Submitted:
10 July 2023
Posted:
12 July 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
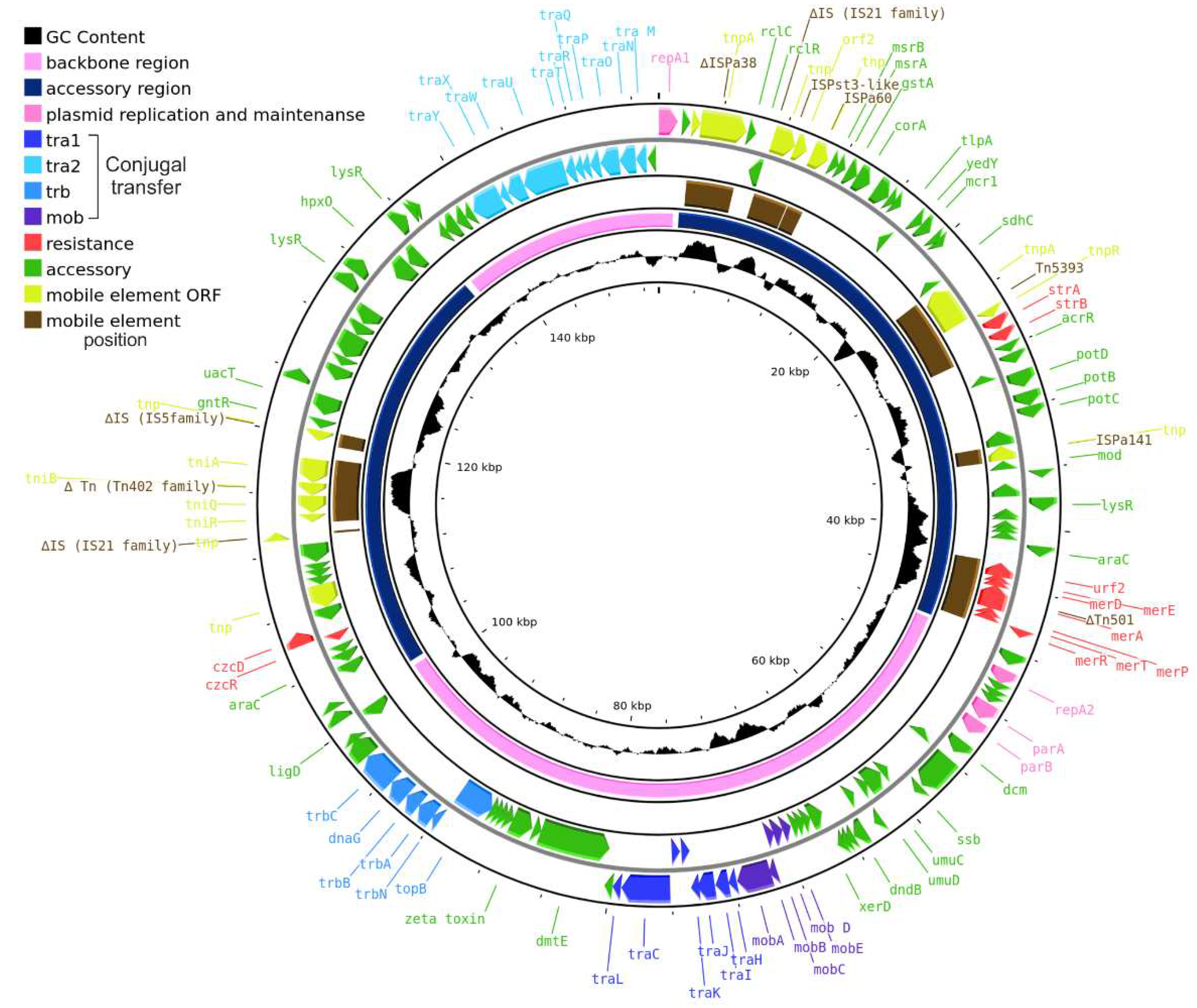
2.1. The prototype plasmid pPPUT-Tik1-1
2.2. Genome of pPPUT-Tik1-1 and related plasmids
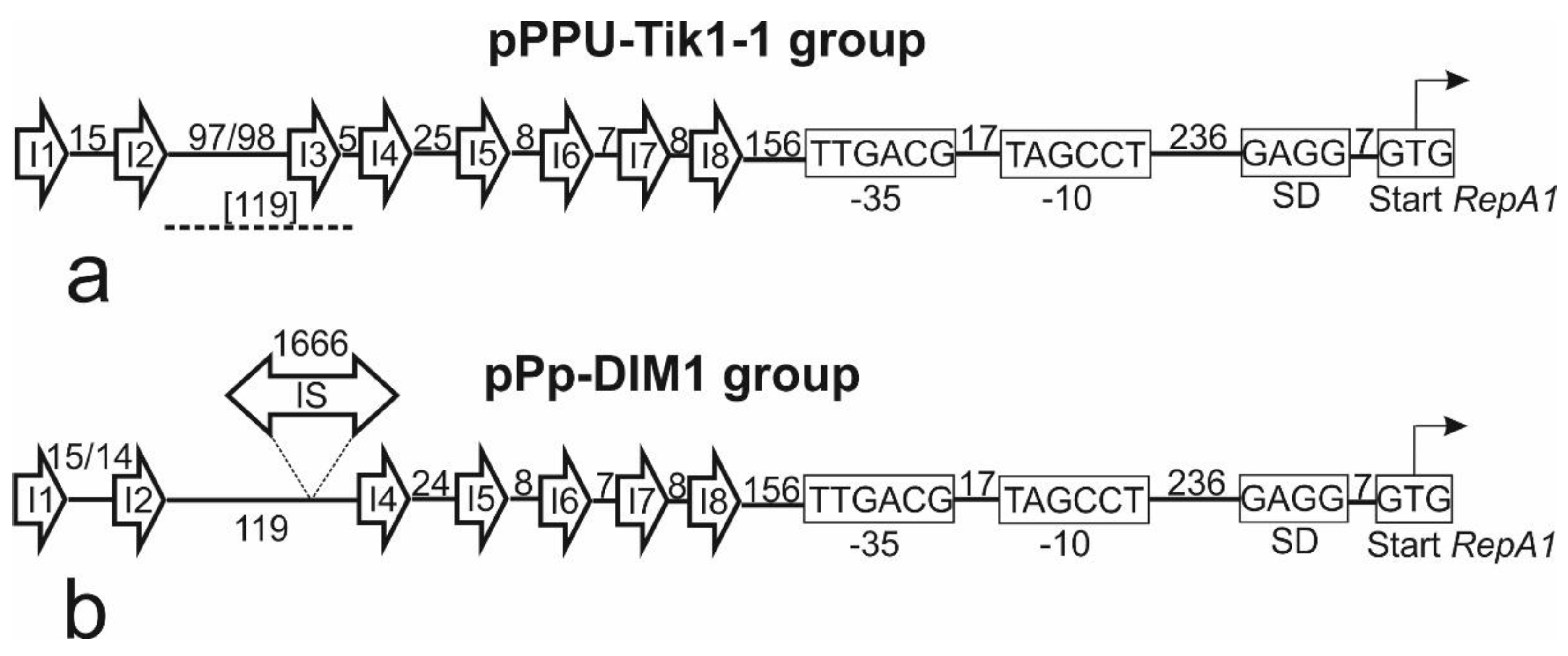
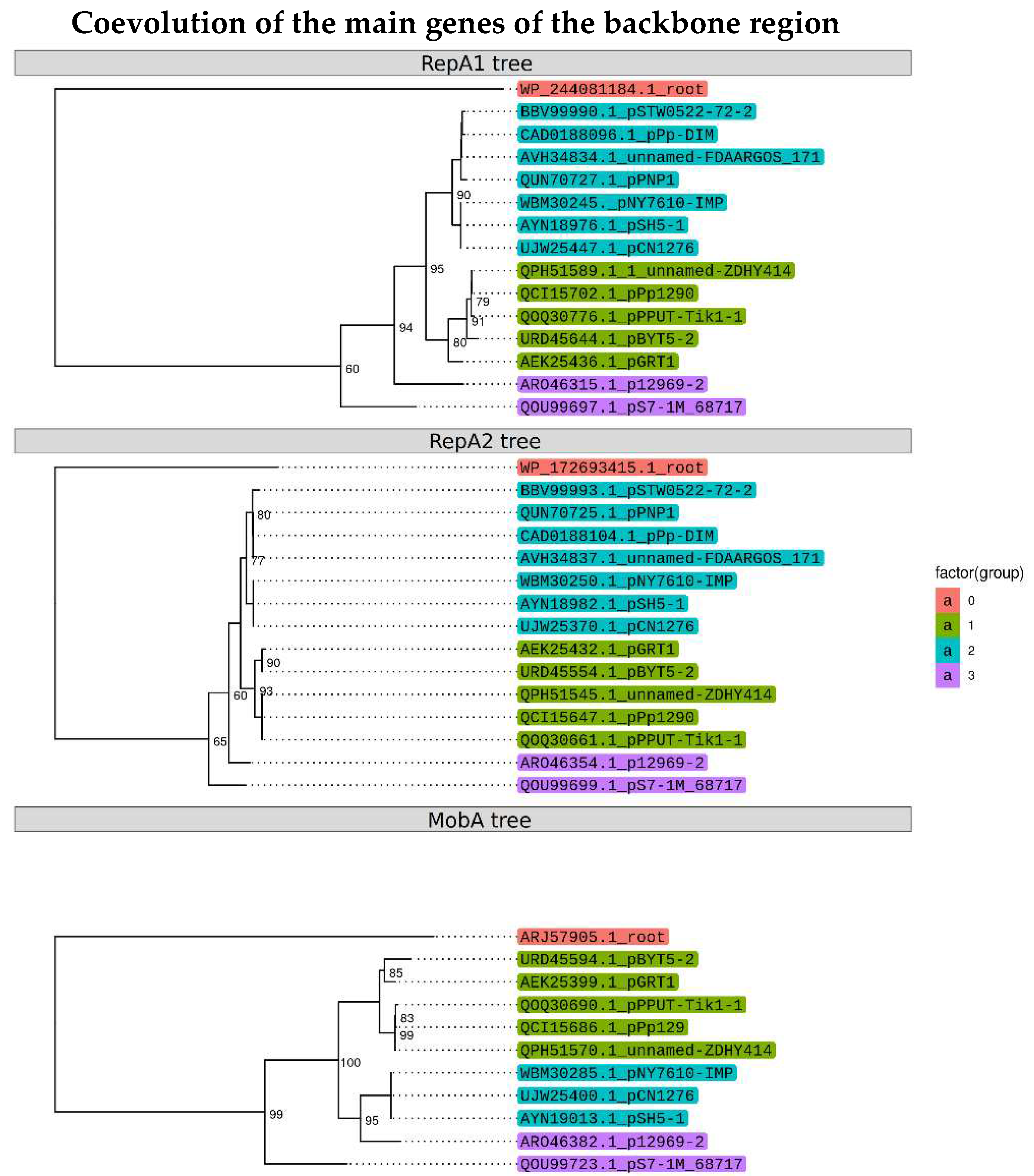
2.3. Identification of the replicon and relaxase types of pPPUT-Tik1-1 and related plasmids
2.4. Determination of the host range of Tik1-1 and its stability in different hosts
2.5. The accessory region of pPPUT-Tik1-1 and related plasmids
2.5.1. Resistance to heavy metals
2.5.2. Biodegradation
2.5.3. Antibiotic resistance
3. Discussion
4. Materials and Methods
4.1. Media and growth conditions
4.2. Bacterial strains
4.3. Matings
4.4. Plasmid stability assays
4.5. Identification of the backbone region of pPPUT-Tik1-1
4.6. Search for plasmids related to pPPUT-Tik1-1 in GenBank
4.7. Whole-genome sequencing and plasmid assembly
4.8. Bioinformatic analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wackett, LP. Pseudomonas putida – a versatile biocatalyst. Nat Biotechnol. 2003, 21(2), 136–8. [Google Scholar] [CrossRef]
- Gutiérrez-Barranquero, JA, Cazorla, FM, de Vicente, A, Sund, GW. Complete sequence and comparative genomic analysis of eight native Pseudomonas syringae plasmids belonging to the pPT23A family. BMC Genomics 2017, 18, 365. [CrossRef]
- Li, Z, Cai, Z, Cai, Z, Zhang, Y, Fu, T, Jin, Y, Cheng, Z, Jin, S, Wu, W, Yang, L, Ba,i F.J Molecular genetic analysis of an XDR Pseudomonas aeruginosa ST664 clone carrying multiple conjugal plasmids. J Molecular genetic analysis of an XDR Pseudomonas aeruginosa ST664 clone carrying multiple conjugal plasmids. Antimicrob Chemother. 2020, 75(6), 1443–52. [CrossRef]
- Paul, D, Chanda, DD, Chakravarty, A, Bhattacharjee, A. An insight into analysis and elimination of plasmids encoding metallo-β-lactamases in Pseudomonas aeruginosa. J Glob Antimicrob Resist. 2020, 21, 3–7. [CrossRef]
- Novick, R. P. Plasmid incompatibility. Microbiol. Rev. 1987, 51, 381–95. [Google Scholar] [CrossRef] [PubMed]
- Shintani, M, Sanchez, ZK, Kimbara, K. Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front Microbiol. 2015, 6, 242. [CrossRef]
- Bardaji, L, Añorga, M, Ruiz-Masó, JA, Del Solar, G, Murillo, J. Plasmid replicons from Pseudomonas are natural chimeras of functional, exchangeable modules. Front Microbiol. 2017, 8, 190. [CrossRef]
- Sentchilo, VS, Perebituk, AN, Zehnder, AJ, van der Meer, JR. Molecular diversity of plasmids bearing genes that encode toluene and xylene metabolism in Pseudomonas strains isolated from different contaminated sites in Belarus. Appl Environ Microbiol. 2000, 66(7), 2842–52. [CrossRef]
- Bonnin, RA, Bogaerts, P, Girlich, D, Huang, TD, Dortet, L, Glupczynski, Y, Naas, T. Molecular characterization of OXA-198 carbapenemase-producing Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 2018, 62, 02496-17. [CrossRef]
- Jiang, X, Yin, Z, Yuan, M, Cheng, Q, Hu, L et al. Plasmids of novel incompatibility group IncpRBL16 from Pseudomonas species. J Antimicrob Chemother. 2020, 75(8), 2093–2100. [CrossRef]
- Garcillán-Barcia, MP, Francia, MV, de la Cruz, F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 2009, 33, 657–87. [CrossRef] [PubMed]
- Garcillán-Barcia, MP, de la Cruz, F. Ordering the bestiary of genetic elements transmissible by conjugation. Mob Genet Elements. 2013, 3(1), 24263. [CrossRef]
- Smillie, C, Garcillán-Barcia, MP, Francia, MV, Rocha, EP, de la Cruz, F. Mobility of plasmids. Microbiol Mol Biol Rev. 2010, 74(3), 434–52. [CrossRef]
- Orlek, A, Phan, H, Sheppard, AE, Doumith, M, Ellington, M, et al. Ordering the mob: Insights into replicon and MOB typing schemes from analysis of a curated dataset of publicly available plasmids. Plasmid 2017, 91, 42–52. [CrossRef] [PubMed]
- Chen, F, Wang, P, Yin, Z, Yang, H, Hu, L et al. VIM-encoding IncpSTY plasmids and chromosome-borne integrative and mobilizable elements (IMEs) and integrative and conjugative elements (ICEs) in Pseudomonas. Ann Clin Microbiol Antimicrob 2022, 21, 10. [CrossRef]
- Moura, A, Oliveira, C, Henriques, I, Smalla, K, Correia, A. Broad diversity of conjugative plasmids in integron-carrying bacteria from wastewater environments. FEMS Microbiol Lett. 2012, 330, 157–164. [CrossRef]
- Mindlin, S, Beletsky, A, Rakitin, A, Mardanov, A, Petrova, M. Acinetobacter plasmids: diversity and Development of classification strategies. Front Microbiol. 2020, 11, 588410. [CrossRef] [PubMed]
- Mindlin, SZ, Petrova, M., Gorlenko, ZM, Soina, VS, Khachikian, NA, Karaevskaya, EA. Multidrug-resistant bacteria in permafrost: Isolation, biodiversity, phenotypic and genotypic analysis. In New Permafrost and Glacier Research, 1st ed.; Krugger, M.I., Stern, H.P., Eds.; Nova Science: Hauppauge, NY, USA, 2009; pp. 89–105. [Google Scholar]
- Gerdes, K, Moller-Jensen, J, Bugge Jensen, R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 2000, 37, 455–466. [CrossRef]
- Xu,Y, Niu,Y, Sun, F, Yang,Y, Luo, W, Wang, Z. The novel Pseudomonas putida plasmid p12969-2 harbors an In127-carrying multidrug-resistant region. Future Microbiol. 2017, 12, 573–84. [CrossRef]
- Mutschler, H, Meinhart, AJ, ε/ζ systems: their role in resistance, virulence, and their potential for antibiotic development. Mol Med (Berl). 2011, 89(12), 1183–94. [CrossRef]
- Carattoli, A, Zankari, E, García-Fernández, A, Voldby Larsen, M, Lund O, Villa, L, Møller Aarestrup, F, Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014, 58(7), 3895–903. [CrossRef]
- Cabezón, E, Ripoll-Rozada, J, Peña, A, de la Cruz, F, Arechaga, I. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev. 2015, 39(1), 81–95. [CrossRef]
- Hesse, C, Schulz, F, Bull, CT, Shaffer, BT, Yan, Q, Shapiro, N et al. Genome-based evolutionary history of Pseudomonas spp. Environ Microbiol 2018, 20, 2142–2159. [CrossRef] [PubMed]
- Passarelli-Araujo, H, Franco, GR, Venancio, TM. Network analysis of ten thousand genomes shed light on Pseudomonas diversity and classification. Microbiol Res. 2022, 254, 126919. [CrossRef] [PubMed]
- Virolle, C., Goldlust, K., Djermoun, S, Bigot, S, Lesterlin, C. Plasmid transfer by conjugation in gram-negative bacteria: from the cellular to the community level. Genes 2020, 11, 1239. [CrossRef]
- Gelder, LD, Ponciano, JM, Joyce, P, Top, EM. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 2007, 153, 452–463. [CrossRef] [PubMed]
- Yeo, CC, Tham, JM, Kwong, SM, Yiin, S, Poh, CL. Tn5563, a transposon encoding putative mercuric ion transport proteins located on plasmid pRA2 of Pseudomonas alcaligenes. Tn 5563, 165(2), 253–60. [CrossRef]
- Molina, L, Duque, E, Gomez, MJ, Krell, T, Lacal, J, García-Puente, A et al. The pGRT1 plasmid of Pseudomonas putida DOT-T1E encodes functions relevant for survival under harsh conditions in the environment. Environ. Microbiol. 2011, 13(8), 2315-27. [CrossRef]
- Lalucat, J, Mulet, M, Gomila, M, García-Valdés, E. Genomics in Bacterial Taxonomy: Impact on the Genus Pseudomonas. Genes (Basel) 2020, 11(2), 139. [CrossRef]
- Volkova, OV, Panov, AV, Kosheleva, IA, Boronin, AM. IncP-7 plasmids classification based on structural diversity of their basic replicon. Mol Biol (Mosk). 2013, 47(2), 232-42. [CrossRef]
- Osborn, AM, Bruce, KD, Strike, P, Ritchie, DA. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol Rev. 1997, 19(4), 239-62. [CrossRef]
- Liebert, CA, Hal,l RM, Summers, AO. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 1999, 63(3), 507-22. [CrossRef]
- Schneiker, S, Keller, M, Dröge, M, Lanka, E, Pühler, A, Selbitschka, W. The genetic organization and evolution of the broad host range mercury resistance plasmid pSB102 isolated from a microbial population residing in the rhizosphere of alfalfa. Nucleic Acids Res. 2001, 29(24), 5169-81. 24. [CrossRef]
- Mindlin, S, Minakhin, L, Petrova, M, Kholodi,i G, Minakhina, S, Gorlenko, Zh, Nikiforov, V. Present-day mercury resistance transposons are common in bacteria preserved in permafrost grounds since the Upper Pleistocene. Res Microbiol. 2005, 156(10), 994-1004. [CrossRef]
- Barkay, T, Miller, SM, Summers, AO. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol Rev. 2003, 27(2-3), 355-84. [CrossRef]
- Driscoll, CT, Mason, RP, Chan, HM, Jacob, DJ, Pirrone, N. Mercury as a global pollutant: sources, pathways, and effects. Environ Sci Technol. 2013, 47(10), 4967-83. [CrossRef]
- Obrist, D, Kirk, JL, Zhang, L, Sunderland, EM, Jiskra, M, Selin, NE. A review of global environmental mercury processes in response to human and natural perturbations: Changes of emissions, climate, and land use. Ambio. 2018, 47(2), 116-140. [CrossRef]
- Raj, D, Maiti, SK. Sources, toxicity, and remediation of mercury: an essence review. Environ Monit Assess. 2019, 191(9), 566. [CrossRef]
- Christakis, CA, Barkay, T, Boyd, ES. Expanded diversity and phylogeny of mer genes broadens mercury resistance paradigms and reveals an origin for MerA among thermophilic archaea. Front Microbiol. 2021, 12, 682605. [CrossRef]
- Babich, K, Engle, M, JS, Laddaga, RA. Deletion mutant analysis of the Staphylococcus aureus plasmid pI258 mercury-resistance determinant. Can J Microbiol. 1991, 7(8), 624-31. [CrossRef]
- Hobman, JL, Brown, NL. Overexpression of MerT, the mercuric ion transport protein of transposon Tn501, and genetic selection of mercury hypersensitivity mutations. Mol Gen Genet. 1996, 250(1), 129-34. [CrossRef]
- Juan, C, Zamorano, L, Mena, A, Albertí, S, Pérez, JL, Oliver, A. Metallo-beta-lactamase-producing Pseudomonas putida as a reservoir of multidrug resistance elements that can be transferred to successful Pseudomonas aeruginosa clones. J Antimicrob Chemother. 2010, 65(3), 474-8. [CrossRef]
- Peter, S, Oberhettinger, P, Schuele, L, Dinkelacker, A, Vogel, W. Genomic characterisation of clinical and environmental Pseudomonas putida group strains and determination of their role in the transfer of antimicrobial resistance genes to Pseudomonas aeruginosa. BMC Genomics 2017, 18(1), 859. [CrossRef]
- Mindlin, S, Maslova, O, Beletsky, A, Nurmukanova, V, Zong, Z, Mardanov, A, Petrova M. Ubiquitous conjugative mega-plasmids of Acinetobacter species and their role in horizontal transfer of multi-drug resistance. Front Microbiol. 2021,12, 728644. [CrossRef]
- Sambrook, J, Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA, 3d edition.
- Deane, S., Rawlings, D. Plasmid evolution and interaction between the plasmid addiction stability systems of two related broad-host-range IncQ-like plasmids. J Bacteriol. 2004, 186(7), 2123-33. [CrossRef]
- Hamidian, M., Ambrose, SJ, Hall, RM. A large conjugative Acinetobacter baumannii plasmid carrying the sul2 sulphonamide and strAB streptomycin resistance genes. Plasmid 2016, 87-88, 43-50. [CrossRef]
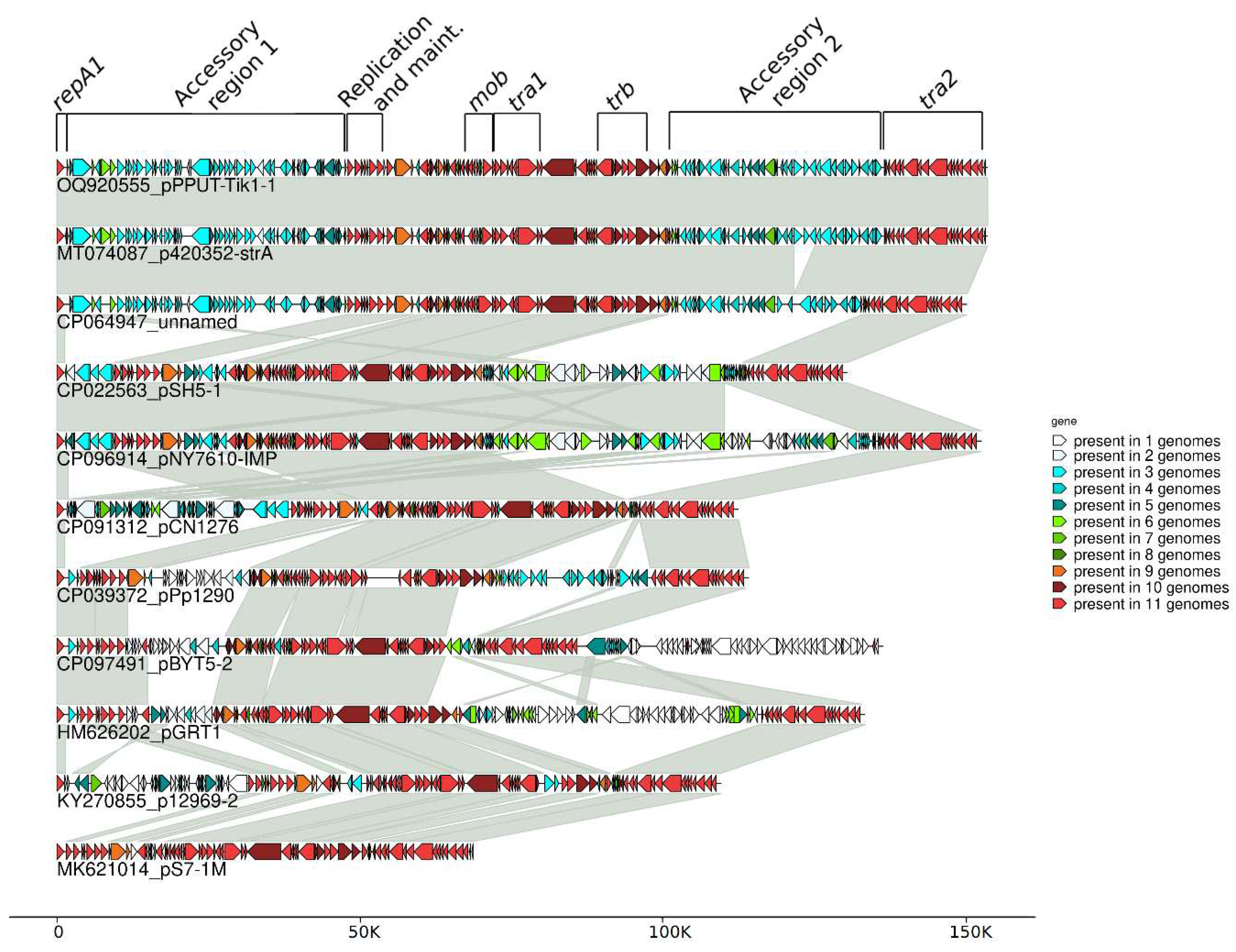
| Strain | Plasmid | Size (bp) | Source | Accessory region1 | Accession Number |
|---|---|---|---|---|---|
| Conjugative plasmids (Tra+) | |||||
| P. putida Tik1 | pPPUT-Tik1-1 | 153663 | permafrost, Russia | Hg, Str | OQ920555 |
| P. putida 15420352 | p420352-strA | 153678 | urine, China | Hg, Str | MT074087 |
| P.fulva ZDHY414 | Unnamed | 150273 | clinic, China | Hg, Str | CP064947 |
| P. putida 12969 | p12969-2 | 109708 | clinic, China | Hg, Str, Sul | KY270855 |
| P. putida 1290 | pPp1290 | 114265 | pear pyllosphere | Aromatic compounds degradation | CP039372 |
| P. putida DOT-T1E | pGRT1 | 133451 | unknown | Tolerance to toluene, UV-resistance | HM626202 |
| P. juntendi 18091276 | pCN1276 | 112529 | urine, China | Hg | CP091312 |
| P. monteilii B5 | pSH5-1 | 130536 | soil | Str | CP022562 |
| Pseudomonas sp. BYT-5 | pBYT5-2 | 136400 | soil | Molybdate absorption | CP097489 |
| P. aeruginosa NY7610 | pNY7610-IMP | 152682 | clinic, China | Str, Gm, Km, Cb, Cf | CP096914 |
| P. syringae pv. actinidiae S7-1M | pS7-1M | 68717 | kiwifruit orchard, New Zealand | - | MK621014 |
| Non-conjugative plasmids (Tra-) | |||||
| P. putida | pPp-DIM | 69823 | water, Spain | Sul, Km/Gm, Cb, Tc | LR822050 |
| P.monteilii | unnamed | 60588 | sputum, USA | - | CP014061 |
| P.monteilii STW0522-72 | pSTW0522-72-2 | 59057 | hospital sewage, Japan | - | AP022475 |
| Pseudomonas sp | pPNP1 | 125508 | activated sludge, USA | Sul, 4-Nitrophenol degradation | CP073662 |
| Group1 | Strain | Species (Collection2) | Transconjugants frequency (per recipient)3 |
|---|---|---|---|
| P. aeruginosa | PAO Type | P. aeruginosa (IMG) | <10-9 |
| B-6643 | P. aeruginosa (VKPM) | <10-9 | |
| B-5807 Type | P. aeruginosa (VKPM) | <10-9 | |
| TA37-4 | P. aeruginosa (IMG) | <10-9 | |
| FA8-1 | P. aeruginosa (IMG) | <10-9 | |
| TA40-1 | P. aeruginosa (IMG) | <10-9 | |
| P. resinovorans | B-14151 Type | P. resinovorans (VKPM) | 1.9*10-3 |
| P. stutzeri | B-975 Type | P. stutzeri (VKM) | 2.4*10-6 |
| B-903 | P. stutzeri (VKM) | 3.4*10-4 | |
| P. oleovorans | B-8623 | P. oleovorans (VKPM) | 9.8*10-5 |
| P. straminea | B-14152 Type | P. seleniipraecipitans (VKPM) | <10-9 |
| P.putida | B-4589 Type | P.putida (VKPM) | 3.7*10-3 |
| P. syringae | B-1546 | P. savastanoi (VKM) | 1.2*10-3 |
| P. fluorescens | B-6735 | P. fluorescens (VKPM) | 5.7*10-4 |
| P22-1-2 | P. fluorescens (IMG) | 2.6*10-4 | |
| B14148 Type | P. fluorescens (VKPM) | 4.7*10-3 |
| Strain | Plasmid | mer-operon | Transposon |
|---|---|---|---|
| P. putida Tik1-1 | pPPUT-Tik1-1 | merRTPADE | ΔTn501 |
| P. putida 12969 | p12969-2 | merRTPABGD | ΔTn5041D |
| merRTPCADE | Tn5046 | ||
| P. juntendii 18091276 | pCN1276 | merRTPFADE | Tn512 |
| merRTPCADE | Tn5046 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





