Submitted:
27 June 2023
Posted:
28 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Cell culture and treatments
2.2. Western blots
2.3. mRNA and miR quantification by RT-qPCR
2.4. Statistical analysis
3. Results
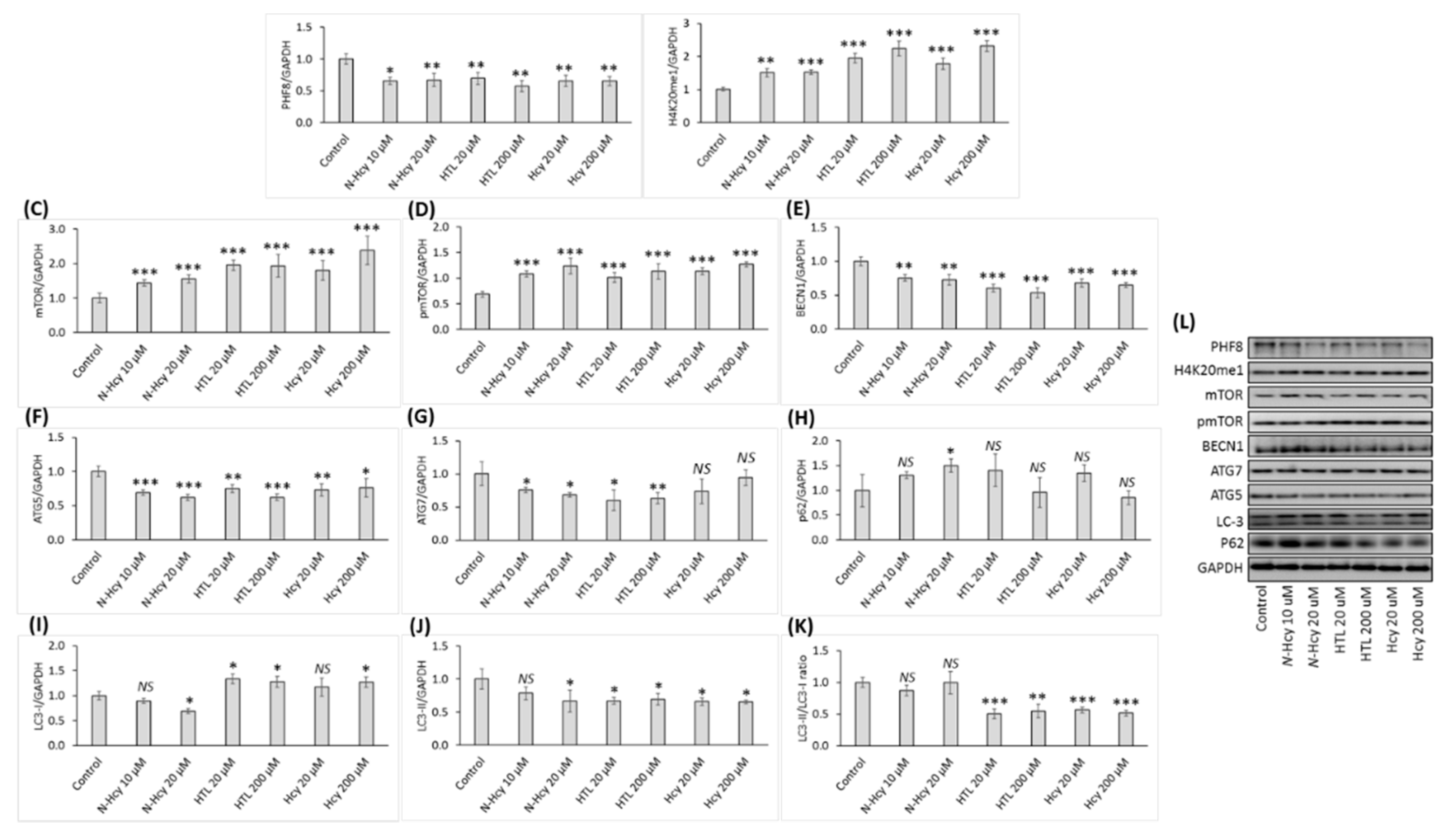
3.1. Hcy-thiolactone, N-Hcy-protein, and Hcy downregulate the histone demethylase Phf8, upregulate the H4K20me1 epigenetic mark and mTOR, and impair autophagy in HUVEC
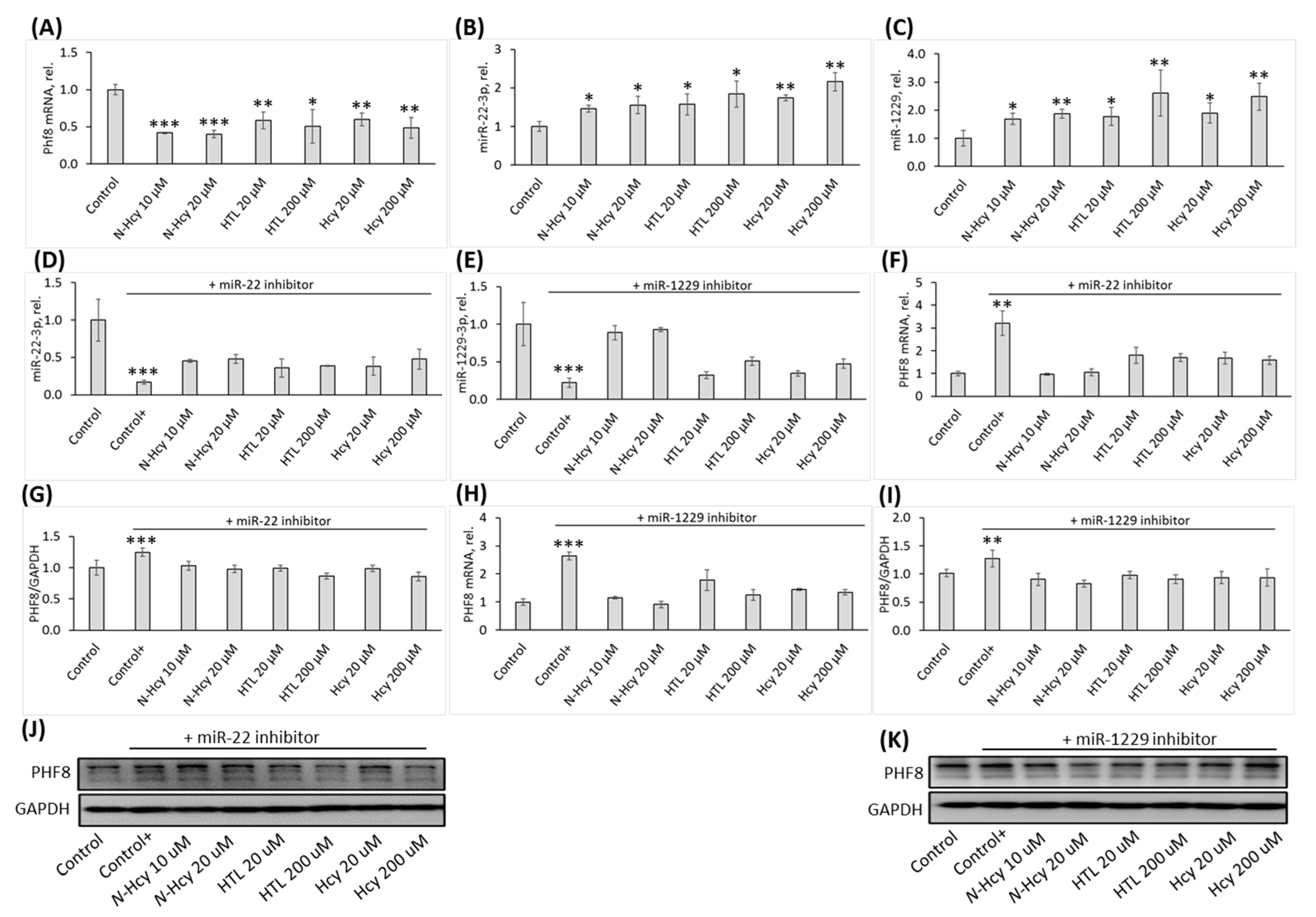
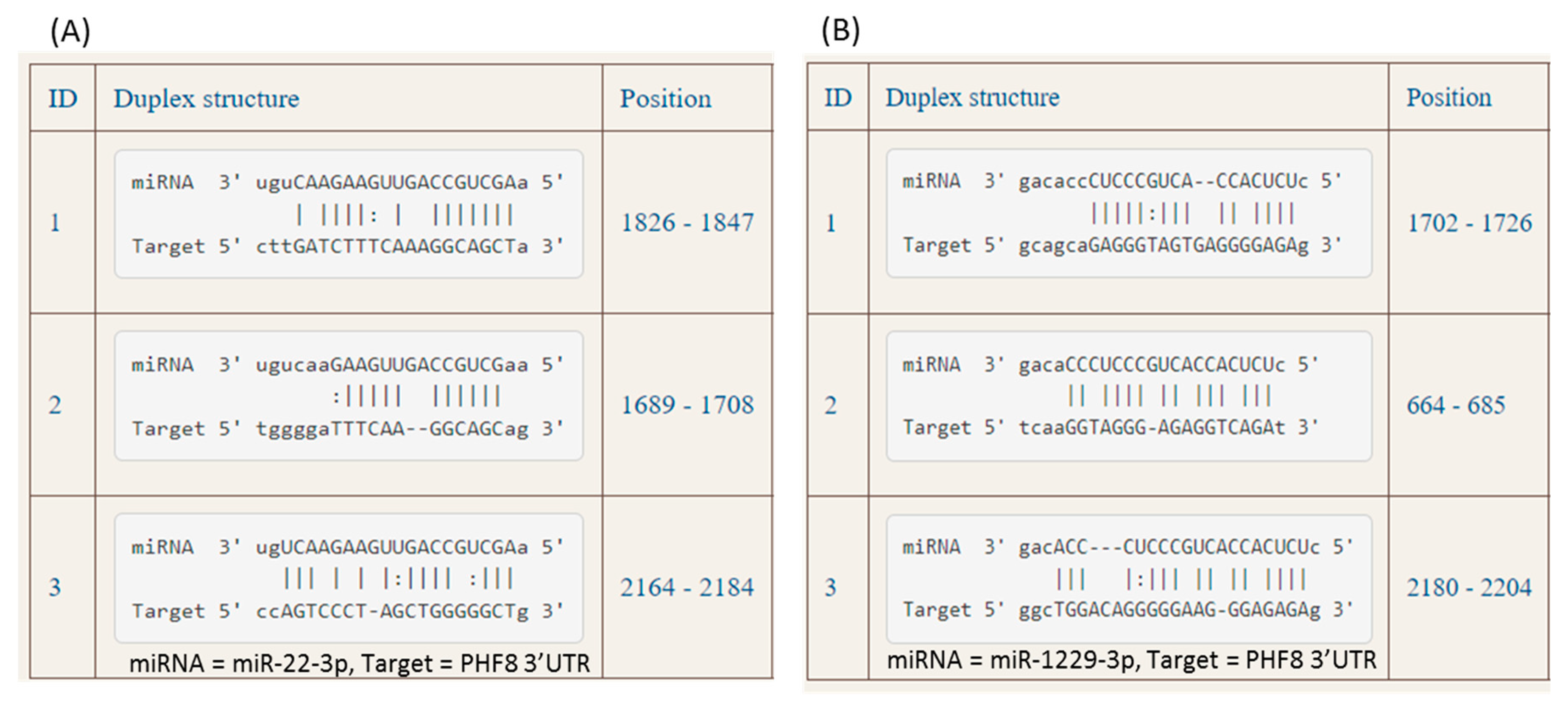
3.2. Hcy-thiolactone, N-Hcy-protein, and Hcy downregulate PHF8 by upregulating the expression of miR-22-3p and miR-1229-3p targeting PHF8
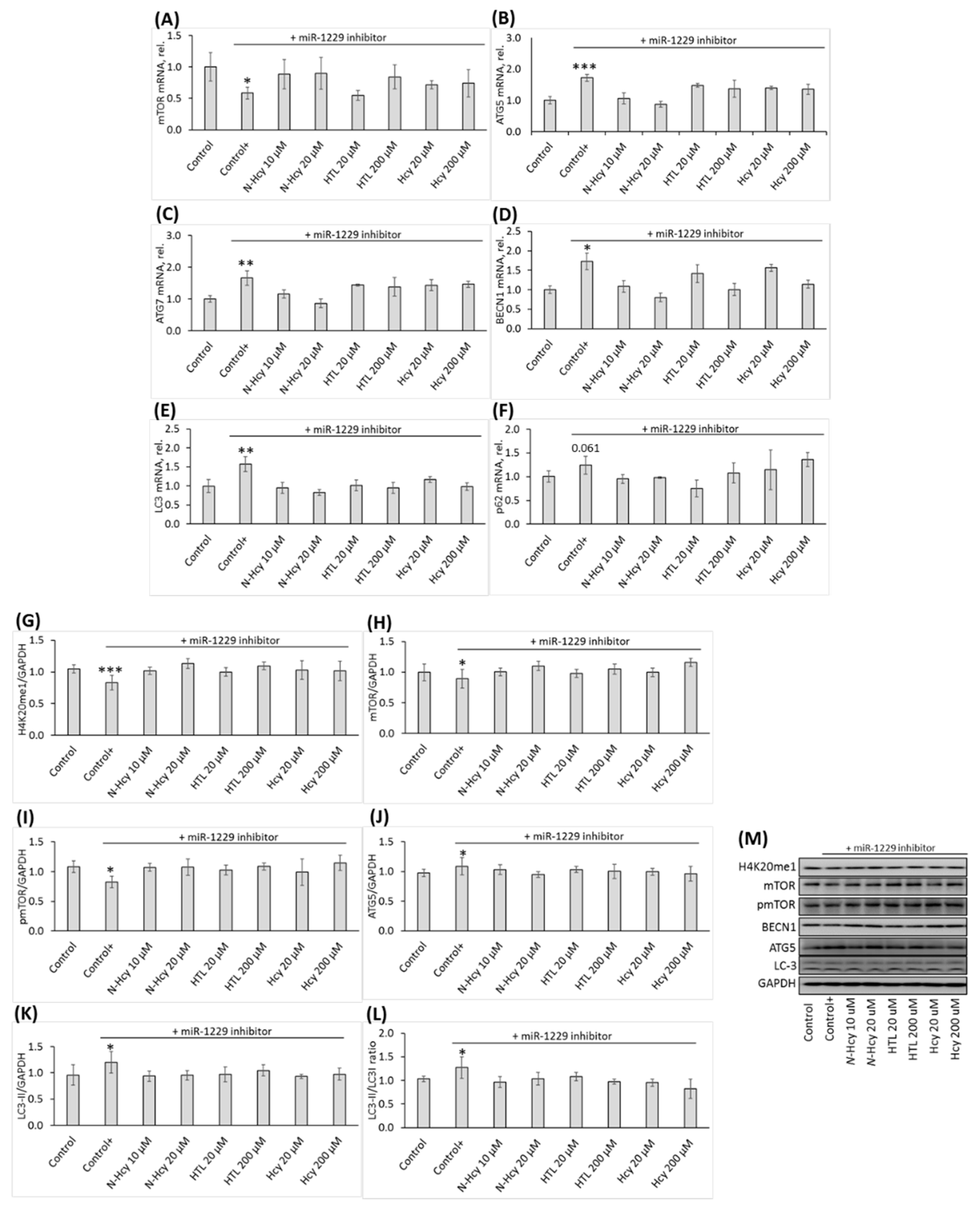
3.3. Treatments with miR inhibitors ameliorate effects of Hcy-thiolactone, N-Hcy-protein, and Hcy on miR-22-3p, miR-1229, and PHF8 expression
Effects of miR inhibitors are propagated to downstream targets of PHF8
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Libby, P. Inflammation in atherosclerosis. Nature 2002, 420, (6917), 868-74.
- Ross, R. Atherosclerosis--an inflammatory disease. N Engl J Med 1999, 340, (2), 115-26.
- Lentz, S. R. Mechanisms of homocysteine-induced atherothrombosis. J Thromb Haemost 2005, 3, (8), 1646-54.
- Dayal, S.; Lentz, S. R. Murine models of hyperhomocysteinemia and their vascular phenotypes. Arterioscler Thromb Vasc Biol 2008, 28, (9), 1596-605.
- Esse, R.; Barroso, M.; Tavares de Almeida, I.; Castro, R. The Contribution of Homocysteine Metabolism Disruption to Endothelial Dysfunction: State-of-the-Art. Int J Mol Sci 2019, 20, (4).
- Poddar, R.; Sivasubramanian, N.; DiBello, P. M.; Robinson, K.; Jacobsen, D. W. Homocysteine induces expression and secretion of monocyte chemoattractant protein-1 and interleukin-8 in human aortic endothelial cells: implications for vascular disease. Circulation 2001, 103, (22), 2717-23.
- Kerkeni, M.; Tnani, M.; Chuniaud, L.; Miled, A.; Maaroufi, K.; Trivin, F. Comparative study on in vitro effects of homocysteine thiolactone and homocysteine on HUVEC cells: evidence for a stronger proapoptotic and proinflammative homocysteine thiolactone. Mol Cell Biochem 2006, 291, (1-2), 119-26.
- Jakubowski, H. Homocysteine Modification in Protein Structure/Function and Human Disease. Physiol Rev 2019, 99, (1), 555-604.
- Sikora, M.; Lewandowska, I.; Marczak, L.; Bretes, E.; Jakubowski, H. Cystathionine beta-synthase deficiency: different changes in proteomes of thrombosis-resistant Cbs(-/-) mice and thrombosis-prone CBS(-/-) humans. Sci Rep 2020, 10, (1), 10726.
- Sikora, M.; Jakubowski, H. Changes in redox plasma proteome of Pon1-/- mice are exacerbated by a hyperhomocysteinemic diet. Free Radic Biol Med 2021, 169, 169–180. [Google Scholar]
- Jakubowski, H.; Zhang, L.; Bardeguez, A.; Aviv, A. Homocysteine thiolactone and protein homocysteinylation in human endothelial cells: implications for atherosclerosis. Circ Res 2000, 87, (1), 45-51.
- Gurda, D.; Handschuh, L.; Kotkowiak, W.; Jakubowski, H. Homocysteine thiolactone and N-homocysteinylated protein induce pro-atherogenic changes in gene expression in human vascular endothelial cells. Amino Acids 2015, 47, 1319–39. [Google Scholar]
- Olejniczak, M.; Urbanek, M. O.; Jaworska, E.; Witucki, L.; Szczesniak, M. W.; Makalowska, I.; Krzyzosiak, W. J. Sequence-non-specific effects generated by various types of RNA interference triggers. Biochim Biophys Acta 2016, 1859, 306–14. [Google Scholar] [CrossRef]
- Starega-Roslan, J.; Krzyzosiak, W. J. Analysis of microRNA length variety generated by recombinant human Dicer. Methods Mol Biol 2013, 936, 21–34. [Google Scholar] [PubMed]
- Starega-Roslan, J.; Galka-Marciniak, P.; Krzyzosiak, W. J. Nucleotide sequence of miRNA precursor contributes to cleavage site selection by Dicer. Nucleic Acids Res 2015, 43, 10939–51. [Google Scholar] [CrossRef] [PubMed]
- Starega-Roslan, J.; Koscianska, E.; Kozlowski, P.; Krzyzosiak, W. J. The role of the precursor structure in the biogenesis of microRNA. Cell Mol Life Sci 2011, (17), 2859–71. [Google Scholar]
- Stroynowska-Czerwinska, A.; Fiszer, A.; Krzyzosiak, W. J. The panorama of miRNA-mediated mechanisms in mammalian cells. Cell Mol Life Sci 2014, 71, 2253–70. [Google Scholar] [PubMed]
- Minjares, M.; Wu, W.; Wang, J. M. Oxidative Stress and MicroRNAs in Endothelial Cells under Metabolic Disorders. Cells 2023, 12, (9).
- Zhou, S. S.; Jin, J. P.; Wang, J. Q.; Zhang, Z. G.; Freedman, J. H.; Zheng, Y.; Cai, L. miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges. Acta Pharmacol Sin 2018, 39, 1073–1084. [Google Scholar] [PubMed]
- Mens, M. M. J.; Heshmatollah, A.; Fani, L.; Ikram, M. A.; Ikram, M. K.; Ghanbari, M. Circulatory MicroRNAs as Potential Biomarkers for Stroke Risk: The Rotterdam Study. Stroke 2021, 52, 945–953. [Google Scholar]
- Sobering, A. K.; Bryant, L. M.; Li, D.; McGaughran, J.; Maystadt, I.; Moortgat, S.; Graham, J. M., Jr.; van Haeringen, A.; Ruivenkamp, C.; Cuperus, R.; Vogt, J.; Morton, J.; Brasch-Andersen, C.; Steenhof, M.; Hansen, L. K.; Adler, E.; Lyonnet, S.; Pingault, V.; Sandrine, M.; Ziegler, A.; Donald, T.; Nelson, B.; Holt, B.; Petryna, O.; Firth, H.; McWalter, K.; Zyskind, J.; Telegrafi, A.; Juusola, J.; Person, R.; Bamshad, M. J.; Earl, D.; University of Washington Center for Mendelian, G.; Tsai, A. C.; Yearwood, K. R.; Marco, E.; Nowak, C.; Douglas, J.; Hakonarson, H.; Bhoj, E. J. Variants in PHF8 cause a spectrum of X-linked neurodevelopmental disorders and facial dysmorphology. HGG Adv 2022, 3, 100102. [Google Scholar]
- Laumonnier, F.; Holbert, S.; Ronce, N.; Faravelli, F.; Lenzner, S.; Schwartz, C. E.; Lespinasse, J.; Van Esch, H.; Lacombe, D.; Goizet, C.; Phan-Dinh Tuy, F.; van Bokhoven, H.; Fryns, J. P.; Chelly, J.; Ropers, H. H.; Moraine, C.; Hamel, B. C.; Briault, S. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J Med Genet 2005, (10), 780–6. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, S.; Zhou, Y.; Han, Y.; Li, S.; Xu, Q.; Xu, L.; Zhu, Z.; Deng, Y.; Yu, L.; Song, L.; Chen, A. P.; Song, J.; Takahashi, E.; He, G.; He, L.; Li, W.; Chen, C. D. Phf8 histone demethylase deficiency causes cognitive impairments through the mTOR pathway. Nat Commun 2018, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Cai, M. Z.; Wen, S. Y.; Wang, X. J.; Liu, Y.; Liang, H. MYC Regulates PHF8, Which Promotes the Progression of Gastric Cancer by Suppressing miR-22-3p. Technol Cancer Res Treat 2020, 19, 1533033820967472. [Google Scholar] [CrossRef]
- Ghanbari, M.; Ikram, M. A.; de Looper, H. W. J.; Hofman, A.; Erkeland, S. J.; Franco, O. H.; Dehghan, A. Genome-wide identification of microRNA-related variants associated with risk of Alzheimer's disease. Sci Rep 2016, 6, 28387. [Google Scholar] [PubMed]
- Witucki, L.; Jakubowski, H. Depletion of Paraoxonase 1 (Pon1) Dysregulates mTOR, Autophagy, and Accelerates Amyloid Beta Accumulation in Mice. Cells 2023, 12, (5).
- Kaldirim, M.; Lang, A.; Pfeiler, S.; Fiegenbaum, P.; Kelm, M.; Bonner, F.; Gerdes, N. Modulation of mTOR Signaling in Cardiovascular Disease to Target Acute and Chronic Inflammation. Front Cardiovasc Med 2022, 9, 907348. [Google Scholar]
- Samidurai, A.; Kukreja, R. C.; Das, A. Emerging Role of mTOR Signaling-Related miRNAs in Cardiovascular Diseases. Oxid Med Cell Longev 2018, 2018, 6141902. [Google Scholar]
- Kim, K. A.; Shin, D.; Kim, J. H.; Shin, Y. J.; Rajanikant, G. K.; Majid, A.; Baek, S. H.; Bae, O. N. Role of Autophagy in Endothelial Damage and Blood-Brain Barrier Disruption in Ischemic Stroke. Stroke 2018, 49, 1571–1579. [Google Scholar] [CrossRef]
- Khayati, K.; Antikainen, H.; Bonder, E. M.; Weber, G. F.; Kruger, W. D.; Jakubowski, H.; Dobrowolski, R. The amino acid metabolite homocysteine activates mTORC1 to inhibit autophagy and form abnormal proteins in human neurons and mice. FASEB J 2017, (2), 598–609. [Google Scholar] [CrossRef]
- Witucki, L., Jakubowski, H. Homocysteine Metabolites Inhibit Autophagy and Elevate Amyloid Beta by Impairing Phf8/H4K20me1-dependent Epigenetic Regulation of mTOR in Cystathionine β-Synthase-Deficient Mice. bioRxiv 2023. [CrossRef]
- Witucki, L. Witucki, L., Borowczyk, K., Suszyńska-Zajczyk, J., Warzych, E., Pawlak, P., Jakubowski, H., Depletion of bleomycin hydrolase (Blmh) downregulates histone demethylase Phf8, impairs mTOR signaling/autophagy, accelerates amyloid beta accumulation, and induces neurological deficits in mice bioRxiv 2023. [CrossRef]
- Witucki, L.; Kurpik, M.; Jakubowski, H.; Szulc, M.; Lukasz Mikolajczak, P.; Jodynis-Liebert, J.; Kujawska, M. Neuroprotective Effects of Cranberry Juice Treatment in a Rat Model of Parkinson's Disease. Nutrients, 2022; 14. [Google Scholar]
- Livak, K. J.; Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–8. [Google Scholar] [CrossRef]
- Smith, A. D.; Refsum, H. Homocysteine - from disease biomarker to disease prevention. J Intern Med 2021, 290, 826–854. [Google Scholar] [CrossRef]
- Mameli, E.; Martello, A.; Caporali, A. Autophagy at the interface of endothelial cell homeostasis and vascular disease. FEBS J 2022, 289, 2976–2991. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H. Metabolism of homocysteine thiolactone in human cell cultures. Possible mechanism for pathological consequences of elevated homocysteine levels. J Biol Chem 1997, 272, 1935–42. [Google Scholar] [CrossRef] [PubMed]
- Chwatko, G.; Boers, G. H.; Strauss, K. A.; Shih, D. M.; Jakubowski, H. Mutations in methylenetetrahydrofolate reductase or cystathionine beta-synthase gene, or a high-methionine diet, increase homocysteine thiolactone levels in humans and mice. Faseb J 2007, 21, 1707–13. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H.; Perla-Kajan, J.; Finnell, R. H.; Cabrera, R. M.; Wang, H.; Gupta, S.; Kruger, W. D.; Kraus, J. P.; Shih, D. M. Genetic or nutritional disorders in homocysteine or folate metabolism increase protein N-homocysteinylation in mice. Faseb J 2009, 23, 1721–7. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, H.; Boers, G. H.; Strauss, K. A. Mutations in cystathionine {beta}-synthase or methylenetetrahydrofolate reductase gene increase N-homocysteinylated protein levels in humans. FASEB J 2008, 22, 4071–6. [Google Scholar] [CrossRef]
- Perla-Kajan, J.; Stanger, O.; Luczak, M.; Ziolkowska, A.; Malendowicz, L. K.; Twardowski, T.; Lhotak, S.; Austin, R. C.; Jakubowski, H. Immunohistochemical detection of N-homocysteinylated proteins in humans and mice. Biomed Pharmacother 2008, 62, 473–9. [Google Scholar] [CrossRef]
- Borowczyk, K.; Piechocka, J.; Glowacki, R.; Dhar, I.; Midtun, O.; Tell, G. S.; Ueland, P. M.; Nygard, O.; Jakubowski, H. Urinary excretion of homocysteine thiolactone and the risk of acute myocardial infarction in coronary artery disease patients: the WENBIT trial. J Intern Med 2019, 285, 232–244. [Google Scholar] [CrossRef]
- Qi, H. H.; Sarkissian, M.; Hu, G. Q.; Wang, Z.; Bhattacharjee, A.; Gordon, D. B.; Gonzales, M.; Lan, F.; Ongusaha, P. P.; Huarte, M.; Yaghi, N. K.; Lim, H.; Garcia, B. A.; Brizuela, L.; Zhao, K.; Roberts, T. M.; Shi, Y. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 2010, 466, 503–7. [Google Scholar] [CrossRef]
- Feinberg, M. W.; Moore, K. J. MicroRNA Regulation of Atherosclerosis. Circ Res 2016, 118, 703–20. [Google Scholar]
- Lu, W.; Liu, X.; Zhao, L.; Yan, S.; Song, Q.; Zou, C.; Li, X. MiR-22-3p in exosomes increases the risk of heart failure after down-regulation of FURIN. Chem Biol Drug Des 2023, 101, 550–567. [Google Scholar] [CrossRef]

| Gene | Primer sequence |
| ATG5 | Forward: 5′-AGTAGTTGCCTGGAGGAGCG-3′ |
| Reverse: 5′-GTCTCGCCAACTCCACCTTG-3′ | |
| ATG7 | Forward: 5′-GATGAAGCTCCCAAGGACAT-3′ |
| Reverse: 5′-GTGGGAGCACTCATGTCAAA-3′ | |
| BECN1 | Forward: 5′-GGGCTCCCGAGGGATGG-3′ |
| Reverse: 5′-TTCCTCCTGGGTCTCTCCTG-3′ | |
| GAPDH | Forward: 5′-AGCCACATCGCTCAGACAC-3′ |
| Reverse: 5′-GCCAATACGACCAAATCC-3′ | |
| LC3 | Forward: 5′-CATGAGCGAGTTGGTCAAGA-3′ |
| Reverse: 5′-CCATGCTGTGCTGGTTCA-3′ | |
| mTOR | Forward: 5′-GCAGCTGCATGGGGTTTA-3′ |
| Reverse: 5′-CCCGAGGGATCATACAGGT-3′ | |
| PHF8 | Forward: 5′-TGGGAGCATGCTTCAAGG-3′ |
| Reverse: 5′-GATTTCAAAGCAGGGTCATCA-3′ | |
| P62 | Forward: 5′-GGTCGCGCTCACCTTTCT-3′ |
| Reverse: 5′-TCCTTTCTCAAGCCCCATGTT-3′ | |
| 18s rRNA | Reverse: 5′-AGGAATTCCCAGTAAGTGCG-3′ |
| Reverse: 5′-GCCTCACTAAACCATCCAA-3′ | |
| U6 snRNA | Reverse: 5′-CTCGCTTCGGCAGCACA-3′ |
| Reverse: 5′-AACGCTTCACGAATTTGCGT-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





