological malignancies; those who use drugs known to affect the complete blood count; those with a hemoglobin value of less than 10 g/dl, those with active bleeding, those with acute or chronic infections, those with acute myocardial infarction, severe renal impairment (estimated glomerular filtration rate (eGFR) <15 ml/min/1.73 m²), severe liver impairment or with chronic obstructive pulmonary disease; those with chronic inflammatory diseases. The study was conducted in accordance with the Declaration of Helsinki, and approved by the Samsun University Clinical Research Ethics Committee (protocol code 2023/10/10 and 24.05.2023).
Age, gender, underlying diseases, LVEF percentage, clinical and echocardiographic characteristics were obtained from the medical records of the patients and the control group at the first admission to the hospital. At the same time, the hematological and biochemical laboratory results obtained from the venous blood taken at the first application to the emergency department of both groups; urea, creatinine, c-reactive protein (CRP), aspartate transaminase (AST), alkaline phosphatase (ALT), hemoglobin (Hbg) values, hematocrit (Hct) values, mean corpuscular volume (MCV) values, mean corpuscular hemoglobin (MCH) values, MCH concentration (MCHC) values, red-cell distribution width-standard deviation (RDW-SD) values, RDW-coefficient of variation (RDW-CV) values, mean platelet volume (MPV) values, platelet width of distribution (PDW) values, procalcitonin (PCT) values, erythrocyte (RBC) platelet (PLT) counts and white blood cell (WBC), neutrophil (NEU), lymphocyte (LY), neutrophil-lymphocyte ratio (NLR), basophil (BA), monocytes (MO) and eosinophil (EO) counts were reached (The dataset is available from
Supplementary Materials).
2.2. Statistical Analysis
The conformity of the variables to the normal distribution was examined by visual (histogram and probability graphs) and analytical (Shapiro-Wilk Test) methods. The assumption of homogeneity of variances was examined with the Levene test. Descriptive statistics are expressed as median, interquartile range for non-normally distributed variables and mean ± standard deviation for normally distributed variables. Independent Samples t-test was used in the comparison of the variables that met the parametric test assumptions of the two groups. The Mann-Whitney U test was used to compare the two groups in terms of variables that did not meet the parametric test assumptions. Frequency (n) and percentage (%) values were calculated for the qualitative variables, and the relationships between the two qualitative variables were examined using the Chi-square test. A p-value of <0.05 was considered statistically significant in all results. Statistical analyzes were performed using the SPSS 28.0 (IBM Corp., Armonk, NY, United States) package program.
2.3. ML and XAI Approach
The Least Absolute Shrinkage and Selection Operator (LASSO) feature selection algorithm was employed in the research to ascertain the primary risk factors associated with Acute Heart Failure. Following the implementation of LASSO, a predictive model was constructed utilizing the variables that were chosen during the selection process [
17] The utilization of the XGBoost algorithm, renowned for its exceptional performance, scalability, and adaptability, was employed in the prognostication of Acute Heart Failure. XGBoost is widely acknowledged as a robust algorithm for handling structured data and employs a boosting technique that progressively incorporates new models derived from the collective knowledge of the community. During each iteration, the algorithm assesses the performance of the current models and proceeds to train a new model with the objective of minimizing errors made by the ensemble [
18,
19]. The validation method employed in this study was 10-fold cross-validation. The model's performance was assessed using several evaluation metrics, including Accuracy, PPV, NPV, F1-score, Area Under the Curve (AUC), and the Receiver Operating Characteristic (ROC) curve was plotted. The Brier score was computed in order to assess the calibration of the model. The Brier score is a metric that quantifies the calibration of a model, with values ranging from 0 to 1. A lower Brier score indicates a higher level of calibration, indicating a better performance of the model [
20]. The importance of the variables incorporated in the model was assessed using both permutation-based importance and SHAP, which is an explainable artificial intelligence (XAI) technique [
21].
3. Results
A total of 855 people participated in the study, of which 430 (50.3%) were healthy controls and 425 (49.7%) were patients with Acute Heart Failure. Of the patients with AHF, 392 (92.2%) had an HFrEF, 18 (4.3%) had an HFmrEF and 15 (3.5%) had an HFpEF. All of the healthy group, 430 (100) had an LVEF 50% and above. Of the participants, 369 (43.2%) were female and 486 (56.8%) were male.
Table 1 presents the descriptive statistics of the sociodemographic data of the patients. Males had more Acute Heart Failure than females (
p<0.001) and patients were significantly older than controls (
p<0.001).
Descriptive statistics for complete blood count parameters in acute heart failure patients and healthy controls are presented in
Table 2. According to
Table 2, the results of WBC, MO, NEU, NLR, RDW-SD, RDW-CV and PDW increased significantly in patients with acute heart failure (
p<0.05). RBC, HGB, BA, LY, MPV, PLT, HCT, MCH, and PCT results were found to be significantly lower in the patient group compared to healthy controls (
p<0.05).
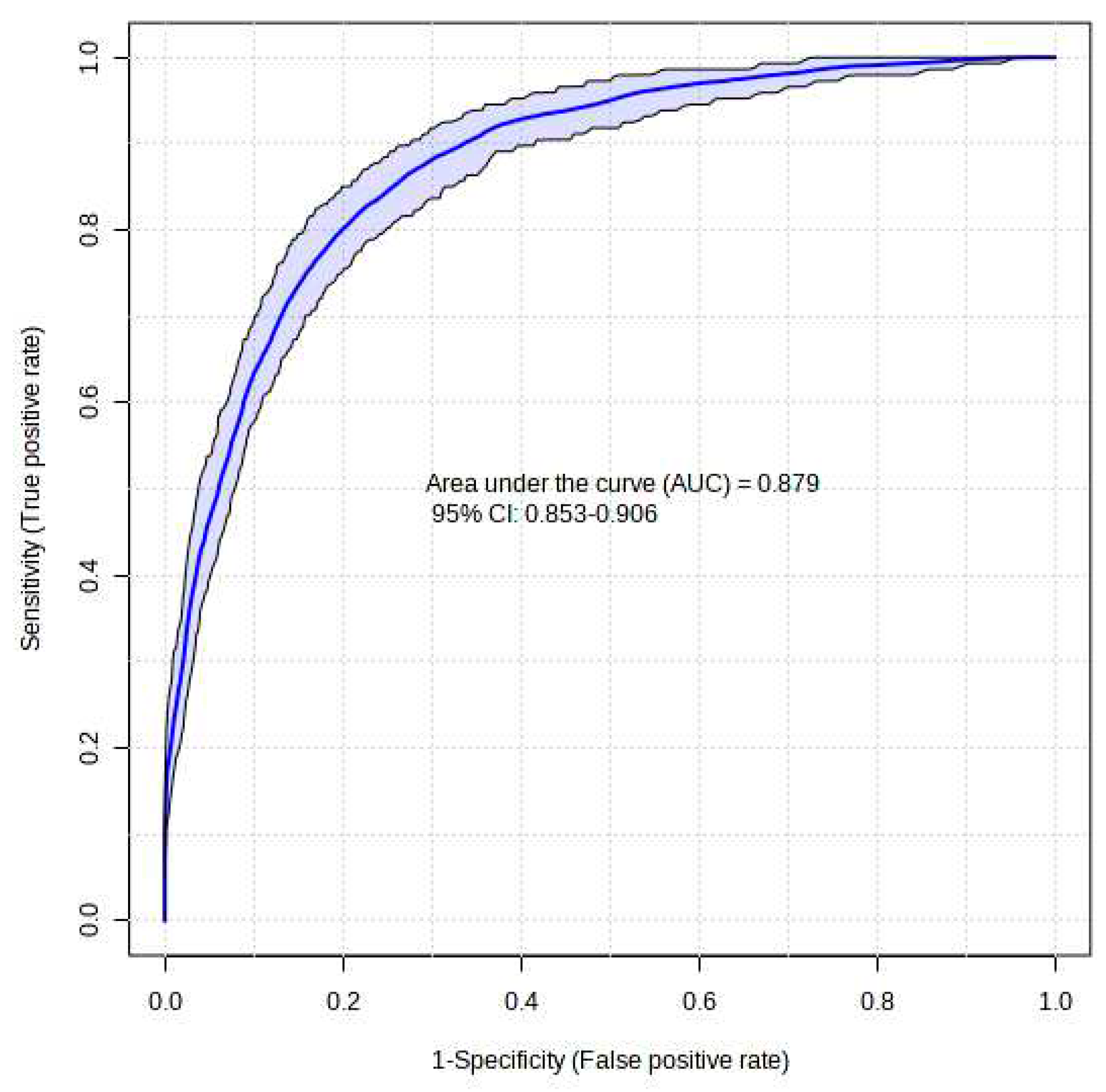
The XGBoost model that was proposed demonstrated a high level of success in accurately predicting instances of Acute Heart Failure. The XGBoost prediction model achieved an accuracy of 87.5%, an F1-score of 87.4%, and an AUC value of 87.9%. The Brier score was computed to assess the calibration of the model. A model is considered to be well-calibrated when its Brier score approaches zero. Based on the Brier score, the XGBoost model demonstrated a high level of quality, with a value of 0.036 (refer to
Table 3 and
Figure 1).
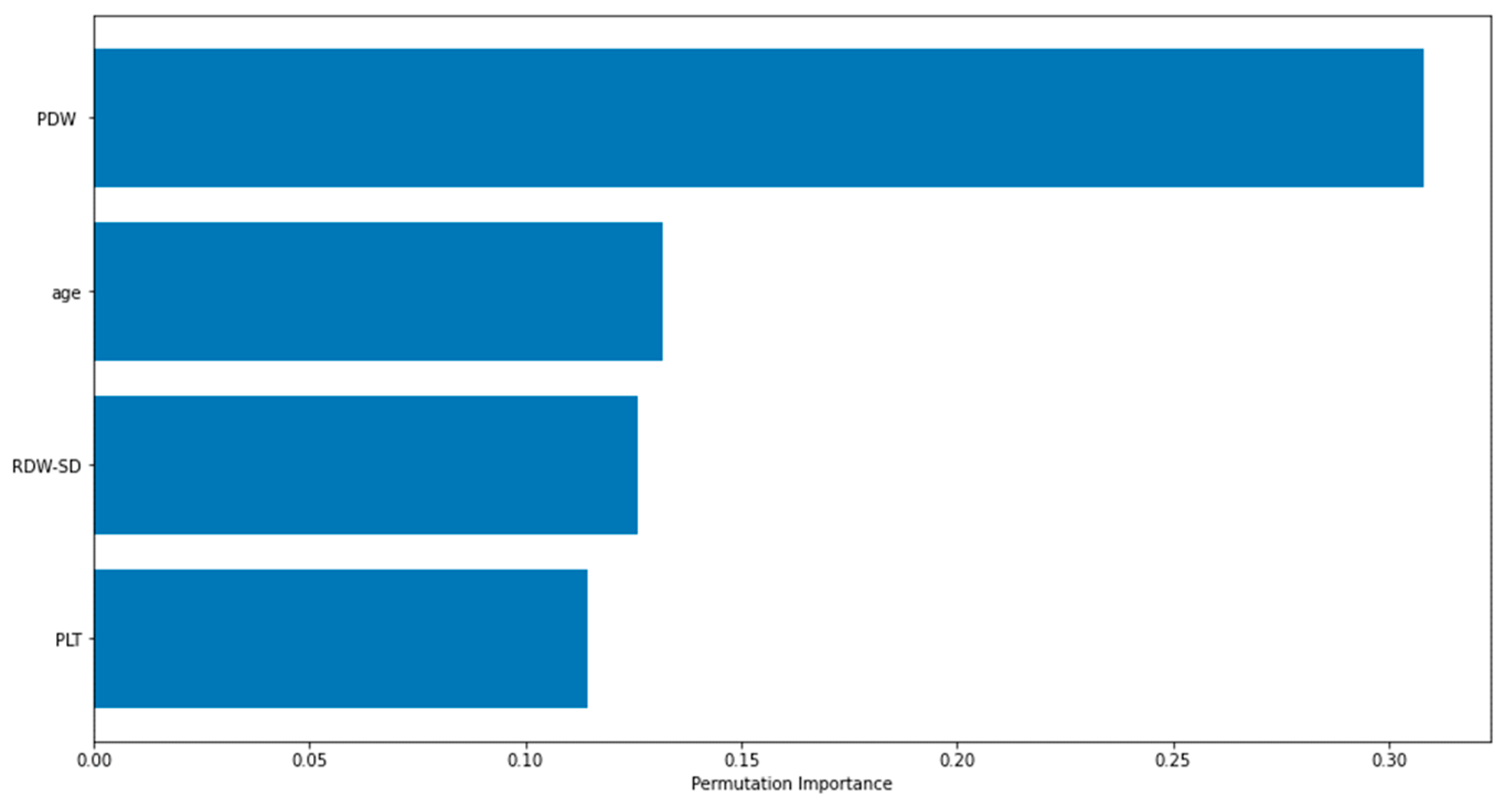
Figure 2 displays the importance graphs of the variables incorporated in the XGBoost model, which have been identified as important risk factors through the utilization of the Lasso feature selection method. The Lasso method successfully identified several key risk factors, including PDW, age, RDW-SD, and PLT, that play a significant role in distinguishing cases of Acute Heart Failure. The findings particularly demonstrated the significance of platelet distribution width (PDW) in distinguishing the disease.
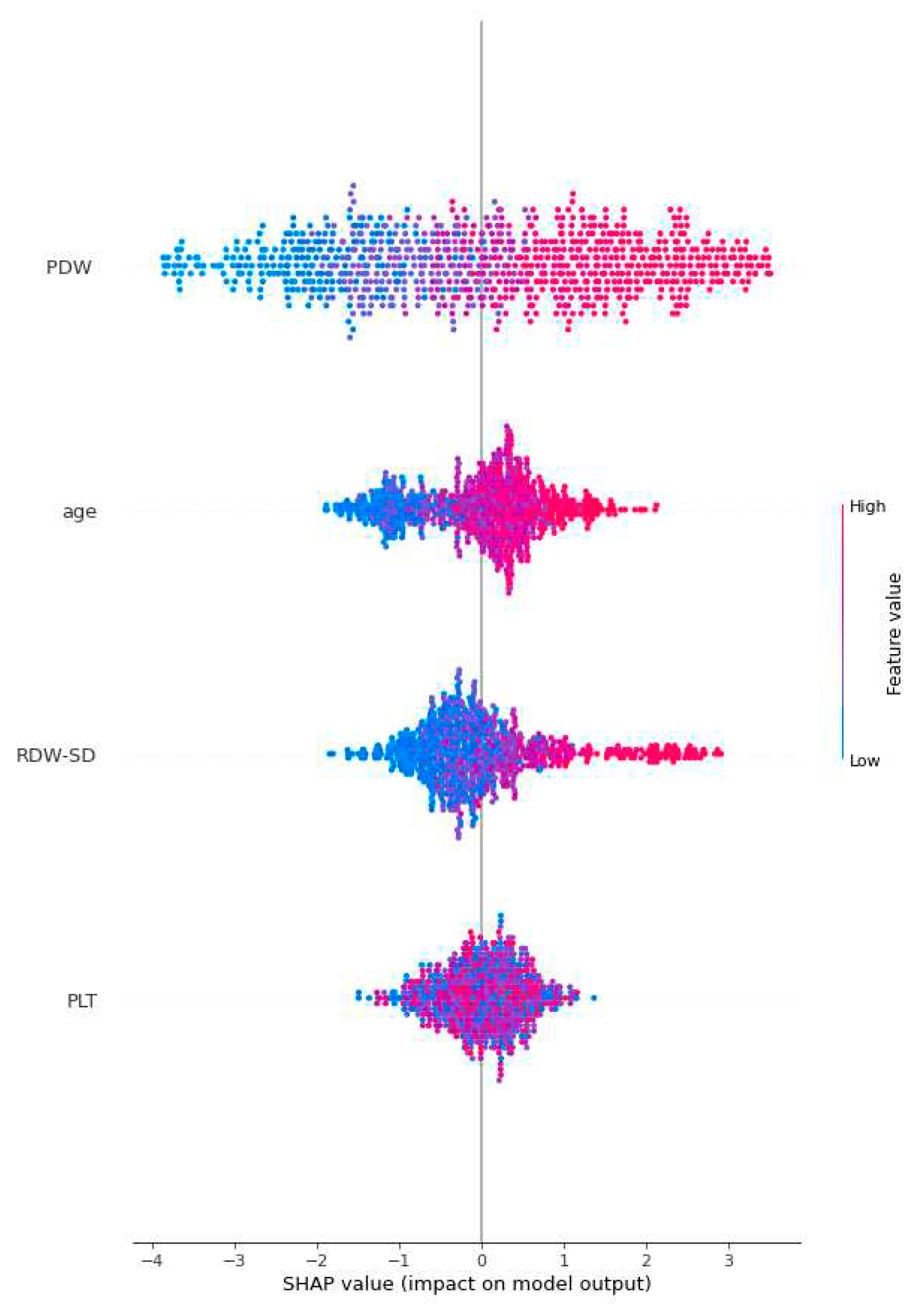
Figure 3 employs global SHAP values, which depict the positive or negative impact of biomarker candidates on the XGBoost model's prediction. This visual representation serves to highlight the importance of biomarker candidates in influencing model decisions. A positive SHAP value indicates a positive contribution to the target variable, while a negative SHAP value indicates a negative contribution. Furthermore, the data points on the graph are shaded based on the normalized values of the variables. The value of the variable increases as it approaches the color pink and decreases as it approaches the color blue. Consequently, it was found that older age is associated with an elevated risk of Acute Heart Failure, as well as higher values of Platelet Distribution Width (PDW) and Red Cell Distribution Width-Standard Deviation (RDW-SD).
4. Discussion
Predictive models of post-AHF death and disability are limited by sample size, the breadth of the clinical and risk factor range, and general clinical applicability, yet the quality of these models varies widely. Based on the patient's predicted lifespan and clinical history, healthcare providers can personalize the provision of intensive therapy or palliative care upon release from an AHF. This study used biostatistical analysis and an ML model combined with XAI to investigate potential hematological predictors of AHF. The observed results indicated that the WBC, MO, NEU, NLR, RDW-SD, RDW-CV, and PDW values of patients with AHF were statistically greater than those of the healthy group (p<0.05). However, compared to healthy controls, AHF patients had significantly reduced RBC, HGB, BA, LY, MPV, PLT, HCT, MCH, and PCT levels (p<0.05). PDW, age, RDW-SD, and PLT were found to be the most relevant risk factors for differentiating AHF by one of the machine learning models using the Lasso approach. The results especially revealed the importance of PDW in differentiating the disease under question.
In this study, the proposed XGBoost model performed quite successfully for the prediction of AHF. With the XGBoost prediction model, 87.5% accuracy, 87.4% F1-score and 87.9% AUC value were obtained. The XGBoost model as important risk factors by the Lasso feature selection method are presented. The proposed XGBoost model gave us clinicians much more specific hematological parameters than the descriptive statistical method in predicting acute heart failure. The artificial intelligence may help clinicians focus on the more important risk factors in predicting AHF. In the next stage, clinical studies on the disease may also help us to do more specific research.
Platelets are cellular fragments that serve a crucial function in the processes of blood coagulation and wound healing [
22,
23,
24]. Prior research has indicated that an unexplained decrease in platelet count is linked to mortality in acute heart failure, particularly among individuals with advanced-stage cardiac dysfunction [
25,
26,
27]. The aetiology of thrombocytopenia in individuals with heart failure remains elusive. Several theories have been proposed in relation to this matter. Examples of physiological conditions that can occur include dysfunction of bone marrow, reduced blood flow, and programmed cell death [
28]. In the present investigation, the descriptive statistics findings indicate a statistically significant decrease in platelet count among patients diagnosed with AHF compared to the control group of individuals in good health (
p<0.05). Furthermore, the artificial intelligence models employed in our present investigation have demonstrated that a diminished count of platelets serves as a significant hematological parameter for the prediction of acute heart failure. The potential cause of the decreased platelet count observed in patients with AHF in comparison to the healthy control group could be attributed to two factors: the reduction in platelet lifespan resulting from hypervolemia, and the impairment of platelet structure due to compromised blood oxygenation, potentially leading to apoptosis. This can potentially lead to hemorrhaging and result in the development of anemia. The presence of this condition may lead to unfavorable outcomes in individuals affected by it. Nevertheless, the available information on this subject is insufficient. Further research and investigation are required to address this topic.
In developed as well as developing countries, where there has been a notable rise in average life expectancy, heart failure has emerged as a significant factor contributing to hospitalizations. Heart failure is a condition that is influenced by various factors, including hypertension, chronic ischemic heart disease, diabetes mellitus, and obesity. These causative factors have been observed to exhibit an increased prevalence with advancing age, thereby contributing to the higher incidence of heart failure. This phenomenon results in significant rates of both mortality and morbidity [
1]. In the present investigation, it was observed that the average age of individuals who sought medical attention from the cardiology department for acute heart failure was significantly greater compared to the control group (
p<0.05). Furthermore, within the context of artificial intelligence modeling, age has been identified as a significant parameter in the prediction of acute heart failure (AHF). The incidence of heart failure with preserved ejection fraction, a condition that is more prevalent among females, demonstrates an upward trend as individuals advance in age [
29,
30]. When kidney functions decline due to the natural aging process, there is a potential for acute heart failure to occur as a result of both increased mortality related to heart failure and reduced urinary excretion. This phenomenon has the potential to result in a higher incidence of hospital emergency department admissions for acute heart failure.
Red-cell Distribution Width (RDW) is a laboratory test that measures the heterogeneity of the size distribution of erythrocytes. Previous research has demonstrated that elevated RDW values serve as a significant predictor of mortality in individuals diagnosed with heart failure [
11]. In another study, patients with HF may have elevated RDW due to secondary causes. The main secondary causes are renal failure, malnutrition, chronic inflammation, and inadequate erythropesis. In patients with HF, causes such as renal hypoperfusion due to low cardiac output, the effect of diuretics used, and comorbidities (for example, diabetes mellitus, hypertension, atherosclerotic vascular diseases) may cause chronic renal failure. This can cause both anemia and high RDW by reducing erythropoietin production. In addition, malnutrition in heart failure patients can cause iron deficiency anemia. In this case, it can indirectly increase the RDW value [
31,
32]. In our current study, according to the descriptive statistics results, RDW-SD was found to be significantly higher in patients with AHF than in the healthy control group (
p<0.05). In addition, artificial intelligence models used in our current study showed that RDW-SD is one of the important hematological parameter in predicting AHF. In our study, patients with a hemoglobin value of less than 10 g/dl, a high CRP, and a creatinine value above 2 mg/dl were not included in the study. AHF patients may have dilutional anemia due to hypervolemia. Intravascular volume increase may indirectly cause RDW-SD elevation. However, there is not enough information on this subject. More work needs to be done on this.
The Platelet Distribution Width is a laboratory assay utilized to quantify the degree of variability in platelet size distribution [
33]. Several studies have indicated an increased presence of PDW in individuals diagnosed with malignancies, cardiovascular disease, diabetes mellitus, and respiratory diseases [
34,
35,
36,
37,
38,
39,
40,
41]. Furthermore, certain studies have demonstrated a positive correlation between elevated platelet distribution width and heightened rates of mortality and morbidity among individuals diagnosed with ischemic heart diseases, pulmonary thromboembolism, and advanced cancer [
42,
43,
44,
45]. Nevertheless, the precise etiology of elevated PDW in these pathological conditions remains unclear. A recent study has revealed that oxidative stress has a detrimental effect on platelet functions. Furthermore, certain enzymes, such as chemokines and cytokines, which are elevated in patients with high platelet distribution width, have been found to enhance platelet activation. These enzymes promote the release of substances that initiate the process of clot formation from platelets. Other studies in the literature has established that this particular circumstance is associated with an elevated susceptibility to cardiovascular thrombosis, as well as an augmented 90-day morbidity and mortality rates [
39,
40,
41,
42,
43,
44,
45,
46,
47,
48,
49,
50,
51]. There is a lack of prior research examining the correlation between elevated PDW levels and their predictive and prognostic value in patients with Acute Heart Failure. Based on the findings of our present investigation, the descriptive statistics indicate a statistically significant elevation in PDW among individuals diagnosed with AHF compared to the healthy control cohort (
p<0.05). Furthermore, the artificial intelligence models employed in our present investigation have demonstrated that PDW is the foremost hematological parameter for predicting acute heart failure. In the context of acute heart failure, the hypervolemic condition, particularly observed in cases of total heart failure or right heart failure, can lead to spleen congestion. This congestion has the potential to disrupt the structure of platelets, resulting in an elevation of the Platelet Distribution Width value. Furthermore, it is worth noting that the hypervolemic condition has the potential to elevate the values of PDW and Red Cell Distribution Width due to the induction of intracellular edema in both erythrocytes and platelets. Consequently, this leads to a decline in the oxygen-carrying capability of compromised erythrocytes and an escalation in tissue hypoxia, thereby initiating a detrimental cycle. This phenomenon has the potential to exacerbate the prognosis in individuals diagnosed with heart failure. Similarly, the degradation of platelets can lead to both bleeding and thrombosis. Nevertheless, a dearth of information pertaining to this subject has persisted until the present day. Therefore, it is imperative to conduct thorough clinical research in order to gain a deeper understanding of this matter.
5. Conclusions
This study demonstrates that explainable artificial intelligence has identified advanced age, low platelet count, high RDW-SD, and PDW as the primary hematological parameters for predicting acute heart failure. The complete blood count (CBC) is a readily available and cost-effective diagnostic technique. It is advisable to direct attention toward these parameters in individuals displaying indications of acute heart failure. Additionally, we propose the utilization of interpretable artificial intelligence in research pertaining to the discipline of cardiology, with the aim of discerning more precise risk factors.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Table S1: Data file.
Author Contributions
“Conceptualization R.Y. and F.H.Y.; methodology, R.Y., F.H.Y., and C.C.; software, F.H.Y., and C.C.; validation R.Y., F.H.Y., C.C., and K.T.; formal analysis, F.H.Y.; investigation, R.Y., F.H.Y., C.C., and K.T.; resources, R.Y., F.H.Y., C.C., and K.T.; data curation, R.Y.; writing—original draft preparation, R.Y., F.H.Y., C.C., K.T., N.A.S., and N.F.M; writing—review and editing, R.Y., F.H.Y., C.C., K.T., N.A.S., and N.F.M; visualization, R.Y., F.H.Y., C.C., K.T., N.A.S., and N.F.M; supervision, R.Y., F.H.Y., C.C., K.T., N.A.S., and N.F.M. All authors have read and agreed to the published version of the manuscript.”.
Funding
Princess Nourah bint Abdulrahman University Researchers Supporting Project Number PNURSP2023R206, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Samsun University Clinical Research Ethics Committee (protocol code 2023/10/10 and 24.05.2023).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The dataset used in this study is provided as a link in the Supplementary Materials of the article.
Acknowledgments
The authors express their gratitude to Princess Nourah bint Abdulrahman University Researchers Supporting Project Number PNURSP2023R206, Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shirazi, L.F.; Bissett, J.; Romeo, F.; Mehta, J.L. Role of inflammation in heart failure. Current atherosclerosis reports 2017, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Böhm, M.; Burri, H.; Butler, J.; Čelutkienė, J.; Chioncel, O. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: Developed by the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) With the special contribution of the Heart Failure Association (HFA) of the ESC. European heart journal 2021, 42, 3599–3726. [Google Scholar] [PubMed]
- Nieminen, M.; Brutsaert, D.; Dickstein, K.; Drexler, H.; Follath, F.; Harjola, V.; Hochadel, M.; Komajda, M.; Lassus, J.; Lopez-Sendon, J. EuroHeart Survey Investigators; Heart Failure Association, European Society of Cardiology. EuroHeart Failure Survey II (EHFS II): a survey on hospitalized acute heart failure patients: description of population. Eur Heart J 2006, 27, 2725–2736. [Google Scholar] [CrossRef] [PubMed]
- Januzzi, J.L.; van Kimmenade, R.; Lainchbury, J.; Bayes-Genis, A.; Ordonez-Llanos, J.; Santalo-Bel, M.; Pinto, Y.M.; Richards, M. NT-proBNP testing for diagnosis and short-term prognosis in acute destabilized heart failure: an international pooled analysis of 1256 patients: the International Collaborative of NT-proBNP Study. European heart journal 2006, 27, 330–337. [Google Scholar] [CrossRef]
- Januzzi, J.L.; Chen-Tournoux, A.A.; Christenson, R.H.; Doros, G.; Hollander, J.E.; Levy, P.D.; Nagurney, J.T.; Nowak, R.M.; Pang, P.S.; Patel, D. N-terminal pro–B-type natriuretic peptide in the emergency department: the ICON-RELOADED study. Journal of the American College of Cardiology 2018, 71, 1191–1200. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.H.; Tabas, J.; Stein, J.; Potocki, M.; Mueller, C.; McCord, J.; Richards, M.; Hartmann, O.; Nowak, R.; Peacock, W.F. The effect of diabetes on the diagnostic and prognostic performance of mid-region pro-atrial natriuretic peptide and mid-region pro-adrenomedullin in patients with acute dyspnea. Biomarkers 2012, 17, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Mueller, C.; McDonald, K.; de Boer, R.A.; Maisel, A.; Cleland, J.G.; Kozhuharov, N.; Coats, A.J.; Metra, M.; Mebazaa, A.; Ruschitzka, F. Heart Failure Association of the European Society of Cardiology practical guidance on the use of natriuretic peptide concentrations. European journal of heart failure 2019, 21, 715–731. [Google Scholar] [CrossRef]
- Núñez, J.; Núñez, E.; Miñana, G.; Sanchis, J.; Bodí, V.; Rumiz, E.; Palau, P.; Olivares, M.; Merlos, P.; Bonanad, C. Effectiveness of the relative lymphocyte count to predict one-year mortality in patients with acute heart failure. The American journal of cardiology 2011, 107, 1034–1039. [Google Scholar] [CrossRef]
- Cikrikcioglu, M.A.; Soysal, P.; Dikerdem, D.; Cakirca, M.; Kazancioglu, R.; Yolbas, S.; Erkal, H.; Hursitoglu, M.; Karakose, T.K.; Kiskac, M. Absolute blood eosinophil count and 1-year mortality risk following hospitalization with acute heart failure. European Journal of Emergency Medicine 2012, 19, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Groenveld, H.F.; Januzzi, J.L.; Damman, K.; van Wijngaarden, J.; Hillege, H.L.; van Veldhuisen, D.J.; van der Meer, P. Anemia and mortality in heart failure patients: a systematic review and meta-analysis. Journal of the American College of Cardiology 2008, 52, 818–827. [Google Scholar] [CrossRef]
- Pascual-Figal, D.A.; Bonaque, J.C.; Redondo, B.; Caro, C.; Manzano-Fernandez, S.; Sánchez-Mas, J.; Garrido, I.P.; Valdes, M. Red blood cell distribution width predicts long-term outcome regardless of anaemia status in acute heart failure patients. European journal of heart failure 2009, 11, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Silverberg, D.S.; Wexler, D.; Iaina, A. The importance of anemia and its correction in the management of severe congestive heart failure. European Journal of Heart Failure 2002, 4, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Varat, M.A.; Adolph, R.J.; Fowler, N.O. Cardiovascular effects of anemia. American heart journal 1972, 83, 415–426. [Google Scholar] [CrossRef] [PubMed]
- PAKSOY, N.; YAĞIN, F.H. Artificial intelligence-based colon cancer prediction by identifying genomic biomarkers. Medical Records 2022, 4, 196–202. [Google Scholar] [CrossRef]
- Koulaouzidis, G.; Jadczyk, T.; Iakovidis, D.K.; Koulaouzidis, A.; Bisnaire, M.; Charisopoulou, D. Artificial intelligence in cardiology—a narrative review of current status. Journal of Clinical Medicine 2022, 11, 3910. [Google Scholar] [CrossRef] [PubMed]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G* Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behavior research methods 2007, 39, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Du, L.-X.; He, Y.-L.; Yuan, J.-P.; Wang, P.; Ye, B.-L.; Wang, C.; Hou, Z. Dictionary learning LASSO for feature selection with application to hepatocellular carcinoma grading using contrast enhanced magnetic resonance imaging. Frontiers in Oncology 2023, 13, 1123493. [Google Scholar] [CrossRef]
- Cansel, N.; Yagin, F.H.; Akan, M.; Ilkay Aygul, B. Interpretable estimation of suicide risk and severity from complete blood count parameters with explainable artificial intelligence methods. Psychiatria Danubina 2023, 35, 62–72. [Google Scholar] [CrossRef]
- Han, Y.; Kim, J.; Enke, D. A machine learning trading system for the stock market based on N-period Min-Max labeling using XGBoost. Expert Systems with Applications 2023, 211, 118581. [Google Scholar] [CrossRef]
- Yagin, F.H.; Gülü, M.; Gormez, Y.; Castañeda-Babarro, A.; Colak, C.; Greco, G.; Fischetti, F.; Cataldi, S. Estimation of Obesity Levels with a Trained Neural Network Approach optimized by the Bayesian Technique. Applied Sciences 2023, 13, 3875. [Google Scholar] [CrossRef]
- Yao, L.; Ni, H. Prediction of patent grant and interpreting the key determinants: an application of interpretable machine learning approach. Scientometrics 2023, 1–37. [Google Scholar] [CrossRef]
- Kamath, S.; Blann, A.D.; Lip, G. Platelet activation: assessment and quantification. European heart journal 2001, 22, 1561–1571. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.T.; Corken, A.; Ware, J. Platelets at the interface of thrombosis, inflammation, and cancer. Blood, The Journal of the American Society of Hematology 2015, 126, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Manne, B.K.; Xiang, S.C.; Rondina, M.T. Platelet secretion in inflammatory and infectious diseases. Platelets 2017, 28, 155–164. [Google Scholar] [CrossRef]
- Hui, P.; Cook, D.J.; Lim, W.; Fraser, G.A.; Arnold, D.M. The frequency and clinical significance of thrombocytopenia complicating critical illness: a systematic review. Chest 2011, 139, 271–278. [Google Scholar] [CrossRef]
- Moreau, D.; Vesin, A.; Garrouste-Orgeas, M.; de Lassence, A.; Zahar, J.-R.; Adrie, C.; Cohen, Y.; Schlemmer, B.; Azoulay, E. Platelet count decline: an early prognostic marker in critically ill patients with prolonged ICU stays. Chest 2007, 131, 1735–1741. [Google Scholar] [CrossRef] [PubMed]
- Akca, S.; Haji-Michael, P.; De Mendonça, A.; Suter, P.; Levi, M.; Vincent, J.-L. Time course of platelet counts in critically ill patients. Critical care medicine 2002, 30, 753–756. [Google Scholar] [CrossRef] [PubMed]
- Westenbrink, B.D.; Voors, A.A.; de Boer, R.A.; Schuringa, J.J.; Klinkenberg, T.; van der Harst, P.; Vellenga, E.; van Veldhuisen, D.J.; van Gilst, W.H. Bone marrow dysfunction in chronic heart failure patients. European Journal of Heart Failure 2010, 12, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, B.A.; Zhao, X.; Heidenreich, P.A.; Peterson, E.D.; Bhatt, D.L.; Cannon, C.P.; Hernandez, A.F.; Fonarow, G.C. Trends in patients hospitalized with heart failure and preserved left ventricular ejection fraction: prevalence, therapies, and outcomes. Circulation 2012, 126, 65–75. [Google Scholar] [CrossRef]
- Redfield, M.M.; Jacobsen, S.J.; Burnett, J.C.; Mahoney, D.W.; Bailey, K.R.; Rodeheffer, R.J. Burden of systolic and diastolic ventricular dysfunction in the community: appreciating the scope of the heart failure epidemic. Jama 2003, 289, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.; Adamson, J.W. The anemia of chronic disorders: studies of marrow regulation and iron metabolism. 1975.
- Weiss, G.; Goodnough, L.T. Anemia of chronic disease. New England Journal of Medicine 2005, 352, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Vagdatli, E.; Gounari, E.; Lazaridou, E.; Katsibourlia, E.; Tsikopoulou, F.; Labrianou, I. Platelet distribution width: a simple, practical and specific marker of activation of coagulation. Hippokratia 2010, 14, 28. [Google Scholar] [PubMed]
- Jindal, S.; Gupta, S.; Gupta, R.; Kakkar, A.; Singh, H.V.; Gupta, K.; Singh, S. Platelet indices in diabetes mellitus: indicators of diabetic microvascular complications. Hematology 2011, 16, 86–89. [Google Scholar] [CrossRef]
- Yu, Y.; Li, N.; Yun, Z.; Niu, Y.; Xu, J.; Liu, Z.; Liu, T.; Wang, R.; Yu, K. Preoperative mean platelet volume and platelet distribution associated with thyroid cancer. Neoplasma 2017, 64, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Liu, L.; Zhang, X.; Liu, Z.-P.; Wang, R.-T. Platelet indices in laryngeal cancer. Cancer Biomarkers 2018, 21, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Khandekar, M.; Khurana, A.; Deshmukh, S.; Kakrani, A.; Katdare, A.; Inamdar, A. Platelet volume indices in patients with coronary artery disease and acute myocardial infarction: an Indian scenario. Journal of clinical pathology 2006, 59, 146–149. [Google Scholar] [CrossRef]
- Kamisli, O.; Kamisli, S.; Kablan, Y.; Gonullu, S.; Ozcan, C. The prognostic value of an increased mean platelet volume and platelet distribution width in the early phase of cerebral venous sinus thrombosis. Clinical and Applied Thrombosis/Hemostasis 2013, 19, 29–32. [Google Scholar] [CrossRef]
- Sevuk, U.; Bahadir, M.V.; Altindag, R.; Baysal, E.; Yaylak, B.; Ay, N.; Ayaz, F.; Demirtas, E. Value of serial platelet indices measurements for the prediction of pulmonary embolism in patients with deep venous thrombosis. Therapeutics and Clinical Risk Management 2015, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Sezgi, C.; Taylan, M.; Kaya, H.; Selimoglu Sen, H.; Abakay, O.; Demir, M.; Abakay, A.; Tanrikulu, A.C. Alterations in platelet count and mean platelet volume as predictors of patient outcome in the respiratory intensive care unit. The clinical respiratory journal 2015, 9, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Bülbül, Y.; Aydın Özgür, E.; Örem, A. Platelet indices in obstructive sleep apnea: the role of mean platelet volume, platelet distribution widht and plateletcrit. Tuberk Toraks 2016, 64, 206–210. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cui, Y.-L.; Diao, M.-Y.; Chen, D.-C.; Lin, Z.-F. Use of platelet indices for determining illness severity and predicting prognosis in critically ill patients. Chinese medical journal 2015, 128, 2012–2018. [Google Scholar] [CrossRef] [PubMed]
- Rechciński, T.; Jasińska, A.; Foryś, J.; Krzemińska-Pakuła, M.; Wierzbowska-Drabik, K.; Plewka, M.; Peruga, J.Z.; Kasprzak, J.D. Prognostic value of platelet indices after acute myocardial infarction treated with primary percutaneous coronary intervention. Cardiology Journal 2013, 20, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Bae, M.H.; Lee, J.H.; Yang, D.H.; Park, H.S.; Cho, Y.; Chae, S.C. White blood cell, hemoglobin and platelet distribution width as short-term prognostic markers in patients with acute myocardial infarction. Journal of Korean Medical Science 2014, 29, 519–526. [Google Scholar] [CrossRef]
- Ulucan, Ş.; Keser, A.; Kaya, Z.; Katlandur, H.; Özdil, H.; Bilgi, M.; Ateş, İ.; Ülgen, M.S. Association between PDW and long term major adverse cardiac events in patients with acute coronary syndrome. Heart, Lung and Circulation 2016, 25, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Kern, A.; Gil, R.J.; Bojko, K.; Sienkiewicz, E.; Januszko-Giergielewicz, B.; Górny, J.; Bil, J. Platelet distribution width as the prognostic marker in coronary bifurcation treatment. European Journal of Clinical Investigation 2017, 47, 524–530. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, L.; Fu, S.; Liu, Y.-S.; Wang, C.; Liu, T.; Liu, Z.-P.; Wang, R.-T.; Yu, K.-J. Higher platelet distribution width predicts poor prognosis in laryngeal cancer. Oncotarget 2017, 8, 48138. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Zhu, H.; Pei, Q.; Tan, F.; Li, C.; Zhou, Z.; Zhou, Y.; Yu, N.; Li, Y.; Pei, H. Significance of inflammation-based indices in the prognosis of patients with non-metastatic colorectal cancer. Oncotarget 2017, 8, 45178. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Abe, M.; Takumi, Y.; Hashimoto, T.; Kobayashi, R.; Osoegawa, A.; Miyawaki, M.; Okamoto, T.; Sugio, K. The prognostic impact of the platelet distribution width-to-platelet count ratio in patients with breast cancer. PloS one 2017, 12, e0189166. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zeng, X.; Cao, S.; Hu, X.; Shi, Q.; Li, D.; Zhou, S.; Gu, P.; Zhang, Z. Elevated pretreatment platelet distribution width and platelet count predict poor prognosis in nasopharyngeal carcinoma. Oncotarget 2017, 8, 106089. [Google Scholar] [CrossRef] [PubMed]
- Araz, O.; Albez, F.S.; Ucar, E.Y.; Kerget, B.; Yılmaz, N.; Akgun, M. Predictive value of mean platelet volume for pulmonary embolism recurrence. Lung 2017, 195, 497–502. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).