Submitted:
29 June 2023
Posted:
30 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Organelle characteristics and comparative analysis
2.2. Foreign sequences in the organelle
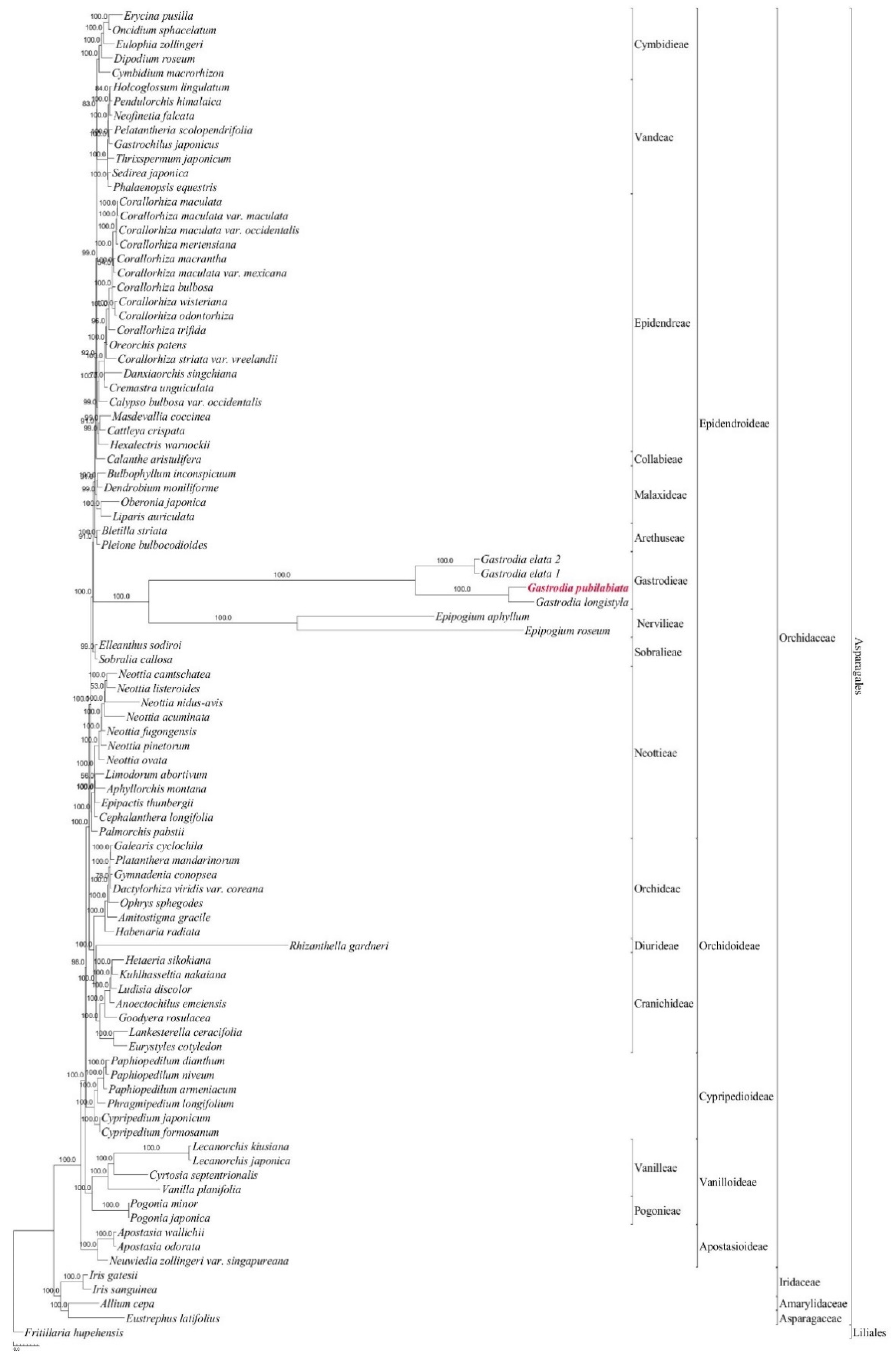
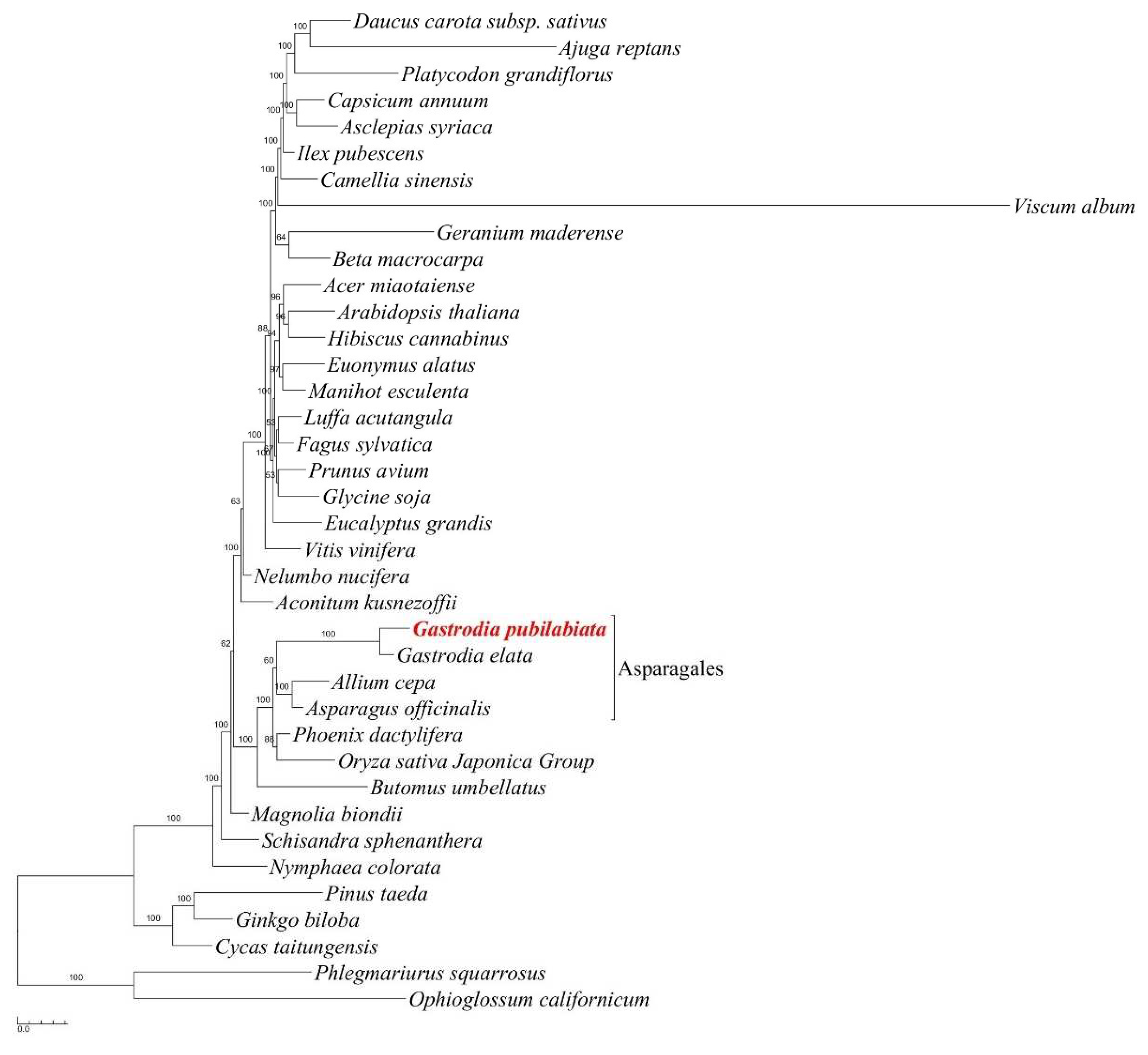
2.3. Phylogenetic position of Gastrodia pubilabiata
3. Discussion
3.1. Organelle genome evolution
3.2. Putative gene transfer
3.3. Phylogenetic implications
4. Materials and Methods
4.1. Plant material and DNA extraction
4.2. Sequencing, Assembly and Annotations
4.3. Phylogenetic analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LSC | Large Single Copy |
| SSC | Small Single Copy |
| IR | Inverted Repeat |
| ML | Maximum Likelihood |
| HGT | Horizontal Gene Transfer |
| IGT | Intracellular Gene Transfer |
| SSR | Simple Sequence Repeat |
| MTPT | Mitochondrial Plastome sequences |
References
- Goremykin, V. V.; Hirsch-Ernst, K.I.; Wölfl, S.; Hellwig, F.H. Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol. Biol. Evol. 2003, 20, 1499–1505. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Penaflor, C.; Kuehl, J. V.; Leebens-Mack, J.; Carlson, J.E.; DePamphilis, C.W.; Boore, J.L.; Jansen, R.K. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: Implications for the phylogenetic relationships of magnoliids. BMC Evol. Biol. 2006, 6, 1–20. [Google Scholar] [CrossRef]
- Givnish, T.J.; Spalink, D.; Ames, M.; Lyon, S.P.; Hunter, S.J.; Zuluaga, A.; Iles, W.J.D.; Clements, M.A.; Arroyo, M.T.K.; Leebens-Mack, J.; et al. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B Biol. Sci. 2015, 282, 20151553. [Google Scholar] [CrossRef]
- Sun, M.; Naeem, R.; Su, J.X.; Cao, Z.Y.; Burleigh, J.G.; Soltis, P.S.; Soltis, D.E.; Chen, Z.D. Phylogeny of the Rosidae: A dense taxon sampling analysis. J. Syst. Evol. 2016, 54, 363–391. [Google Scholar] [CrossRef]
- DePamphilis, C.W.; Palmer, J.D.; Rounsley, S.; Sankoff, D.; Schuster, S.C.; Ammiraju, J.S.S.; Barbazuk, W.B.; Chamala, S.; Chanderbali, A.S.; Determann, R.; et al. The Amborella genome and the evolution of flowering plants. Science (80-. ). 2013, 342. [Google Scholar] [CrossRef]
- Chen, J.; Huang, Q.; Gao, D.; Wang, J.; Lang, Y.; Liu, T.; Li, B.; Bai, Z.; Luis Goicoechea, J.; Liang, C.; et al. Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef]
- Liu, Z.J. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015, 47, 65–72. [Google Scholar] [CrossRef]
- Mayer, K.F.X.; Marcussen, T.; Sandve, S.R.; Heier, L.; Pfeifer, M.; Kugler, K.G.; Zhan, B.; Spannagl, M.; Pfeifer, M.; Jakobsen, K.S.; et al. A chromosome-based draft sequence of the hexaploid bread wheat ( Triticum aestivum ) genome Ancient hybridizations among the ancestral genomes of bread wheat Genome interplay in the grain transcriptome of hexaploid bread wheat Structural and functional pa. Science 2014, 345, 1250092. [Google Scholar] [CrossRef]
- Wu, Z.; Sloan, D.B. Recombination and intraspecific polymorphism for the presence and absence of entire chromosomes in mitochondrial genomes. Heredity (Edinb). 2019, 122, 647–659. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Li, P.; Zhang, Y.; Akhter, D.; Pan, R.; Fu, Z.; Huang, M.; Li, X.; Feng, Y. Unprecedented organelle genomic variations in morning glories reveal independent evolutionary scenarios of parasitic plants and the diversification of plant mitochondrial complexes. BMC Biol. 2022, 20, 1–16. [Google Scholar] [CrossRef]
- Skippingtona, E.; Barkmanb, T.J.; Ricea, D.W.; Palmera, J.D. Miniaturized mitogenome of the parasitic plant viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, E3515–E3524. [Google Scholar] [CrossRef]
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012, 10. [Google Scholar] [CrossRef]
- Sloan, D.B.; Wu, Z. History of plastid DNA insertions reveals weak deletion and AT mutation biases in angiosperm mitochondrial genomes. Genome Biol. Evol. 2014, 6, 3210–3221. [Google Scholar] [CrossRef]
- Wynn, E.L.; Christensen, A.C. Repeats of unusual size in plant mitochondrial genomes: Identification, incidence and evolution. G3 Genes, Genomes, Genet. 2019, 9, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Gandini, C.L.; Garcia, L.E.; Abbona, C.C.; Sanchez-Puerta, M.V. The complete organelle genomes of Physochlaina orientalis: Insights into short sequence repeats across seed plant mitochondrial genomes. Mol. Phylogenet. Evol. 2019, 137, 274–284. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, J.S. Structural mutations in the organellar genomes of Valeriana sambucifolia f. dageletiana (Nakai. ex Maekawa) hara show dynamic gene transfer. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef]
- Choi, K.S.; Park, S. Complete plastid and mitochondrial genomes of Aeginetia indica reveal intracellular gene transfer (IGT), horizontal gene transfer (HGT), and cytoplasmic male sterility (cms). Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef] [PubMed]
- Sinn, B.T.; Barrett, C.F. Ancient Mitochondrial Gene Transfer between Fungi and the Orchids. Mol. Biol. Evol. 2020, 37, 44–57. [Google Scholar] [CrossRef]
- Rice, D.W.; Alverson, A.J.; Richardson, A.O.; Young, G.J.; Sanchez-Puerta, M.V.; Munzinger, J.; Barry, K.; Boore, J.L.; Zhang, Y.; DePamphilis, C.W.; et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science (80-. ). 2013, 342, 1468–1473. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Puerta, M.V.; Edera, A.; Gandini, C.L.; Williams, A. V.; Howell, K.A.; Nevill, P.G.; Small, I. Genome-scale transfer of mitochondrial DNA from legume hosts to the holoparasite Lophophytum mirabile (Balanophoraceae). Mol. Phylogenet. Evol. 2019, 132, 243–250. [Google Scholar] [CrossRef]
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef]
- Chase, M.W.; Cameron, K.M.; Freudenstein, J. V.; Pridgeon, A.M.; Salazar, G.; van den Berg, C.; Schuiteman, A. An updated classification of Orchidaceae. Bot. J. Linn. Soc. 2015, 177, 151–174. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, H.C.; Lin, I.P.; Chow, T.Y.; Chen, H.H.; Chen, W.H.; Cheng, C.H.; Lin, C.Y.; Liu, S.M.; Chang, C.C.; et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): Comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 2006, 23, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.F.; Davis, J.I. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am. J. Bot. 2012, 99, 1513–1523. [Google Scholar] [CrossRef]
- Barrett, C.F.; Wicke, S.; Sass, C. Dense infraspecific sampling reveals rapid and independent trajectories of plastome degradation in a heterotrophic orchid complex. New Phytol. 2018. [Google Scholar] [CrossRef]
- Yang, J.-B.; Tang, M.; Li, H.-T.; Zhang, Z.-R.; Li, D.-Z. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013, 13, 84. [Google Scholar] [CrossRef]
- Niu, Z.; Xue, Q.; Zhu, S.; Sun, J.; Liu, W.; Ding, X. The Complete Plastome Sequences of Four Orchid Species: Insights into the Evolution of the Orchidaceae and the Utility of Plastomic Mutational Hotspots. Front. Plant Sci. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Li, Z.H.; Ma, X.; Wang, D.Y.; Li, Y.X.; Wang, C.W.; Jin, X.H. Evolution of plastid genomes of Holcoglossum (Orchidaceae) with recent radiation. BMC Evol. Biol. 2019, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.L.; Wicke, S.; Li, J.W.; Han, Y.; Lin, C.S.; Li, D.Z.; Zhou, T.T.; Huang, W.C.; Huang, L.Q.; Jin, X.H. Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol. Evol. 2016, 8, 2164–2175. [Google Scholar] [CrossRef]
- Lin, C.S.; Chen, J.J.W.; Huang, Y.T.; Chan, M.T.; Daniell, H.; Chang, W.J.; Hsu, C.T.; Liao, D.C.; Wu, F.H.; Lin, S.Y.; et al. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 2015, 5, 1–10. [Google Scholar] [CrossRef]
- Delannoy, E.; Fujii, S.; Colas Des Francs-Small, C.; Brundrett, M.; Small, I. Rampant Gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 2011, 28, 2077–2086. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, H.T.; Kim, J.-H. The largest plastid genome of monocots: a novel genome type containing AT residue repeats in the slipper orchid Cypripedium japonicum. Plant Mol. Biol. Report. 2015, 33, 1210–1220. [Google Scholar] [CrossRef]
- Kim, Y.K.; Jo, S.; Cheon, S.H.; Joo, M.J.; Hong, J.R.; Kwak, M.H.; Kim, K.J. Extensive losses of photosynthesis genes in the plastome of a mycoheterotrophic orchid, Cyrtosia septentrionalis (Vanilloideae: Orchidaceae). Genome Biol. Evol. 2019, 11, 565–571. [Google Scholar] [CrossRef]
- Freudenstein, J. V.; Chase, M.W.M.; Lanfear, R.; Synergy, B.; Phytol, N.; Page, V.I.; De, A.A.P.; Wu, G.A.; Terol, J.; Ibanez, V.; et al. Phylogenetic relationships in Epidendroideae (Orchidaceae), one of the great flowering plant radiations: Progressive specialization and diversification. PLoS One 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Barrett, C.F.; Kennedy, A.H. Plastid genome degradation in the endangered, mycoheterotrophic, North American orchid Hexalectris warnockii. Genome Biol. Evol. 2018, 10, 1657–1662. [Google Scholar] [CrossRef]
- Guo, Y.Y.; Yang, J.X.; Bai, M.Z.; Zhang, G.Q.; Liu, Z.J. The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC Plant Biol. 2021, 21, 1–14. [Google Scholar] [CrossRef]
- Merckx, V.S.F.T.; Freudenstein, J. V; Kissling, J.; Christenhusz, M.J.M.; Stotler, R.E.; Crandall-Stotler, B.; Wickett, N.; Rudall, P.J.; Maas-van de Kamer, H.; Maas, P.J.M. Taxonomy and Classification BT - Mycoheterotrophy: The Biology of Plants Living on Fungi. In; Merckx, V., Ed.; Springer New York: New York, NY, 2013; pp. 19–101 ISBN 978-1-4614-5209-6.
- Barrett, C.F.; Freudenstein, J. V.; Li, J.; Mayfield-Jones, D.R.; Perez, L.; Pires, J.C.; Santos, C. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol. 2014, 31, 3095–3112. [Google Scholar] [CrossRef]
- Schelkunov, M.I.; Shtratnikova, V.Y.; Nuraliev, M.S.; Selosse, M.A.; Penin, A.A.; Logacheva, M.D. Exploring the limits for reduction of plastid genomes: A case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol. Evol. 2015, 7, 1179–1191. [Google Scholar] [CrossRef]
- Yuan, Y.; Jin, X.; Liu, J.; Zhao, X.; Zhou, J.; Wang, X.; Wang, D.; Lai, C.; Xu, W.; Huang, J.; et al. The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Kim, Y.K.; Jo, S.; Cheon, S.H.; Joo, M.J.; Hong, J.R.; Kwak, M.; Kim, K.J. Plastome Evolution and Phylogeny of Orchidaceae, With 24 New Sequences. Front. Plant Sci. 2020, 11, 1–27. [Google Scholar] [CrossRef]
- Lam, V.K.Y.; Darby, H.; Merckx, V.S.F.T.; Lim, G.; Yukawa, T.; Neubig, K.M.; Abbott, J.R.; Beatty, G.E.; Provan, J.; Soto Gomez, M.; et al. Phylogenomic inference in extremis: A case study with mycoheterotroph plastomes. Am. J. Bot. 2018, 105, 480–494. [Google Scholar] [CrossRef]
- Yang, F.X.; Gao, J.; Wei, Y.L.; Ren, R.; Zhang, G.Q.; Lu, C.Q.; Jin, J.P.; Ai, Y.; Wang, Y.Q.; Chen, L.J.; et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef]
- Zhang, G.-Q.; Xu, Q.; Bian, C.; Tsai, W.-C.; Yeh, C.-M.; Liu, K.-W.; Yoshida, K.; Zhang, L.-S.; Chang, S.-B.; Chen, F. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Chen, G.Z.; Chen, L.J.; Zhai, J.W.; Huang, J.; Wu, X.Y.; Li, M.H.; Peng, D.H.; Rao, W.H.; Liu, Z.J.; et al. Phylogenetic incongruence in Cymbidium orchids. Plant Divers. 2021, 43, 452–461. [Google Scholar] [CrossRef]
- Li, Y.X.; Li, Z.H.; Schuitman, A.; Chase, M.W.; Li, J.W.; Huang, W.C.; Hidayat, A.; Wu, S.S.; Jin, X.H. Phylogenomics of Orchidaceae based on plastid and mitochondrial genomes. Mol. Phylogenet. Evol. 2019, 139, 106540. [Google Scholar] [CrossRef]
- Committee, F. of K.E; Park, C.-W. The genera of vascular plants of Korea; Academy Publ., 2007; ISBN 897616380X. [Google Scholar]
- Chen, X.Q.; Liu, Z.J.; Zhu, G.H.; Lang, K.Y.; Ji, Z.H.; Luo, Y.B.; Jin, X.H.; Cribb, P.J.; Wood, J.J.; Gale, S.W. Flora of China: Orchidaceae. Chen XQ, Wood JJ 2009, 175, 477. [Google Scholar]
- Kim, H.T.; Shin, C.H.; Sun, H.; Kim, J.H. Sequencing of the plastome in the leafless green mycoheterotroph Cymbidium macrorhizon helps us to understand an early stage of fully mycoheterotrophic plastome structure. Plant Syst. Evol. 2018, 304, 245–258. [Google Scholar] [CrossRef]
- Wicke, S.; Muller, K.F.; de Pamphilis, C.W.; Quandt, D.; Wickett, N.J.; Zhang, Y.; Renner, S.S.; Schneeweiss, G.M. Mechanisms of Functional and Physical Genome Reduction in Photosynthetic and Nonphotosynthetic Parasitic Plants of the Broomrape Family. Plant Cell 2013, 25, 3711–3725. [Google Scholar] [CrossRef]
- Sanderson, M.J.; Copetti, D.; Burquez, A.; Bustamante, E.; Charboneau, J.L.M.; Eguiarte, L.E.; Kumar, S.; Lee, H.O.; Lee, J.; McMahon, M.; et al. Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): loss of the ndh gene suite and inverted repeat. Am. J. Bot. 2015, 102, 1115–1127. [Google Scholar] [CrossRef]
- Guisinger, M.M.; Kuehl, J. V; Boore, J.L.; Jansen, R.K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol. Biol. Evol. 2011, 28, 583–600. [Google Scholar] [CrossRef]
- Blazier, J.C.; Jansen, R.K.; Mower, J.P.; Govindu, M.; Zhang, J.; Weng, M.L.; Ruhlman, T.A. Variable presence of the inverted repeat and plastome stability in Erodium. Ann. Bot. 2016, 117, 1209–1220. [Google Scholar] [CrossRef]
- Palmer, J.D.; Thompson, W.F. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 1982, 29, 537–550. [Google Scholar] [CrossRef]
- Guo, W.; Grewe, F.; Cobo-Clark, A.; Fan, W.; Duan, Z.; Adams, R.P.; Schwarzbach, A.E.; Mower, J.P. Predominant and substoichiometric isomers of the plastid genome coexist within Juniperus plants and have shifted multiple times during cupressophyte evolution. Genome Biol. Evol. 2014, 6, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Raubeson, L.A.; Jansen, R.K. A rare chloroplast-DNA structural mutation is shared by all conifers. Biochem. Syst. Ecol. 1992, 20, 17–24. [Google Scholar] [CrossRef]
- Moghaddam, M.; Ohta, A.; Shimizu, M.; Terauchi, R.; Kazempour-Osaloo, S. The complete chloroplast genome of Onobrychis gaubae (Fabaceae-Papilionoideae): comparative analysis with related IR-lacking clade species. BMC Plant Biol. 2022, 22, 75. [Google Scholar] [CrossRef]
- Wu, S.; Chen, J.; Li, Y.; Liu, A.; Li, A.; Yin, M.; Shrestha, N.; Liu, J.; Ren, G. Extensive genomic rearrangements mediated by repetitive sequences in plastomes of Medicago and its relatives. BMC Plant Biol. 2021, 21, 1–16. [Google Scholar] [CrossRef]
- Haberle, R.C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 2008, 66, 350–361. [Google Scholar] [CrossRef]
- Kim, H.T.; Kim, J.S.; Moore, M.J.; Neubig, K.M.; Williams, N.H.; Whitten, W.M.; Kim, J.H. Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across orchids and associated instability of the inverted repeat/small single-copy region boundaries. PLoS One 2015, 10. [Google Scholar] [CrossRef]
- Wicke, S.; Müller, K.F.; dePamphilis, C.W.; Quandt, D.; Bellot, S.; Schneeweiss, G.M. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc. Natl. Acad. Sci. 2016, 113, 9045–9050. [Google Scholar] [CrossRef]
- Suetsugu, K.; Ohta, T.; Tayasu, I. Partial mycoheterotrophy in the leafless orchid Cymbidium macrorhizon. Am. J. Bot. 2018, 105, 1595–1600. [Google Scholar] [CrossRef] [PubMed]
- Blumenfeld, H. Beitrage zur Physiologie des Wurzepilzes von Limodorum abortivum (L.) Sw; Universitat Basel., 1935.
- Girlanda, M.; Selosse, M.A.; Cafasso, D.; Brilli, F.; Delfine, S.; Fabbian, R.; Ghignone, S.; Pinelli, P.; Segreto, R.; Loreto, F.; et al. Inefficient photosynthesis in the Mediterranean orchid Limodorum abortivum is mirrored by specific association to ectomycorrhizal Russulaceae. Mol. Ecol. 2006, 15, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B. One ring to rule them all? Genome sequencing provides new insights into the “master circle” model of plant mitochondrial DNA structure. New Phytol. 2013, 200, 978–985. [Google Scholar] [CrossRef]
- Niu, Y.; Gao, C.; Liu, J. Complete mitochondrial genomes of three Mangifera species, their genomic structure and gene transfer from chloroplast genomes. BMC Genomics 2022, 23, 1–8. [Google Scholar] [CrossRef]
- Kim, Y.; Cheon, S.; Hong, J.; Kim, K. Evolutionary Patterns of the Chloroplast Genome in Vanilloid Orchids ( Vanilloideae , Orchidaceae ). 2023. [CrossRef]
- Fonseca, L.H.M.; Lohmann, L.G. Exploring the potential of nuclear and mitochondrial sequencing data generated through genome-skimming for plant phylogenetics: A case study from a clade of neotropical lianas. J. Syst. Evol. 2020, 58, 18–32. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: a quality control tool for high throughput sequence data 2010.
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Chin, C.-S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. HybridSPAdes: An algorithm for hybrid assembly of short and long reads. Bioinformatics 2016, 32, 1009–1015. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Bock, R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 gateway computing environments workshop (GCE); Ieee, 2010; pp. 1–8.
- Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Stöver, B.C.; Müller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 2010, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C. Phobos Version 3.3. 12. A tandem repeat search Progr. 2010, 20. [Google Scholar]
- Kurtz, S. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Wickham, H.; Chang, W.; Wickham, M.H. Package ‘ggplot2. ’ Creat. Elegant Data Vis. Using Gramm. Graph. Version 2016, 2, 1–189. [Google Scholar]
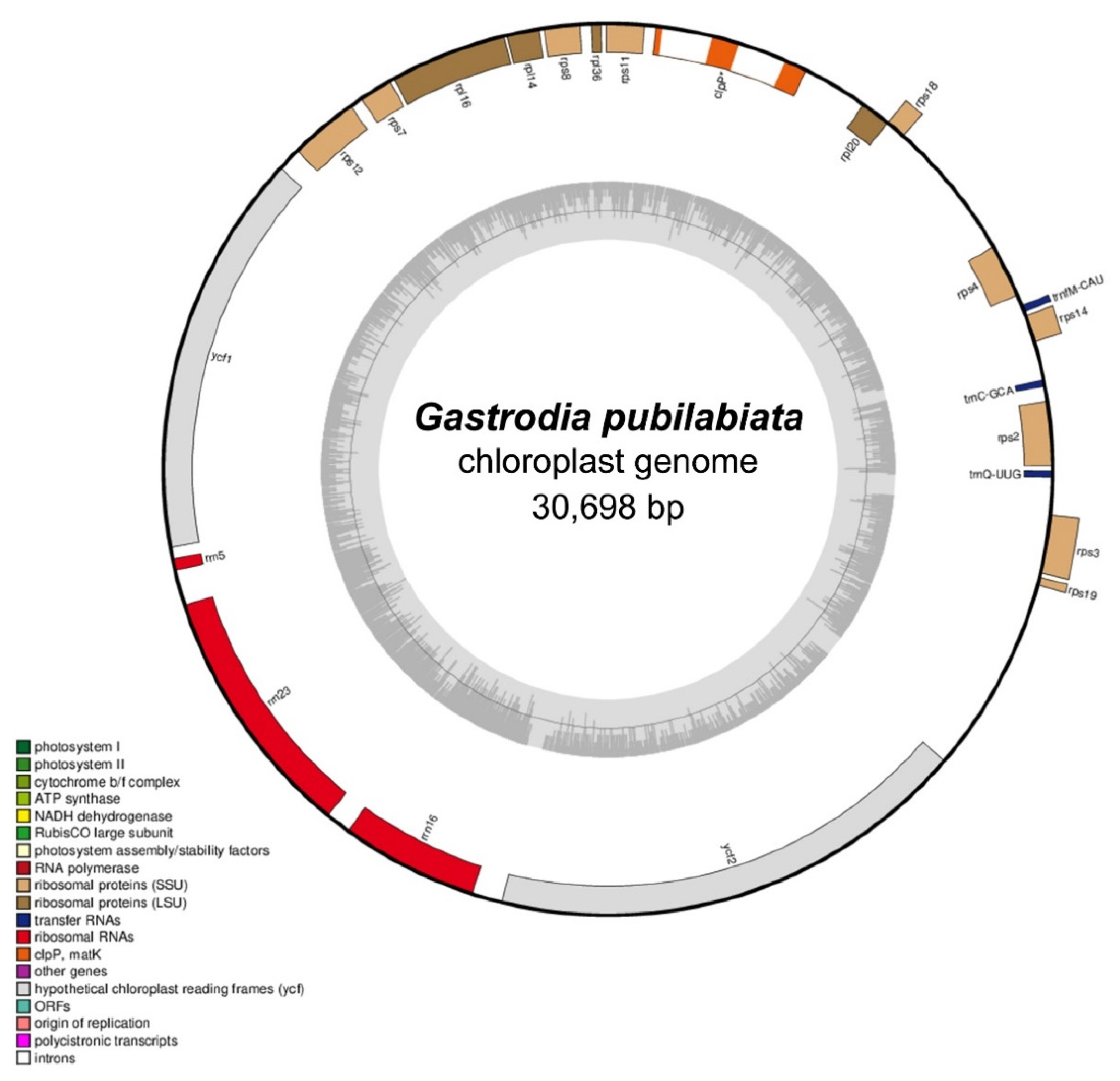
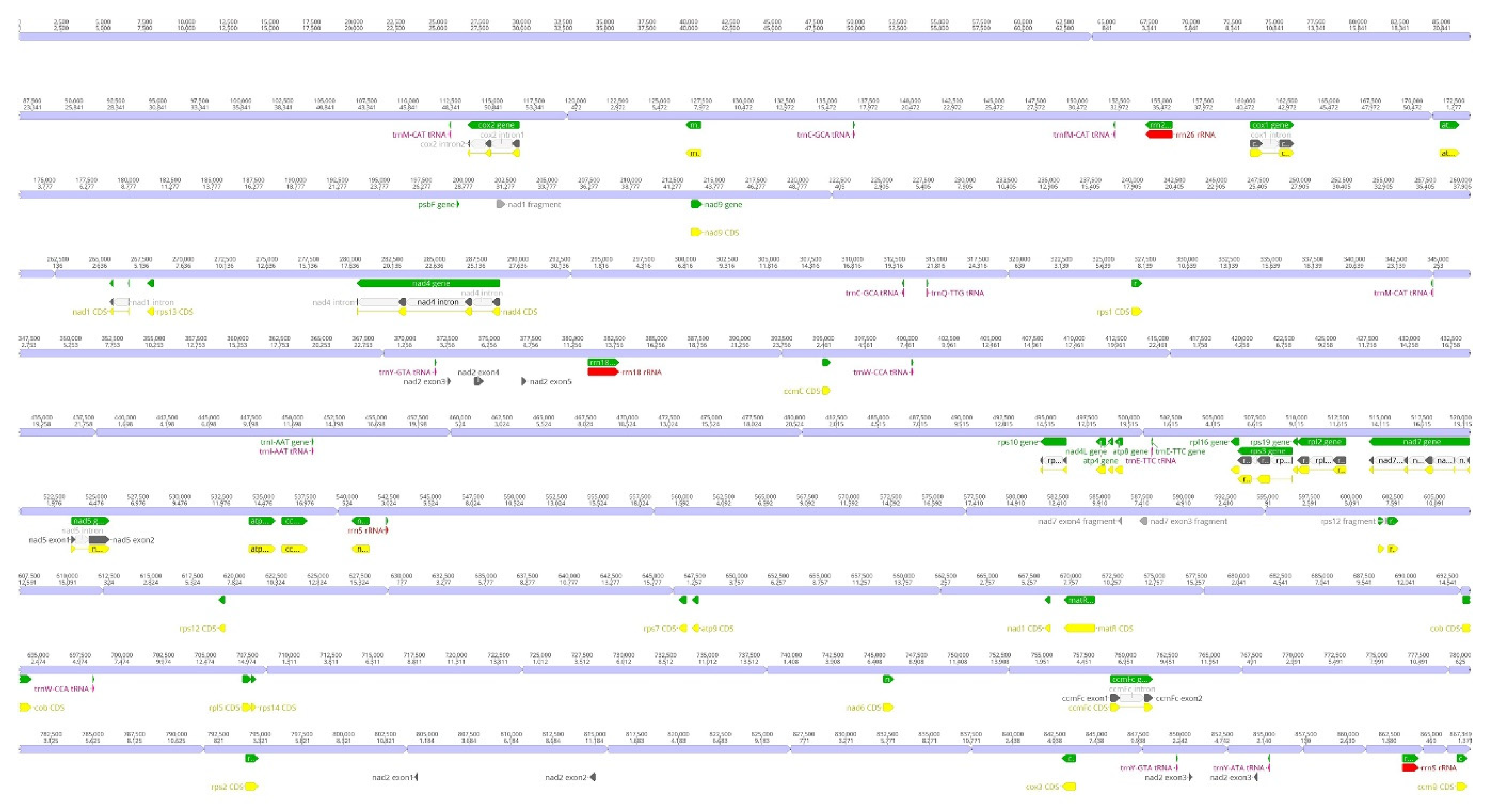
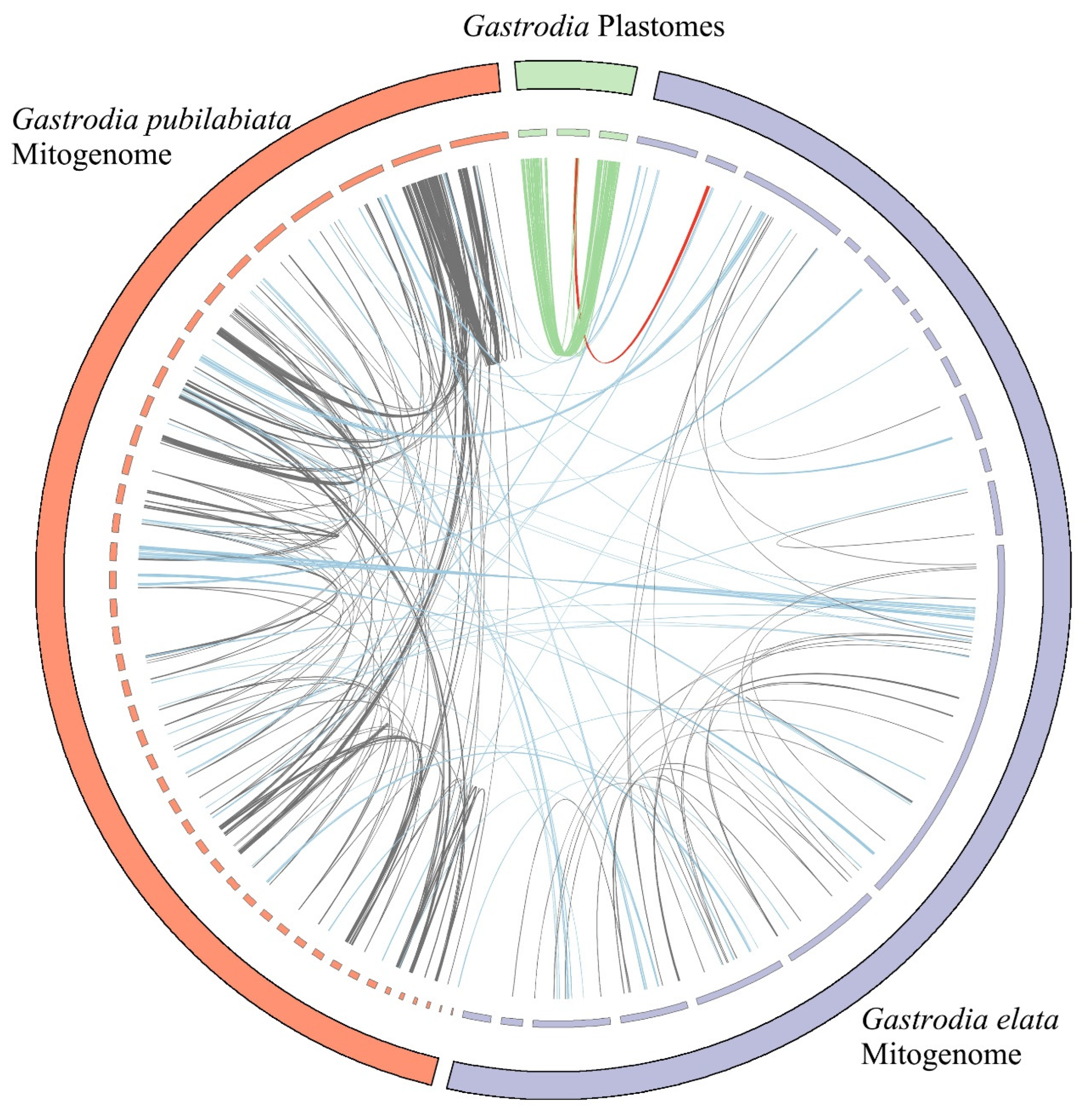
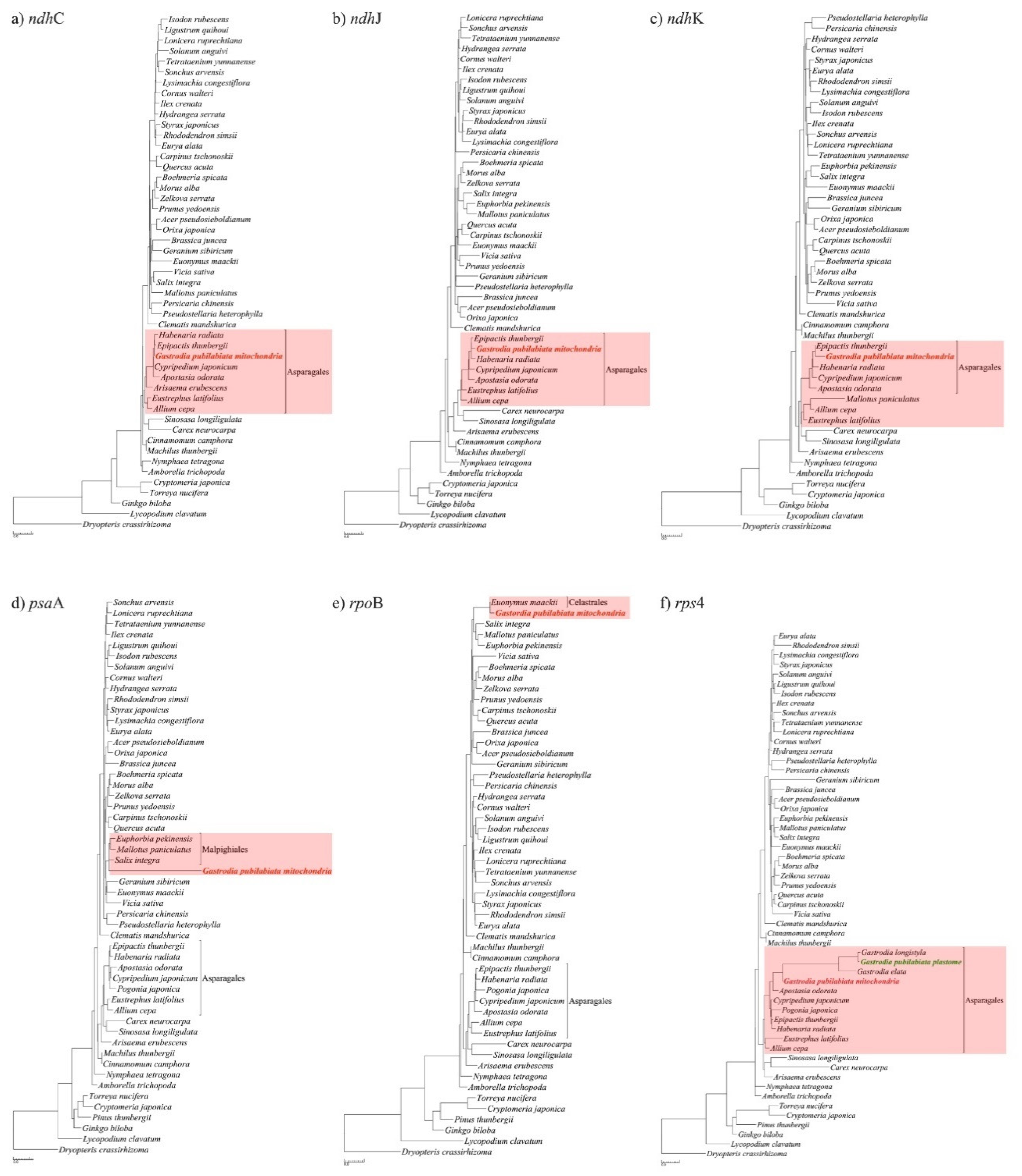

| Plastome | Mitogenome | |
|---|---|---|
| Contigs | 1 | 44 |
| Total Length (bp) | 30,698 | 867,349 |
| GC (%) | 24.9 | 42.1 |
| Depth Coverage (x) | 236.0 | 150.0 |
| Protein Coding Genes | 19 | 38 |
| tRNAs | 3 | 9 |
| rRNAs | 3 | 3 |
| Scientific Name | Accesions | Contigs | Total Length (bp) |
Protein Coding Genes |
tRNAs | rRNAs |
|---|---|---|---|---|---|---|
| Gastrodia pubilabiata | OR004100-OR004143 | 44 | 867,349 | 38 | 9 | 3 |
| Gastrodia elata | MF070084~ MF070102 |
19 | 1,339,825 | 38 | 14 | 3 |
| Allium cepa | NC030100 | 1 | 316,363 | 24 | 5 | 3 |
| Asparagus officinalis | NC053642 | 1 | 492,062 | 36 | 14 | 3 |
| Butomus umbellatus | NC021399 | 1 | 450,826 | 28 | 12 | 4 |
| Oryza sativa | NC011033 | 1 | 490,520 | 31 | 15 | 3 |
| Phoenix dactylifera | NC016740 | 1 | 715,001 | 37 | 16 | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





