1. Introduction
Plant proteins are generally more readily available, less costly and more environmentally friendly as compared to animal proteins[
1]. Chickpea (
Cicer arietinum L.) is one of the most abundant legume grown in the world and chickpeas are also very nutritious, containing a variety of beneficial compounds including carbohydrates, protein, unsaturated fatty acids, minerals, vitamins, dietary fibre and a range of isoflavones[
2]. Chickpea has a high protein content around 13-31%
[3, 4]. In addition, chickpea proteins have high bioavailability, a balanced amino acid composition and good digestive properties[
5]. Therefore, chickpea has received extensive research interest
[6, 7]. In recent years, plant proteins-derived digestion products have been widely studied due to the potential health implications of dietary peptides[
8]. Chandrasekaran and Gonzalez [
9] showed that chickpea protein hydrolysates produced from germinated chickpeas can be used to generate models to optimize the production of anti-diabetic peptides, and indicated that chickpea peptides can be used as commercial functional food ingredients. Acevedo and Gonzalezde [
10] showed that chickpea hydrolysates contributed to the prevention and management of type 2 diabetes, and the chickpea protein digests of pepsin and trypsin showed good dipeptidyl peptidase IV inhibition potential.
Food irradiation is the process of exposing food to ionising and non-ionising radiation to destroy microorganisms, bacteria, viruses or insects that may be present in the food[
11]. It is considered to be an economical, safe and environmentally friendly technology
[12, 13]. Electron beam irradiation (EBI), which consists of high-energy electrons and does not involve the use of radioactive material, can inactivate microorganisms by damaging critical components of cells[
14]. In recent years, EBI has been widely used in food preservation
[15, 16].
The free radicals generated during EBI processing can also affect the critical components of food matrix. It has been demonstrated that EBI can modify protein structure and therefore alter the protein functionality. Zhang et al.[
17] showed that EBI reduced the content of α-helix and β-sheet in the secondary structure of the okara protein, increased the surface hydrophobicity and solubility, and improved the emulsifying properties of the okara protein. Liu et al.[
18] showed that the S-S bond was broken into-SH bond after EBI, and the α-helix was transformed into β-sheet and unfolded structure, which exposed the buried hydrophobic amino acids of egg white protein, and then the antioxidant activity of egg white protein was improved.
While EBI can be applied in the preservation of chickpea protein, its effects on the functional and nutritional value of chickpea protein is generally lacking. Zhang et al.[
19] claimed that EBI was capable to improve food proteins mainly in four aspects: (1) functional improvement of proteins; (2) promoting crude protein digestibility; (3) enhancing the biological activities of proteins; (4) facilitating the proteolytic effect via the modification of the protein structure. In order to test the effects of EBI on the physicochemical properties of chickpea protein and the subsequent digested products, chickpea protein concentrates (CPC) were subjected to EBI at various doses. Protein solubility, oxidative modifications, advanced structures, and the peptidomic profile of the digested products were characterized.
2. Results
2.1. The effect of EBI on the amino acid residues of CPC
Amino acid composition can be used to evaluate protein modification[
20]. Generally, the amino acid composition of CPC was not affected by EBI (
Table 1). There were only minor changes in serine, arginine, tyrosine, phenylalanine, isoleucine and lysine after EBI. Those amino acids appeared to be increased in groups with higher dose of irradiation.
Sulfhydryl group is an important functional group of proteins and it is susceptible to oxidative modification[
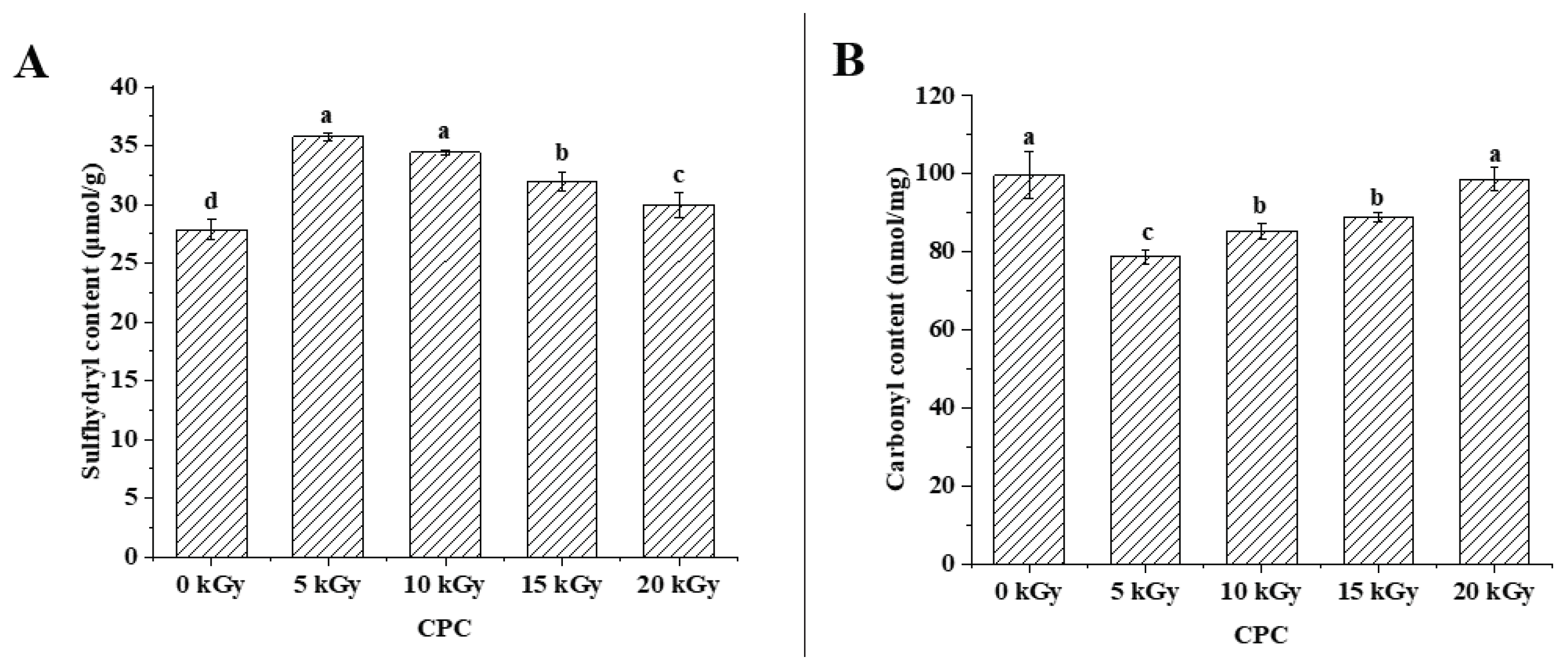
21]. Results showed that the total sulfhydryl content of CPC was higher in irradiated groups than the unirradiated group (
Figure 1A). With increased irradiation dose, sulfhydryl content decreased. Protein carbonyl content has been widely used as an indicator to evaluate protein oxidation[
22]. The carbonyl content increased with increasing irradiation dose, but the control group was higher than groups of 5-15 kGy (
Figure 1B).
2.2. The effect of EBI on the physicochemical properties of proteins in CPC
Solubility is an important physicochemical property of proteins, reflecting the interactions between proteins and between proteins and water, and can influence the functional properties of proteins[
23].
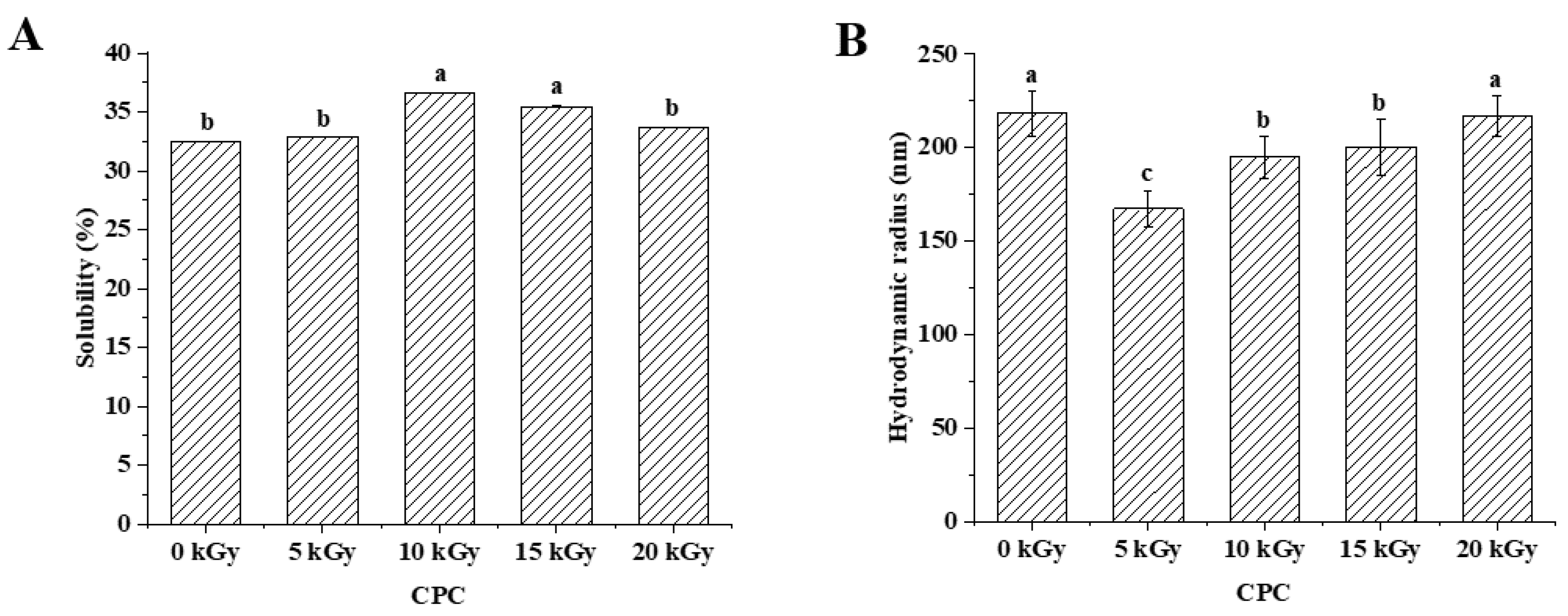
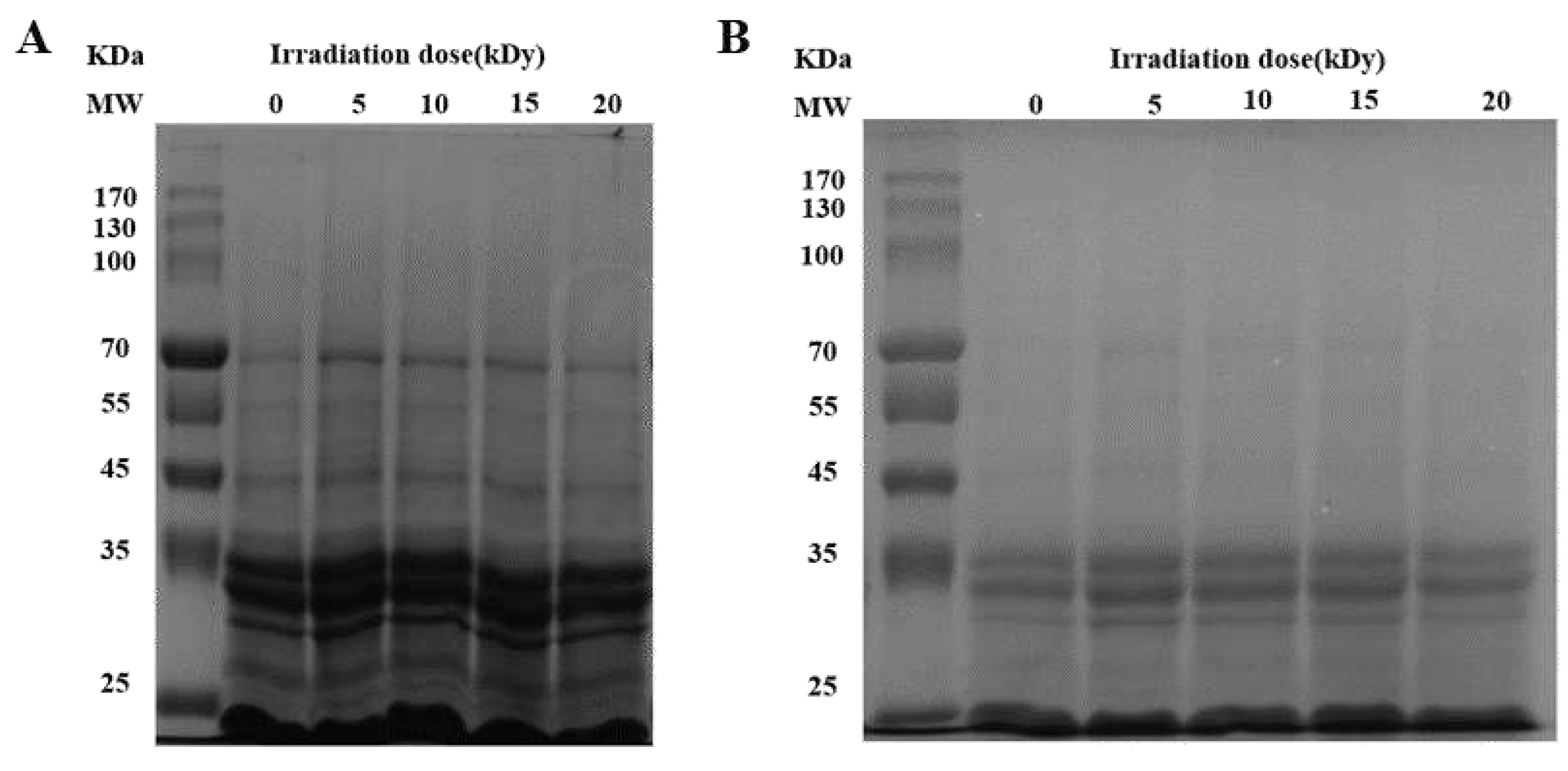
Figure 2A showed that the solubility of CPC increased and then decreased with increasing irradiation dose. The solubility increased significantly at a dose of 10 kGy as compared to the unirradiated samples. There was no further increase when the dose increased to 15 kGy. When the dose reached 20 kGy, the solubility decreased to a similar level of unirradiated. The solubilized proteins were subjected to SDS-PAGE analysis, and the results showed that the intensity of major protein bands was greater in CPC irradiated with 5, 10 and 15 kGy (
Figure 3).
Protein solubility is affected its physiochemical properties including particle size[
24], net charge[
25], surface hydrophobicity[
26], etc. The hydrodynamic diameter of irradiated CPC was generally less than unirradiated (
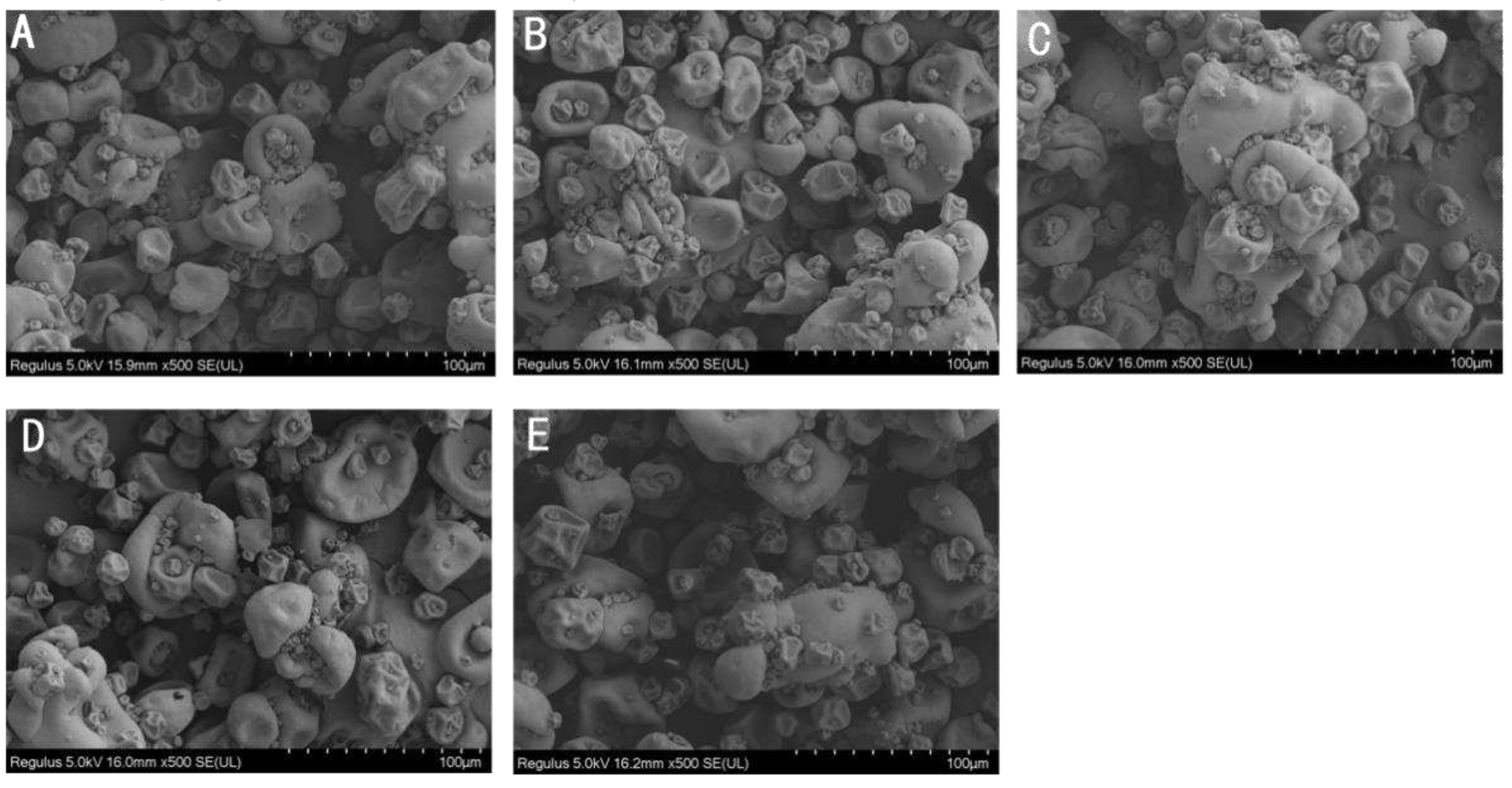
Figure 2B). Increased irradiation generally led to increased hydrodynamic diameter. The SEM image of CPC powder also showed that a higher proportion of large particles in irradiated groups (
Figure 4).
In general, zeta potential of CPC was between -20 and -30 mV, and irradiation at lower doses (5 and 10 kGy) led to a lower absolute value of zeta potential (
Figure 2C). It was observed that irradiation at 10 and 15 kGy led to a higher surface hydrophobicity (
Figure 2D).
Fourier transform infrared spectroscopy (FTIR) was used to study the effect of EBI on the secondary structure of chickpea protein. As shown in
Table 2, the secondary structure of CPC is dominant by β-sheet structure. With increasing irradiation dose, the content of α-helix generally decreased while β-sheet increased.
2.3. The effect of EBI on the peptidomics of CPC
The digestibility of protein is closely related to its nutritional quality. The in vitro digestibility of CPC of unirradiated and irradiated at doses of 5 kGy, 10 kGy, 15 kGy, 20 kGy were 94.35 %, 94.25 %, 93.96 %, 94.28 % and 93.84 %, respectively, all exceeding 93 % but without significant difference among groups.
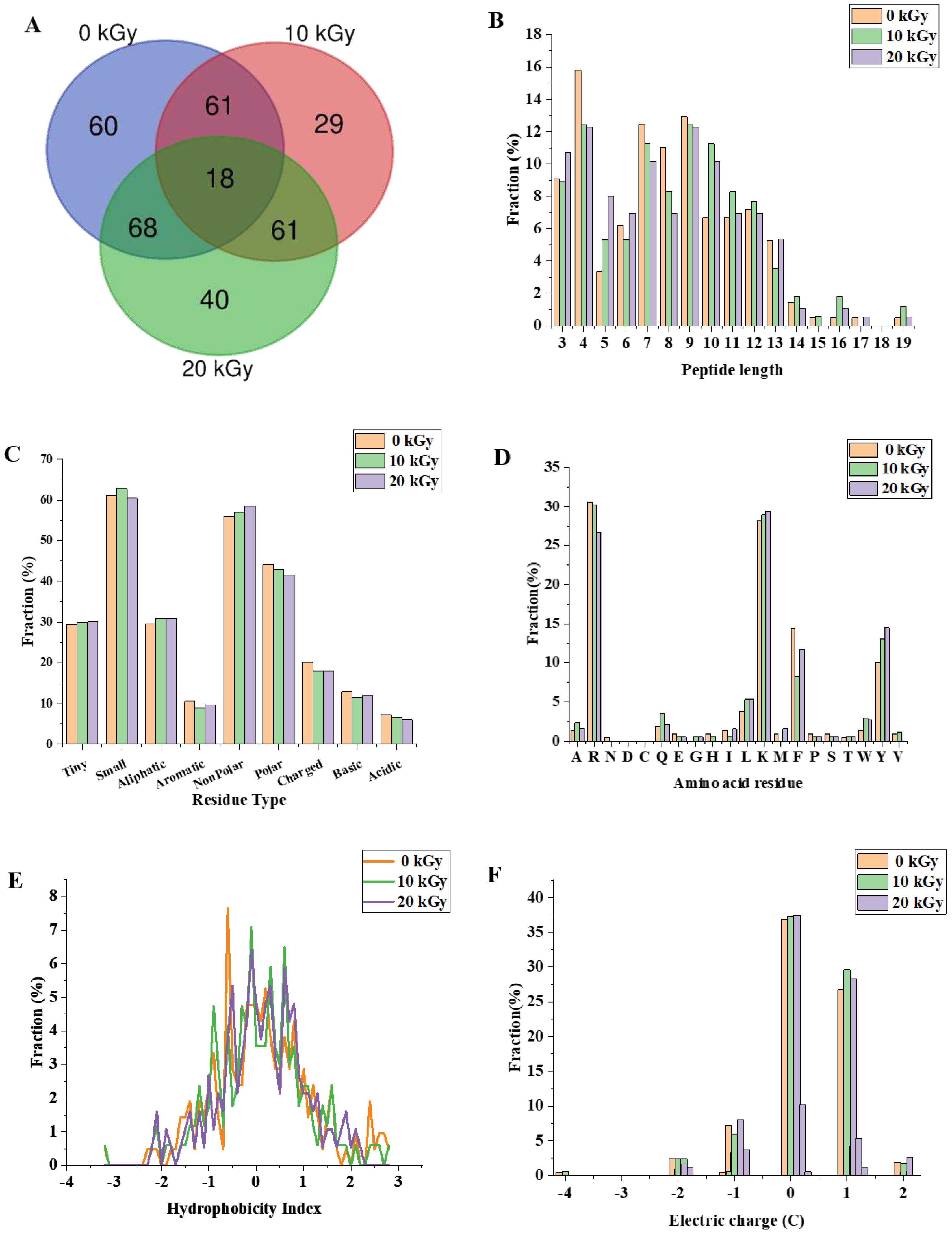
The digested CPC (0, 10 and 20 kGy) was further analysed by peptidomics. A total of 337 peptides were identified from the gastrointestinal digestion products of CPC treated with 0 kGy, 10 kGy and 20 kGy irradiation doses (
Figure 5A), with 18 overlapping peptides and 60, 29 and 40 peptides specific to 0, 10 and 20 kGy irradiation doses, respectively. A full list of those peptides can be found in the supplementary file. The majority of peptides identified had a length of 3-13 residues (
Figure 5B), with only about 2% having more than 14 residues. Irradiation generally led to a higher proportion of larger peptides. In terms of the amino acid composition of the peptides, more non-polar amino acid residues were found in irradiated groups (
Figure 5C). When the C-terminal was considered alone, it was found that the two major residues were lysine and arginine, and irradiation appeared to increase the frequency of lysine while decreased the frequency of arginine in the C-terminal of identified peptides (
Figure 5D). The theoretical hydrophobicity and net charge of the peptides were also estimated (
Figure 5E, F). The overall mean value of hydrophobicity was estimated using the Kyte-Doolittle scale, where scores below 0 indicate hydrophilicity, while values above 0 indicate hydrophobicity
[27, 28]. Overall, there were slightly more hydrophobic than hydrophilic peptides. The net charges of the three sample peptides were mainly 0 C and 1 C. The generated peptides may have various potential biological activity. ABTS radical scavenging test showed that the antioxidant capacity of the digested products decreased slightly with increased irradiation (
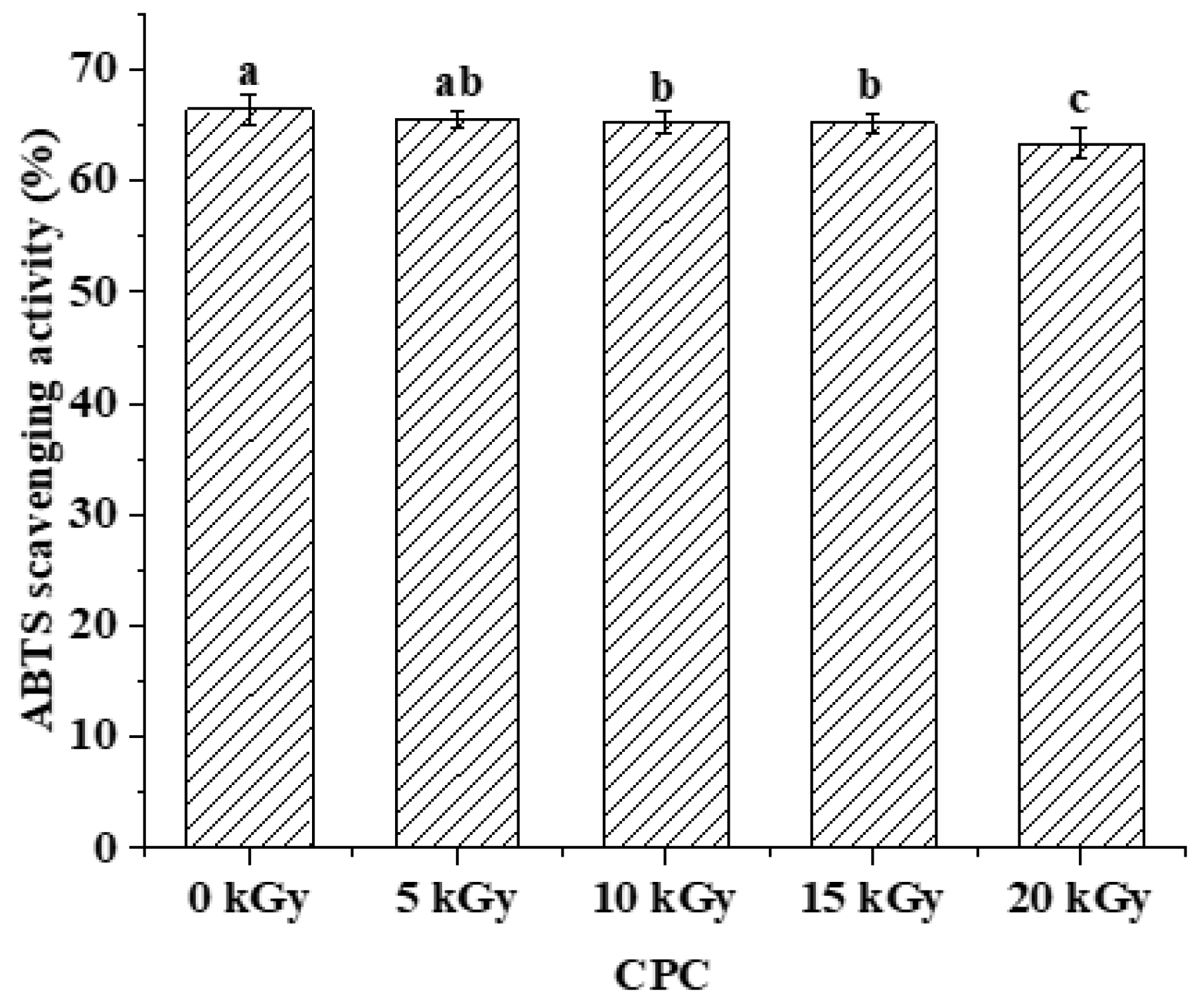
Figure 6).
3. Discussion
3.1. Oxidative modification of chickpea protein
EBI employs high-energy electron beam to damage critical components of microorganisms, and thus extends shelf-life of irradiated foods. However, the free radicals generated in EBI can lead to oxidative modifications of food components. In this study, it was found that increased irradiation from 5 kGy to 20 kGy generally led to enhanced protein oxidation in CPC indicated by decreased sulfhydryl group (
Figure 1A) and increased carbonyl group (
Figure 1B). Similarly, increased irradiation dose promoted protein oxidation were found in various foods
[17, 29, 30]. In the unirradiated CPC, protein oxidation seems to be more severe than irradiated ones, especially at lower doses of 5-15 kGy. This may be attributed to the release of bound phenolics and destruction of the activity of endogenous lipoxygenase by EBI[
31], resulting in the initial decrease in protein oxidation. With increased level of irradiation, the excessive production of harmful free radicals can offset the beneficial effects observed at lower doses, resulting in increased protein oxidation. Loss of parent amino acids has been suggested as another indicator for the quantification of protein oxidation in biological systems[
32]. In this study, the amino acid composition of CPC in different groups did not change much (
Table 1). This suggested that EBI did not lead to severe protein oxidation in CPC. According to Lund et al.[
33], the amino acid analyses is most appropriate for evaluation of large losses of amino acid residues. It is often difficult to detect small losses due to poor method sensitivity. And the hydrolysis during sample preparation may have destroyed some highly oxidizable amino acid residues, such as Met, Trp and Cys.
3.2. Physicochemical properties of irradiated chickpea protein
Protein oxidation is known to affect the physicochemical properties and eventually the functionality of food proteins[
34]. Formation of protein aggregation is one of the general consequences of protein oxidation. In the dry state, SEM images showed that a higher proportion of large particles in irradiated CPC than unirradiated ones (
Figure 4). This was supported by particle size measurement in the solubilized state (
Figure 2B). Liu et al.[
18] also found that the particle size of EBI treated egg white protein was significantly larger than that of untreated egg white protein. The hydrodynamic radius of CPC solution increased with increased irradiation dose and reached a value around 220 nm at 20 kGy, the large particle size at 20 kGy may have caused the reduction in solubility, which was also observed in SDS-PAGE that the intensity of major protein bands in CPC irradiated with 20 kGy was less than that of 5, 10 or 15 kGy. Protein aggregates may be formed through covalent cross-linking or non-covalent interaction. In this study, no evidence of protein cross-linking was observed, as the SDS-PAGE did not show new bands in irradiated CPC. Hydrophobic interaction often leads to protein aggregation. It was observed that the surface hydrophobicity of CPC was greater in 10 and 15 kGy as compared to unirradiated (
Figure 2D). The increase was likely due to protein structural changes which may expose previously buried hydrophobic amino acid residues to the surface. FTIR analysis revealed that with increasing irradiation dose, the content of α-helix generally decreased while β-sheet increased (
Table 2). Similar structural changes were reported in EBI treated myofibrillar proteins[
30]. At 20 kGy surface hydrophobicity showed a trend of decrease, likely due to extensive protein aggregation. It was found EBI up to a dose of 15 kGy increased the solubility of CPC followed by a decrease (
Figure 2A). Similar trend was reported in EBI treated okara protein that the solubility increased at lower dose and decreased at higher dose. The altered solubility was attributed to protein unfolding and deamination during irradiation[
35]. In this study, the increased hydrophobicity (
Figure 2D) and decreased zeta-potential (
Figure 2C) was expected to lower the solubility of CPC irradiated with 10 or 15 kGy, some other factors may have contributed to the observed solubility increase. However, this requires further investigation.
3.3. Characterization of the peptides of in-vitro digested chickpea protein
Amino acid side chain modification and protein structural changes are known to affect protein digestibility. Generally, low degrees of oxidation were found to enhance digestibility, whereas high degrees of oxidation reversed this effect[
27]. In this study, in-vitro digestibility was not significantly affected by EBI though there were significant protein oxidation and structural changes. Despite of similar digestibility, peptidomic analysis revealed significant difference among the three groups of CPC. Peptides from these groups differed in the composition and physicochemical properties (
Figure 5). Irradiation generated new peptides while it also led to a diminish of some other peptides which can only be found in unirradiated CPC. Increased dose from 10 kGy to 20 kGy led to even greater number of new peptides. As there were differentially abundant peptides (For full list, see supplementary file) in each group, it was expected that the physicochemical properties would vary. This expectation was supported by a theoretical analysis of the peptides which showed that the distribution of peptide length, hydrophobicity, net charge, and C-terminal residues were affected by irradiation. The theoretical analysis of peptide mixture was also performed by Boachie et al.[
27] who investigated how the interaction with tannic acid affected the peptides of digested lentil protein. Digestion of irradiated CPC generally led to a higher proportion of large peptides, possibly due to the chemical modification and structural changes. According to Xiong et al.[
36], oxidative modification can change the proteolytic sites, this was supported by our finding that the frequency of top 4 kinds (R, K, Y, F) of C-terminal amino acid residues were affected by irradiation. Of those 4 residues, arginine and lysine are the major cleavage sites of trypsin while tyrosine and phenylalanine correspond to the major cleavage sites of pepsin. Both chickpea protein[
37] and its hydrolysates[
5] have been demonstrated to possess antioxidant properties. In this study, the antioxidant capacity of the digested products was negatively affected by irradiation, but with only a marginal decrease (
Figure 6). In addition to antioxidant activity, the digested products may have a range of biological activity such as antimicrobial activity, ACE inhibitory activity, etc. The peptides identified in this study can be compared to database of biological active peptides, thereby serving as an initial screen step for further studies.4. Materials and Methods
4.1. Materials and electron beam irradiation
Chickpea protein concentrate, CPC (protein: 64.9%, moisture: 4.46%, ash: 3.90%) was purchased from Shandong Jianyuan Biological Engineering Co.. Pepsin (enzyme activity > 3000 units/mg) was purchased from Shanghai Maclean Biochemical Technology Co.. Trypsin (enzyme activity > 2500 units/mg) was purchased from Shanghai Yuanye Biotechnology Co.. All other chemicals were of analytical grade and purchased from Sinopharm Chemical Reagent Co., ltd. (Shanghai, China). For the electron beam irradiation, CPC were exposed to different doses (0, 5, 10, 15 and 20 kGy) of high-energy electron beam (produced from a 10 MeV, 25 kW electron-beam accelerator) at a dosage rate of 2 kGy/h. Different doses were achieved through exposure of different time.
4.2. Amino acid content
CPC (150.0 mg) was weighed into a hydrolysis tube, 8 mL of 6 M HCl was added, nitrogen was used to remove the air in the hydrolysis tube. The tubes were then placed in an oven at 120 ℃ and hydrolyzed for 22 h. The hydrolysates were then transferred to a volumetric flask, neutralized by adding 4.8 mL of 10 M NaOH, and the volume was adjusted to 25 mL with deionized water. The suspensions were centrifugated at 5000 r/min for 5 min at room temperature (Model TGL-16gR, Shanghai Anting Scientific Instrument Factory, China), and the supernatant was subject to amino acid analysis according to the method of Jiang et al.[
38].
4.3. Oxidative modification of amino acids
4.3.1. Sulfhydryl content
Total sulfhydryl content was determined using Ellman's reagent (DTNB) according to the method of Cheng et al.[
39]. The absorbance was measured at 412 nm (Model UV1601, Riley Analytical Instruments, Beijing, China).
A412, the absorbance at 412 nm; D, dilution factor; and C, protein concentration (mg protein/mL).
4.3.2. Carbonyl content
The carbonyl content was measured using 2,4-dinitrophenyl hydra-zine (DNPH). CPC (0.03 g) were suspended with 3 mL 0.1 M Tris-HCl (pH 8.0, 5 % Sodium dodecyl sulfate w/v). The suspension was heated at 80 ℃ for 30 min and then cooled to room temperature. And the suspension was subject to determination of carbonyl content according to the method of Yu et al.[
40].The carbonyl content was calculated according to the equation:
4.4. Solubility of CPC
The protein concentration of CPC was determined with the biuret reagent according to the method of Ji et al.[
41] with some modifications. A 1% (w/v) CPC suspension was first centrifuged (8000 r/min, 4 °C, 10 min), and then 1 mL of the supernatant was mixed with 4 mL of biuret reagent. After a water bath at 37 °C for 20 min, the absorbance at 540 nm was measured to obtain the protein concentration. Protein solubility was defined as protein content in the supernatant divided by total protein in the suspension.
4.5. Particle size and zeta potential of CPC
A 1% (w/v) CPC suspension was first centrifuged (8000 r/min, 4 °C, 10 min). The protein concentration of supernatant was determined with the biuret reagent, and then the protein concentration of the supernatant was adjusted to 1 mg/mL. The particle size and zeta potential of the CPC were then measured at 25 °C using a particle size meter (Model Litesizer 500, Anton Paar Trading Ltd., USA). The parameters of the test were as fol lows: equilibration time of 60 s, and the test temperature of 25 °C. The refractive index was set to 1.46 and the absorption coefficient to 0.01.
4.6. Surface hydrophobicity
The surface hydrophobicity of CPC was determined using the 8-Anilino-1-naphthalenesulfonic Acid (ANS) fluorescent probe method, slightly modified from Gulseren et al.[
42]. CPC (1 mg/mL) was first dispersed in 0.01 M phosphate buffer (Na
2HPO
4 - NaH
2PO
4,pH 7.0). The protein suspension was then centrifuged (8000 r/min, 4 °C, 15 min), and protein concentration of supernatant was determined with the biuret reagent as mentioned above. Next, the supernatant was diluted with phosphate buffers to 0.5, 0.1, 0.025, 0.005, and 0.001 mg/mL respectively. ANS was dissolved in 0.01 M (pH 7.0) phosphate buffer and 40 μL of ANS solution was added to 4 mL protein solution. Absorbance was then measured using a fluorescence spectrophotometer (Model Cary Eclipse, Agilent Technologies Ltd., USA) at an excitation wavelength of 280 nm and an emission wavelength of 355 nm. The H
0 index was calculated as the initial slope of the fluorescence intensity versus protein concentration by linear regression analysis.
4.7. Fourier transform infrared spectroscopy (FTIR) analysis
CPC was mixed with KBr(1:200), ground and then a total of 200 mg of the mixture powder was pressed into a narrow slice. According to the method of Wang et al.[
43], the Fourier transform infrared (FTIR) spectra were recorded by the FTIR spectrometer (Model Nicolet iS10, Thermo Fisher Scientific, USA). The wavelength range was measured from 4000 to 400 cm
-1. The obtained FTIR spectra were further processed using Omnic V8.1 (Version V8.1, Thermo Fisher Scientific, USA) and PeakFit 4.12 (Version 4.12, Systat Software, San Jose, CA). Baseline correction, deconvolution, and second-order derivatives were performed on the original data at the fraction of 1,600-1,700 cm
-1. The following ranges were assigned to each secondary structure: β-Sheet (1,615-1,637 cm
-1, 1682-1700cm
-1), Random coil (1,637-1,645 cm
-1), α-Helix (1,646-1,664 cm
-1), and β-Turn (1,664-1,681 cm
-1). The percentages of each secondary structure were calculated to present the relative content of secondary structures in the CPC.
4.8. SDS polyacrylamide gel electrophoresis (SDS-PAGE)
Electrophoresis was carried out with 12% separating gel and 4% stacking gel according to the method of Wang et al.[
44] under reduced (+DTT) condition. CPC solution (80μL, 4 mg/mL) was mixed with 20 μL 5X sample buffer. Then the mixture was heated under 95 °C for 10 min and then cooled to room temperature. Later, the mixture (10 μL, 8μL) was loaded to the gel. Electrophoresis was first carried out at a voltage of 80 V for 30 min and then 120 V for 110 min. Next, Coomassie brilliant blue R-250 was used to stain the gel for 30 min. After destaining overnight, the gel scanned with a multifunctional gel image analysis system (Model Tanon MINI Space, Shanghai Tianneng Life Science Co., Ltd., China).
4.9. Scanning electron microscopy (SEM)
Chickpea protein powder was freeze-dried (Model FD-1A-50, Boyikang (Beijing) Instrument Co., Ltd., China), and protein powder after freeze-drying was subject to scanning electron microscope observation according to the method of Zhang et al.[
45].
4.10. Intrinsic fluorescence spectroscopy (IFS)
The method of Jiang et al.[
46] was used to determine intrinsic fluorescence spectroscopy, and the method was slightly modified. CPC (1 mg/mL) was first dispersed in 0.01 M phosphate buffer (Na
2HPO
4 - NaH
2PO
4,pH 7.0). The protein suspension was then centrifuged (8000 r/min, 4 °C, 15 min), and protein concentration of supernatant was determined with the biuret reagent as mentioned above. Next, the supernatant was diluted with phosphate buffers to 0.5 mg/mL. The fluorescence intensity of the samples was performed with a fluorescence spectrophotometer (Model Cary Eclipse, Agilent Technologies Ltd., USA) at room temperature. Intrinsic spectra were recorded between 300 and 450 nm with an excitation wavelength of 280 nm (slit = 2.5 nm) at 10 nm s
-1 of scanning speed.
4.11. In vitro digestion of CPC
4.11.1. In vitro digestibility
The method of Wen et al.[
47] was referenced with slight modifications. CPC (0.32 g) was added to deionized water (16 mL) and pH of the suspension was adjusted to 2.0 ± 0.1 with 1 M HCl. Pepsin premix (0.8 mg/mL, 4 mL) was added to each sample. The mixture was shaken uniformly at 37 °C for 1 h. The pH of the suspension was quickly adjusted to 7.5±0.1 with 1 M NaOH. The mixture was added to 4 mL of trypsin premix (0.24 mg/mL), and the mixture was shaken uniformly at 37 ℃ for 2 h. The enzymatic reaction was terminated by heating at 95 ℃ for 5 min, and 4 mL of the mixture was removed as the gastrointestinal digested products. The gastrointestinal digested products were added to 12 mL of anhydrous ethanol at 4 ℃ and left to stand for 12 h. The product was centrifuged (8000 r/min, 15 min, 4 ℃). The precipitate after separation is used to determine the protein content after digestion and the supernatant is used for peptidomic identification.
Determination of in vitro digestibility of proteins:
M1, protein content of the sample before digestion; M2, the protein content of the sample in the precipitate after digestion.
4.11.2. Peptidomic analysis
Unirradiated and irradiated samples (doses of 10 kGy and 20 kGy) were taken from the alcohol-soluble supernatant of the gastrointestinal digest of CPC (described in 4.11.1). The supernatant was freeze-dried (Model FD-1A-50, Boyikang (Beijing) Instrument Co., Ltd., China). The obtained powder of peptides were re-dissolved in 0.1% formic acid and analysed by Q-Exactive Plus coupled to an EASY-nano LC 1200 system (Thermo Fisher Scientific, MA, USA). A1 μL peptide sample was loaded onto a 25 cm analytical column (75 μm inner diameter, 1.9 μm resin) and separated with a 60 min-gradient starting at 2% (80% acetonitrile with 0.1% formic acid) followed by a stepwise increase to 35% in 47 min, 100% in 1 min and stayed there for 12 min. The flow rate was maintained at 300 nL/min, and the column temperature set at 40 °C.
The mass spectrometer was run under data dependent acquisition (DDA) mode. The survey of full scan MS spectra (m/z 200-2000) was acquired in the Orbitrap with 70,000 resolution.
Tandem mass spectra were processed by PEAKS Studio version 10.6 (Bioinformatics Solutions Inc., Waterloo, Canada). The database was Cicer arietinum (24812 entries) download from uniprot. Pepsin and trypsin was the digestion enzyme and semi-specific was the digest type. PEAKS DB were searched with a fragment ion mass tolerance of 0.02 Da and a parent ion tolerance of 10 ppm. The max missed cleavage was set to 2. Oxidation on methionine, Deamidation on asparagine and glutamine were specified as the variable modifications. The peptides with 1% FDR and the proteins with at least 1 unique peptide were filtered. “Match between runs” was enabled with default settings. Label free quantification was also performed using Peaks Studio. Relative abundance of peptide features (precursor peak area) was detected in multiple samples. Feature detection is performed separately on each sample, and then the features of the same peptide from different samples are reliably aligned together using a high-performance retention time alignment algorithm. Normalization was performed on total ion current (TIC) of the samples and normalized abundance is calculated from the raw abundance divided by the normalization factor.
A Venn diagram depicting the degree of similarity of peptides in the gastrointestinal digestion products of the three groups was generated by online tools (
https://bioinformatics.psb.ugent.be/webtools/Venn/). Peptide properties were obtained from the Peptides R package[
48].
4.11.3. ABTS radical scavenging capacity
The 2, 2'-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) in the digestion products of CPC was determined according to the method of Xing et al.[
49]. Results were expressed as ABTS radical scavenging activity, calculated using the following equation:
Ac, the absorbance of ABTS reacted with the sample at OD734; M, the absorbance of ABTS reacted with distilled water at OD734.
4.12. Statistical analysis
All measurements were performed in triplicate. Results were expressed as mean ± standard deviation (SD). Data were analysed using IBM SPSS software (version 20.0, IBM, Chicago, IL, USA). One-way analysis of variance (ANOVA) was done with p < 0.05 set as statistically significant.
5. Conclusions
In this study, it was found that electron beam irradiation induced significant protein oxidation and altered the structure and physicochemical properties of chickpea protein in a dose-dependent manner. Proteins were found to be aggregated, likely due to hydrophobic interaction rather than covalent protein cross-linking. The apparent digestibility of CPC at different doses was similar to that of unirradiated CPC, however, the peptide profile of the digests differed among CPC at 0, 10 and 20 kGy. Peptidomic analysis showed that the distribution of peptide length, hydrophobicity, net charge, and C-terminal residues were all affected by irradiation. Arginine, lysine, tyrosine and phenylalanine, being the major cleavage sites of trypsin or pepsin used in the in-vitro digestion test, their frequency in the C-terminal were all affected, suggesting that irradiation altered the proteolytic behavior of chickpea protein. Antioxidant capacity of the digests was negatively affected by irradiation, it was only a minor effect though. This study provided theoretical guidance in the application of electron beam irradiation technology in chickpea protein concentrate.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, supplementary file 1.
Author Contributions
Conceptualization, Yulong Bao and Yang Zhen; methodology, Yulong Bao, Yaqi Zhang; software, Yulong Bao; validation, Yaqi Zhang and Wanjun Xu; formal analysis, Yaqi Zhang and Yunfei Kong; investigation, Yaqi Zhang; resources, Yulong Bao; writing—original draft preparation, Yaqi Zhang and Yulong Bao; writing—review and editing, Yulong Bao; visualization, Yaqi Zhang; supervision, Yulong Bao; project administration, Yulong Bao; funding acquisition, Yulong Bao. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Senior Talent Program of Jiangsu University (20JDG062).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data will be made available upon reasonable request.
Acknowledgments
The authors acknowledge Ms. Keyu Wang at Jiangnan University for her help in the analysis of amino acid content.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Felix, M.; Cermeno, M.; Romero, A.; FitzGerald, R. J. Characterisation of the bioactive properties and microstructure of chickpea protein-based oil in water emulsions. Food Res Int 2019, 121, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Jukanti, A. K.; Gaur, P. M.; Gowda, C. L.; Chibbar, R. N. Nutritional quality and health benefits of chickpea (Cicer arietinum L.): a review. Br J Nutr 2012, 108 Suppl 1, S11–26. [Google Scholar] [CrossRef] [PubMed]
- Glusac, J.; Isaschar-Ovdat, S.; Fishman, A. Transglutaminase modifies the physical stability and digestibility of chickpea protein-stabilized oil-in-water emulsions. Food Chem 2020, 315, 126301. [Google Scholar] [CrossRef] [PubMed]
- Bi, C. H.; Chi, S. Y.; Zhou, T.; Zhang, J. Y.; Wang, X. Y.; Li, J.; Shi, W. T.; Tian, B.; Huang, Z. G.; Liu, Y. Effect of low-frequency high-intensity ultrasound (HIU) on the physicochemical properties of chickpea protein. Food Res Int 2022, 159, 111474. [Google Scholar] [CrossRef]
- Perovic, M. N.; Pajin, B. S.; Antov, M. G. The effect of enzymatic pretreatment of chickpea on functional properties and antioxidant activity of alkaline protein isolate. Food Chem 2022, 374, 131809. [Google Scholar] [CrossRef]
- Khrisanapant, P.; Kebede, B.; Leong, S. Y.; Oey, I. Effects of Hydrothermal Processing on Volatile and Fatty Acids Profile of Cowpeas (Vigna unguiculata), Chickpeas (Cicer arietinum) and Kidney Beans (Phaseolus vulgaris). Molecules 2022, 27(23). [Google Scholar] [CrossRef]
- Perez-Perez, L. M.; Huerta-Ocampo, J. A.; Ruiz-Cruz, S.; Cinco-Moroyoqui, F. J.; Wong-Corral, F. J.; Rascon-Valenzuela, L. A.; Robles-Garcia, M. A.; Gonzalez-Vega, R. I.; Rosas-Burgos, E. C.; Corella-Madueno, M. A. G.; Del-Toro-Sanchez, C. L. Evaluation of Quality, Antioxidant Capacity, and Digestibility of Chickpea (Cicer arietinum L. cv Blanoro) Stored under N(2) and CO(2) Atmospheres. Molecules 2021, 26(9). [Google Scholar] [CrossRef]
- Peng, L.; Guo, F.; Pei, M.; Tsao, R.; Wang, X.; Jiang, L.; Sun, Y.; Xiong, H. Anti-inflammatory effect of lentil hull (Lens culinaris) extract via MAPK/NF-κB signaling pathways and effects of digestive products on intestinal barrier and inflammation in Caco-2 and Raw264.7 co-culture. J Funct Foods 2022, 92. [Google Scholar] [CrossRef]
- Chandrasekaran, S.; Gonzalez de Mejia, E. Optimization, identification, and comparison of peptides from germinated chickpea (Cicer arietinum) protein hydrolysates using either papain or ficin and their relationship with markers of type 2 diabetes. Food Chem 2022, 374, 131717. [Google Scholar] [CrossRef]
- Acevedo Martinez, K. A.; Gonzalezde Mejia, E. Comparison of five chickpea varieties, optimization of hydrolysates production and evaluation of biomarkers for type 2 diabetes. Food Res Int 2021, 147, 110572. [Google Scholar] [CrossRef]
- Farkas, J.; Mohácsi-Farkas, C. History and future of food irradiation. TRENDS FOOD SCI TECH 2011, 22(2-3), 121–126. [Google Scholar] [CrossRef]
- Li, T.; Wang, L.; Sun, D.; Li, Y.; Chen, Z. Effect of enzymolysis-assisted electron beam irradiation on structural characteristics and antioxidant activity of rice protein. J. Cereal Sci 2019, 89. [Google Scholar] [CrossRef]
- Wang, L.; Ding, Y.; Zhang, X.; Li, Y.; Wang, R.; Luo, X.; Li, Y.; Li, J.; Chen, Z. Effect of electron beam on the functional properties and structure of defatted wheat germ proteins. J Food Eng 2017, 202, 9–17. [Google Scholar] [CrossRef]
- Steele, J. H. Food irradiation: a public health challenge for the 21st century. Clin Infect Dis 2001, 33, 376–7. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, C.; He, J.; Wu, Z.; Sun, Y.; Pan, D.; Tian, H.; Xia, Q. Combining e-beam irradiation and modified atmosphere packaging as a preservation strategy to improve physicochemical and microbiological properties of sauced duck product. Food Control 2022, 136. [Google Scholar] [CrossRef]
- Wei, Q.; Mei, J.; Xie, J. Application of electron beam irradiation as a non-thermal technology in seafood preservation. Lwt 2022, 169. [Google Scholar] [CrossRef]
- Zhang, M.; Feng, X.; Liang, Y.; He, M.; Geng, M.; Huang, Y.; Teng, F.; Li, Y. Effects of electron beam irradiation pretreatment on the structural and functional properties of okara protein. Innov Food Sci Emerg 2022, 79. [Google Scholar] [CrossRef]
- Liu, X.; Liu, J.; Zhang, W.; Han, S.; Zhang, T.; Liu, B. Electron beam irradiation-induced structural changes increase the antioxidant activities of egg white protein. Lwt 2019, 111, 846–852. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Lu, H.; Zong, Z.; Chen, Z.; Li, Y.; Luo, X.; Li, Y. Preservation of hydrogen peroxide-induced oxidative damage in HepG-2 cells by rice protein hydrolysates pretreated with electron beams. Sci Rep 2020, 10, 8415. [Google Scholar] [CrossRef]
- Miron, L.; Montevecchi, G.; Bruggeman, G.; Macavei, L. I.; Maistrello, L.; Antonelli, A.; Thomas, M. Functional properties and essential amino acid composition of proteins extracted from black soldier fly larvae reared on canteen leftovers. Innov Food Sci Emerg 2023, 87. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, J.; He, J.; Xu, Y.; Guo, X. Effects of high-pressure homogenization on the physicochemical, foaming, and emulsifying properties of chickpea protein. Food Res. Int 2023, 170. [Google Scholar] [CrossRef] [PubMed]
- Soglia, F.; Petracci, M.; Ertbjerg, P. Novel DNPH-based method for determination of protein carbonylation in muscle and meat. Food Chem 2016, 197 (Pt A), 670–5. [Google Scholar] [CrossRef]
- Jun, S.; Yaoyao, M.; Hui, J.; Obadi, M.; Zhongwei, C.; Bin, X. Effects of single- and dual-frequency ultrasound on the functionality of egg white protein. J Food Eng 2020, 277. [Google Scholar] [CrossRef]
- Fu, L.; Wang, Z.; He, Z.; Zeng, M.; Qin, F.; Chen, J. Effects of soluble aggregates sizes on rheological properties of soybean protein isolate under high temperature. Lwt 2023, 182. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, N.; Li, Y.; Cheng, S.; Jiang, C.; Lin, S. Effects of electron beam irradiation (EBI) on structure characteristics and thermal properties of walnut protein flour. Food Res Int 2017, 100 (Pt 1), 850–857. [Google Scholar] [CrossRef]
- Huang, X.; Yan, C.; Lin, M.; He, C.; Xu, Y.; Huang, Y.; Zhou, Z. The effects of conjugation of walnut protein isolate with polyphenols on protein solubility, antioxidant activity, and emulsifying properties. Food Res. Int 2022, 161. [Google Scholar] [CrossRef] [PubMed]
- Boachie, R. T.; Okagu, O. D.; Abioye, R.; Huttmann, N.; Oliviero, T.; Capuano, E.; Fogliano, V.; Udenigwe, C. C. Lentil Protein and Tannic Acid Interaction Limits in Vitro Peptic Hydrolysis and Alters Peptidomic Profiles of the Proteins. J Agric Food Chem 2022, 70, 6519–6529. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.; Mirdamadi, S.; Safavi, M.; Hadizadeh, M. In vitro and in silico studies of novel synthetic ACE-inhibitory peptides derived from Saccharomyces cerevisiae protein hydrolysate. Bioorg Chem 2019, 87, 647–654. [Google Scholar] [CrossRef]
- Feng, X.; Jo, C.; Nam, K. C.; Ahn, D. U. Impact of electron-beam irradiation on the quality characteristics of raw ground beef. Innov Food Sci Emerg 2019, 54, 87–92. [Google Scholar] [CrossRef]
- Lv, M.; Mei, K.; Zhang, H.; Xu, D.; Yang, W. Effects of electron beam irradiation on the biochemical properties and structure of myofibrillar protein from Tegillarca granosa meat. Food Chem 2018, 254, 64–69. [Google Scholar] [CrossRef]
- Kumari, S.; Gupta, O. P.; Mishra, C. B.; Thimmegowda, V.; Krishnan, V.; Singh, B.; Sachdev, A.; Dahuja, A. Gamma irradiation, an effective strategy to control the oxidative damage of soy proteins during storage and processing. Radiat Phys Chem 2020, 177. [Google Scholar] [CrossRef]
- Hawkins, C. L.; Morgan, P. E.; Davies, M. J. Quantification of protein modification by oxidants. Free Radic Biol Med 2009, 46, 965–88. [Google Scholar] [CrossRef]
- Lund, M. N.; Heinonen, M.; Baron, C. P.; Estevez, M. Protein oxidation in muscle foods: a review. Mol Nutr Food Res 2011, 55, 83–95. [Google Scholar] [CrossRef]
- Liu, Y.; Mubango, E.; Dou, P.; Bao, Y.; Tan, Y.; Luo, Y.; Li, X.; Hong, H. Insight into the protein oxidation impact on the surface properties of myofibrillar proteins from bighead carp. Food Chem 2023, 411, 135515. [Google Scholar] [CrossRef]
- Dogbevi, M. K.; Vachon, C.; Lacroix, M. Effect of gamma irradiation on the microbiological quality and on the functional properties of proteins in dry red kidney beans (Phaseolus vulgaris). Radiat Phys Chem 2000, 57(3-6), 265–268. [Google Scholar] [CrossRef]
- Xiong, Y. L.; Guo, A. Animal and Plant Protein Oxidation: Chemical and Functional Property Significance. Foods 2020, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Arcan, I.; Yemenicioğlu, A. Antioxidant activity of protein extracts from heat-treated or thermally processed chickpeas and white beans. Food Chem 2007, 103, 301–312. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, B.; Li, B.; Wang, C.; Luo, Y. Preparation and identification of peptides and their zinc complexes with antimicrobial activities from silver carp (Hypophthalmichthys molitrix) protein hydrolysates. Food Res Int 2014, 64, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Sun, J.; Yao, K.; Xu, M.; Zhou, X. Nondestructive detection and visualization of protein oxidation degree of frozen-thawed pork using fluorescence hyperspectral imaging. Meat Sci 2022, 194, 108975. [Google Scholar] [CrossRef]
- Yu, Q.; Shi, T.; Xiong, Z.; Yuan, L.; Hong, H.; Gao, R.; Bao, Y. Oxidation affects dye binding of myofibrillar proteins via alteration in net charges mediated by a reduction in isoelectric point. Food Res Int 2023, 163, 112204. [Google Scholar] [CrossRef]
- Ji, H.; Dong, S.; Han, F.; Li, Y.; Chen, G.; Li, L.; Chen, Y. Effects of Dielectric Barrier Discharge (DBD) Cold Plasma Treatment on Physicochemical and Functional Properties of Peanut Protein. Food Bioprocess Tech 2017, 11, 344–354. [Google Scholar] [CrossRef]
- Gulseren, I.; Guzey, D.; Bruce, B. D.; Weiss, J. Structural and functional changes in ultrasonicated bovine serum albumin solutions. Ultrason Sonochem 2007, 14, 173–83. [Google Scholar] [CrossRef]
- Wang, T.; Liu, F.; Wang, R.; Wang, L.; Zhang, H.; Chen, Z. Solubilization by freeze-milling of water-insoluble subunits in rice proteins. Food Funct 2015, 6, 423–30. [Google Scholar] [CrossRef]
- Wang, H.; Ke, B.; Wang, W.; Guo, J.; Ying, W.; Ma, S.; Jiang, H. A novel method for the component identification of human blood products: Mass spectrometric analysis of human fibrinogen digested after SDS-PAGE in-gel digestion. J Chromatogr B 2023, 1226. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Chen, Z.; Li, Y.; Luo, X.; Li, Y. Effect of electron beam irradiation on the structural characteristics and functional properties of rice proteins. RSC Adv 2019, 9, 13550–13560. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, M.; Liu, H.; Li, Q.; Xue, D.; Nian, Y.; Zhao, D.; Shan, K.; Dai, C.; Li, C. Ultrasound treatment can increase digestibility of myofibrillar protein of pork with modified atmosphere packaging. Food Chem 2022, 377. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Zhou, G.; Li, L.; Xu, X.; Yu, X.; Bai, Y.; Li, C. Effect of cooking on in vitro digestion of pork proteins: a peptidomic perspective. J Agric Food Chem 2015, 63, 250–61. [Google Scholar] [CrossRef] [PubMed]
- Osorio, D.; Rondón-Villarreal, P.; Torres, R. Peptides: A Package for Data Mining of Antimicrobial Peptides. The R Journal 2015, 7(1). [Google Scholar] [CrossRef]
- Xing, Z.; Li, J.; Zhang, Y.; Gao, A.; Xie, H.; Gao, Z.; Chu, X.; Cai, Y.; Gu, C. Peptidomics comparison of plant-based meat alternatives and processed meat after in vitro digestion. Food Res Int 2022, 158, 111462. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).