Submitted:
23 June 2023
Posted:
26 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Thiosemicarbazones as Inhibitors of Topoisomerases
2.1. TSC Ligand or Bis(TSC) Ligand Inhibition of Topoisomerases
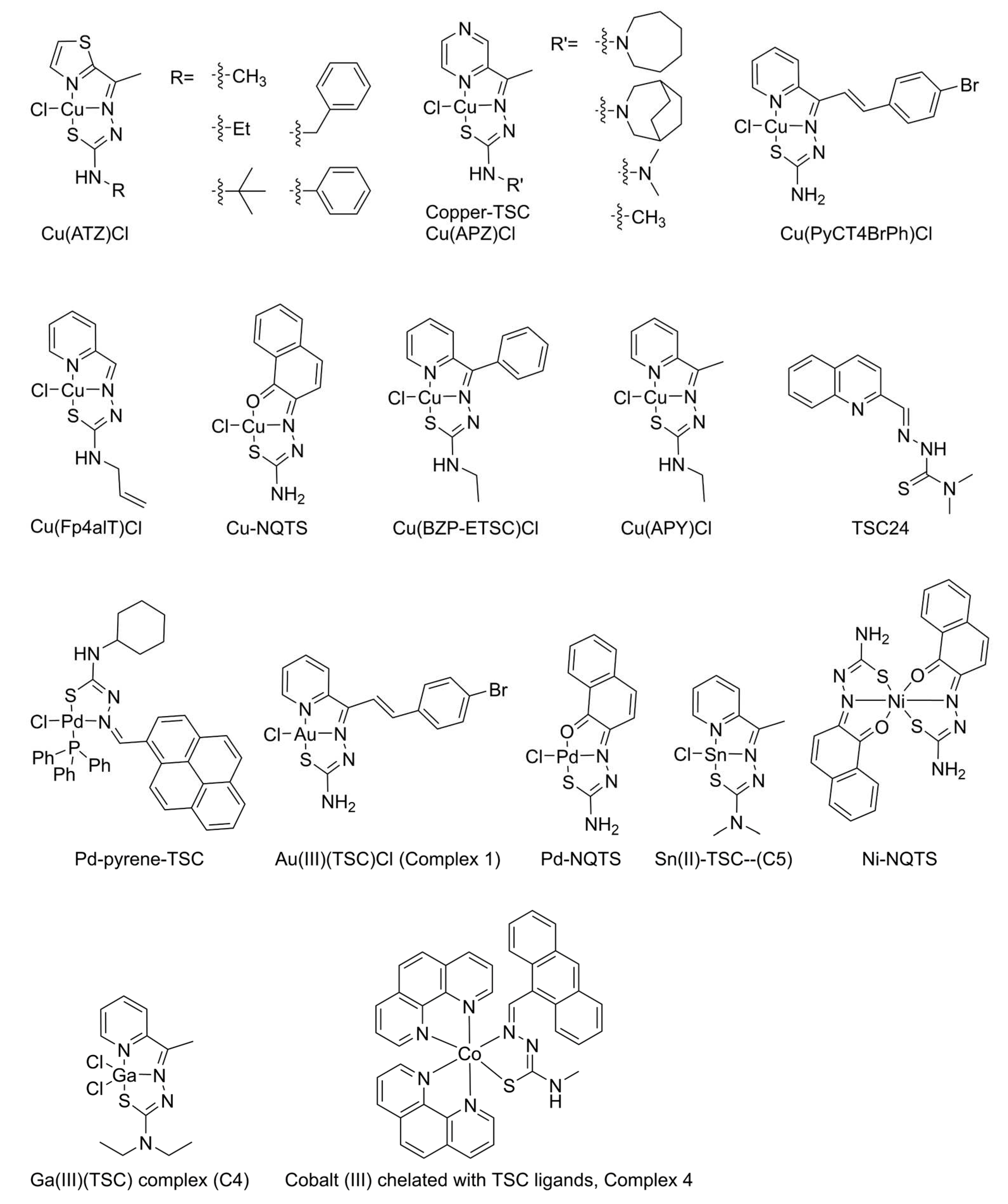
| Name | Inhibition of Top | Reference |
|---|---|---|
| TSC24 | Inhibit Top2α DNA relaxation at 25 µM Inhibit Top2 at decatenation at 100 µM |
[21] |
| Dp44mT | Inhibit human Top2α in relaxation assay with 5’ labeled 161-bp fragment from pBluescript SK phagemid DNA Do not inhibit human Top2β or human TopoI |
[22] |
| Dp44mT | Do not inhibit Top2α in decatenation Do not increase cleavage complex by human Top2α |
[20] |
| Triapine | ||
| Compound 2b | Inhibit human Top2α weakly at 100 µM | [23] |
| Compound 3a | Inhibit human Top2α-mediated DNA relaxation at 100 µM | [24] |
| Compound 3h | ||
| Triapine | Do not inhibit Top2α-mediated DNA relaxation at 50 µM | [19] |
| Dp44mT | ||
| Compound 24 | Do not inhibit isolated Top2 from L1210 | [12] |
| Compound 36 | ||
| NQTS | Do not inhibit Top2α | [30] |
| HFp4mT, HFp4pyrrT |
Inhibit Top2α at 100 µM | [31] |
| HFp4eT, HFp4ipT, HFp4alT, HAp4mT, HAp4-eT, HFp4bzT, HFpT, HAp4alT, and HApz4mT. |
Do not inhibit Top2α at 100 µM | [31] |
| BZP-TSC ligands series | Inhibits Top2α slightly at 50 µM. | [14] |
| ATZ ligand series | Do not inhibit Top2α at 10µM, do not increase cleavage complex | [27] |
| BZP ligands series | ||
| APY | Do not inhibit Top2α at 100 µM | [26] |
| APY | Do not inhibit Top2β at 200 µM | [28] |
| APZ | Inhibit Top2 α at 100µM | [26] |
| BZP | Inhibit Top2β at 200µM weakly | [28] |
| HPyCT4BrPh | Do not inhibit human Top1B at 50 µM | [29] |
2.2. Metal-TSC or Metal-Bis(TSC) Inhibition of Topoisomerases
2.2.1. Inhibition of Type I Top
| Name | Inhibition of TopI | Reference |
|---|---|---|
| Cu(PyCT4BrPh)Cl | Inhibit TopI The inhibition is severe when pre-incubation of the compound to TopI Inhibited the cleavage step and partially inhibited religation. |
[29] |
| Pd-pyrene-TSC | Inhibited human Top1B at 12.5 µM | [38] |
| Ga(III)-TSC complex (C4) | Inhibited TopI | [36] |
| Au(III)(TSC)Cl (complex 1) | Inhibited human Top1B activity starting at 1.5 μM Pre-incubation of Top1B with Complex 1 increased the inhibition |
[25] |
| Ni-bis(TSC) | No inhibition of E. coli TopI | [39] |
| Nine copper complexes | Inhibited TopI | [40] |
| Cobalt (III)-TSC (Complex 4) | Inhibited TopI-induced DNA relaxation | [35] |
2.2.2. Inhibition of Type II Top
- Ni chelated with bis(TSC)
- 2.
- Cu chelated TSCs
| Name | Inhibition of Top2 | Reference |
|---|---|---|
| Nine compounds and their copper complexes | Inhibited human Top2α | [40] |
| Cobalt (III) chelated with TSC ligands. Complex 4 | Inhibited human Topo2α-induced DNA relaxation | [35] |
| Ni-bis(TSC) complex 1 | Did not inhibit isolated Top2 from L1210 cells at 100 µM | [32] |
| Cu-TSCs (Compound 1 and 2) | Inhibited isolated Top2 from L1210 cells | [12] |
| Copper TSC | Inhibited Top2 of L1210 cells with IC50 value of 6.25-12.2 µM. antagonize the DNA break affect by etoposide. |
[42] |
| Ni-NQTS | Inhibited DNA relaxation | [30] |
| Cu-NQTS | Inhibited DNA relaxation (TOPOGEN kit) | [30] |
| Pd-NQTS | ||
| Cu(Fp4alT)Cl and its family of Cu(TSC)Cl |
Inhibited relaxation by Top2 at 10 µM Completely inhibits Top2α without promoting the formation of linear DNA products |
[31] |
| Ni-bis(TSC) | No effect in stabilizing DNA breaks | [45] |
| Cu(TSC) | Stabilizes DNA breaks | [45] |
| Ni(L1)(H1)Cl, Ni(HL2)2Cl2, Ni(L3)2,Ni(L4)2 Ni(L5)2Cl2 |
No inhibition of Top2 (TopoGEN) | [41] |
| Cu(L1)Cl, Cu(L2)Cl, Cu(L3)Cl, Cu(L4)Cl, Cu(L5)Cl2 | Inhibited Top2 (TopoGEN) | [41] |
| complex 1 (CuTSC cation) | inhibited DNA relaxation and increase DNA cleavage | [43] |
| Sn(II)-TSC—(C5) | Inhibited Topo2α at 20 µM. | [34] |
| Cu(S,R)L and Cu(R,S)L | Inhibited Top2α relaxation at 300 µM | [44] |
| Cu(APY)Cl] | Inhibited Top2α from 0.5 µM, Increased DNA cleavage Inhibited Top2α ATP hydrolysis No inhibition of ligation by Top2α Pre-incubate compounds with Top2α inactivate the enzyme |
[26] |
| Cu(APZ)Cl] | ||
| [Cu(APY)Cl] | Inhibited Top2β at 5 µM, Increased DNA cleavage by Top2β, Inhibited Top2β ATP hydrolysis, Inhibited ligation by Top2β Pre-incubate compounds with Top2β inactivate the enzyme Stabilized closure of N-terminal Top2α and Top2β clamp |
[28] |
| [Cu(BZP)Cl] | ||
| Cu(BZP)Cl series | Inhibited Top2α relaxation and increased DNA cleavage |
[27] |
| Cu(ATZ)Cl series | ||
| Pd(BZP)Cl series | Inhibited Top2α relaxation and increased DNA cleavage | [27] |
2.2.3. Inhibition of Type I and Type II Top
3. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pommier, Y.; Sun, Y.; Huang, S.N.; Nitiss, J.L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016, 17, 703-721. [CrossRef]
- Thomas, A.; Pommier, Y. Targeting Topoisomerase I in the Era of Precision Medicine. Clin. Cancer Res. 2019, 25, 6581-6589. [CrossRef]
- McKie, S.J.; Desai, P.R.; Seol, Y.; Allen, A.M.; Maxwell, A.; Neuman, K.C. Topoisomerase VI is a chirally-selective, preferential DNA decatenase. Elife 2022, 11. [CrossRef]
- Vann, K.R.; Oviatt, A.A.; Osheroff, N. Topoisomerase II Poisons: Converting Essential Enzymes into Molecular Scissors. Biochemistry 2021, 60, 1630-1641. [CrossRef]
- Deweese, J.E.; Osheroff, N. The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Res. 2009, 37, 738-749. [CrossRef]
- Murphy, M.B.; Mercer, S.L.; Deweese, J.E. Inhibitors and Poisons of Mammalian Type II Topoisomerases. In Advances in Molecular Toxicology, Fishbein, J.C., Heilman, J., Eds.; Academic Press: Cambridge, MA, 2017; Volume 11, pp. 203-240. [CrossRef]
- Gibson, E.G.; Deweese, J.E. Covalent poisons of topoisomerase II. Curr. Top. Pharm. 2013, 17, 1-12.
- Moorthy, N.S.; Cerqueira, N.M.; Ramos, M.J.; Fernandes, P.A. Aryl- and heteroaryl-thiosemicarbazone derivatives and their metal complexes: a pharmacological template. Recent Pat. Anticancer Drug Discov. 2013, 8, 168-182. [CrossRef]
- Bai, X.G.; Zheng, Y.; Qi, J. Advances in thiosemicarbazone metal complexes as anti-lung cancer agents. Front. Pharmacol. 2022, 13, 1018951. [CrossRef]
- Matesanz, A.I.; Herrero, J.M.; Quiroga, A.G. Chemical and Biological Evaluation of Thiosemicarbazone-Bearing Heterocyclic Metal Complexes. Curr. Top. Med. Chem. 2021, 21, 59-72. [CrossRef]
- Liberta, A.E.; West, D.X. Antifungal and antitumor activity of heterocyclic thiosemicarbazones and their metal complexes: current status. Biometals 1992, 5, 121-126. [CrossRef]
- Miller, M.C., 3rd; Stineman, C.N.; Vance, J.R.; West, D.X.; Hall, I.H. The cytotoxicity of copper(II) complexes of 2-acetyl-pyridyl-4N-substituted thiosemicarbazones. Anticancer Res. 1998, 18, 4131-4139.
- Baruffini, E.; Ruotolo, R.; Bisceglie, F.; Montalbano, S.; Ottonello, S.; Pelosi, G.; Buschini, A.; Lodi, T. Mechanistic insights on the mode of action of an antiproliferative thiosemicarbazone-nickel complex revealed by an integrated chemogenomic profiling study. Sci. Rep. 2020, 10, 10524. [CrossRef]
- Karp, J.E.; Giles, F.J.; Gojo, I.; Morris, L.; Greer, J.; Johnson, B.; Thein, M.; Sznol, M.; Low, J. A phase I study of the novel ribonucleotide reductase inhibitor 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP, Triapine) in combination with the nucleoside analog fludarabine for patients with refractory acute leukemias and aggressive myeloproliferative disorders. Leuk. Res. 2008, 32, 71-77. [CrossRef]
- Ma, B.; Goh, B.C.; Tan, E.H.; Lam, K.C.; Soo, R.; Leong, S.S.; Wang, L.Z.; Mo, F.; Chan, A.T.; Zee, B.; et al. A multicenter phase II trial of 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP, Triapine) and gemcitabine in advanced non-small-cell lung cancer with pharmacokinetic evaluation using peripheral blood mononuclear cells. Invest. New Drugs 2008, 26, 169-173. [CrossRef]
- Nutting, C.M.; van Herpen, C.M.; Miah, A.B.; Bhide, S.A.; Machiels, J.P.; Buter, J.; Kelly, C.; de Raucourt, D.; Harrington, K.J. Phase II study of 3-AP Triapine in patients with recurrent or metastatic head and neck squamous cell carcinoma. Ann. Oncol. 2009, 20, 1275-1279. [CrossRef]
- Finch, R.A.; Liu, M.C.; Cory, A.H.; Cory, J.G.; Sartorelli, A.C. Triapine (3-aminopyridine-2-carboxaldehyde thiosemicarbazone; 3-AP): an inhibitor of ribonucleotide reductase with antineoplastic activity. Adv. Enzyme Regul. 1999, 39, 3-12. [CrossRef]
- Finch, R.A.; Liu, M.; Grill, S.P.; Rose, W.C.; Loomis, R.; Vasquez, K.M.; Cheng, Y.; Sartorelli, A.C. Triapine (3-aminopyridine-2-carboxaldehyde- thiosemicarbazone): A potent inhibitor of ribonucleotide reductase activity with broad spectrum antitumor activity. Biochem. Pharmacol. 2000, 59, 983-991. [CrossRef]
- Posa, V.; Hajdu, B.; Toth, G.; Domotor, O.; Kowol, C.R.; Keppler, B.K.; Spengler, G.; Gyurcsik, B.; Enyedy, E.A. The coordination modes of (thio)semicarbazone copper(II) complexes strongly modulate the solution chemical properties and mechanism of anticancer activity. J. Inorg. Biochem. 2022, 231, 111786. [CrossRef]
- Yalowich, J.C.; Wu, X.; Zhang, R.; Kanagasabai, R.; Hornbaker, M.; Hasinoff, B.B. The anticancer thiosemicarbazones Dp44mT and triapine lack inhibitory effects as catalytic inhibitors or poisons of DNA topoisomerase IIα. Biochem. Pharmacol. 2012, 84, 52-58. [CrossRef]
- Huang, H.; Chen, Q.; Ku, X.; Meng, L.; Lin, L.; Wang, X.; Zhu, C.X.; Wang, Y.; Chen, Z.; Li, M.; et al. A series of α-heterocyclic carboxaldehyde thiosemicarbazones inhibit topoisomerase IIα catalytic activity. J. Med. Chem. 2010, 53, 3048-3064. [CrossRef]
- Rao, V.A.; Klein, S.R.; Agama, K.K.; Toyoda, E.; Adachi, N.; Pommier, Y.; Shacter, E.B. The iron chelator Dp44mT causes DNA damage and selective inhibition of topoisomerase IIalpha in breast cancer cells. Cancer Res. 2009, 69, 948-957. [CrossRef]
- de Oliveira, J.F.; Lima, T.S.; Vendramini-Costa, D.B.; de Lacerda Pedrosa, S.C.B.; Lafayette, E.A.; da Silva, R.M.F.; de Almeida, S.M.V.; de Moura, R.O.; Ruiz, A.; de Carvalho, J.E.; et al. Thiosemicarbazones and 4-thiazolidinones indole-based derivatives: Synthesis, evaluation of antiproliferative activity, cell death mechanisms and topoisomerase inhibition assay. Eur J Med Chem 2017, 136, 305-314. [CrossRef]
- da Silva Filho, F.A.; de Freitas Souza, T.; Ribeiro, A.G.; Alves, J.E.F.; de Oliveira, J.F.; de Lima Souza, T.R.C.; de Moura, R.O.; do Carmo Alves de Lima, M.; de Carvalho Junior, L.B.; de Almeida, S.M.V. Topoisomerase inhibition and albumin interaction studies of acridine-thiosemicarbazone derivatives. Int. J. Biol. Macromol. 2019, 138, 582-589. [CrossRef]
- Samia, L.B.; Parrilha, G.L.; Da Silva, J.G.; Ramos, J.P.; Souza-Fagundes, E.M.; Castelli, S.; Vutey, V.; Desideri, A.; Beraldo, H. Metal complexes of 3-(4-bromophenyl)-1-pyridin-2-ylprop-2-en-1-one thiosemicarbazone: cytotoxic activity and investigation on the mode of action of the gold(III) complex. Biometals 2016, 29, 515-526. [CrossRef]
- Wilson, J.T.; Jiang, X.; McGill, B.C.; Lisic, E.C.; Deweese, J.E. Examination of the Impact of Copper(II) alpha-(N)-Heterocyclic Thiosemicarbazone Complexes on DNA Topoisomerase IIalpha. Chem. Res. Toxicol. 2016, 29, 649-658. [CrossRef]
- Morris, W.H.; Ngo, L.; Wilson, J.T.; Medawala, W.; Brown, A.R.; Conner, J.D.; Fabunmi, F.; Cashman, D.J.; Lisic, E.C.; Yu, T.; et al. Structural and Metal Ion Effects on Human Topoisomerase IIalpha Inhibition by alpha-(N)-Heterocyclic Thiosemicarbazones. Chem. Res. Toxicol. 2019, 32, 90-99. [CrossRef]
- Keck, J.M.; Conner, J.D.; Wilson, J.T.; Jiang, X.; Lisic, E.C.; Deweese, J.E. Clarifying the Mechanism of Copper(II) alpha-(N)-Heterocyclic Thiosemicarbazone Complexes on DNA Topoisomerase IIalpha and IIbeta. Chem. Res. Toxicol. 2019, 32, 2135-2143. [CrossRef]
- Vutey, V.; Castelli, S.; D'Annessa, I.; Samia, L.B.; Souza-Fagundes, E.M.; Beraldo, H.; Desideri, A. Human topoisomerase IB is a target of a thiosemicarbazone copper(II) complex. Arch Biochem Biophys 2016, 606, 34-40. [CrossRef]
- Chen, J.; Huang, Y.; Liu, G.; Afrasiabi, Z.; Sinn, E.; Padhye, S.; Ma, Y. The cytotoxicity and mechanisms of 1,2-naphthoquinone thiosemicarbazone and its metal derivatives against MCF-7 human breast cancer cells. Tox. App. Pharm. 2004, 197, 40-48. [CrossRef]
- Zeglis, B.M.; Divilov, V.; Lewis, J.S. Role of metalation in the topoisomerase IIα inhibition and antiproliferation activity of a series of α-heterocyclic-N4-substituted thiosemicarbazones and their Cu(II) complexes. J. Med. Chem. 2011, 54, 2391-2398. [CrossRef]
- Hall, I.H.; Miller, M.C.; West, D.X. Antineoplastic and Cytotoxic Activities of Nickel(II) Complexes of Thiosemicarbazones. Met Based Drugs 1997, 4, 89-95. [CrossRef]
- Beckford, F.; Thessing, J.; Woods, J.; Didion, J.; Gerasimchuk, N.; Gonzalez-Sarrias, A.; Seeram, N.P. Synthesis and structure of [(eta(6)-p-cymene)Ru(2-anthracen-9-ylmethylene-N-ethylhydrazinecarbothioamide)Cl]Cl; biological evaluation, topoisomerase II inhibition and reaction with DNA and human serum albumin. Metallomics 2011, 3, 491-502. [CrossRef]
- Wu, J.; Yang, T.; Wang, X.; Li, W.; Pang, M.; Sun, H.; Liang, H.; Yang, F. Development of a multi-target anticancer Sn(ii) pyridine-2-carboxaldehyde thiosemicarbazone complex. Dalton Trans 2021, 50, 10909-10921. [CrossRef]
- Beebe, S.J.; Celestine, M.J.; Bullock, J.L.; Sandhaus, S.; Arca, J.F.; Cropek, D.M.; Ludvig, T.A.; Foster, S.R.; Clark, J.S.; Beckford, F.A.; et al. Synthesis, characterization, DNA binding, topoisomerase inhibition, and apoptosis induction studies of a novel cobalt(III) complex with a thiosemicarbazone ligand. J. Inorg. Biochem. 2020, 203, 110907. [CrossRef]
- Qi, J.; Zheng, Y.; Qian, K.; Tian, L.; Zhang, G.X.; Cheng, Z.; Wang, Y. Synthesis, crystal structure and antiproliferative mechanisms of 2-acetylpyridine-thiosemicarbazones Ga(III) with a greater selectivity against tumor cells. J. Inorg. Biochem. 2017, 177, 110-117. [CrossRef]
- Teicher, B.A. Next generation topoisomerase I inhibitors: Rationale and biomarker strategies. Biochem. Pharmacol. 2008, 75, 1262-1271. [CrossRef]
- Oliveira, C.G.; Romero-Canelon, I.; Silva, M.M.; Coverdale, J.P.C.; Maia, P.I.S.; Batista, A.A.; Castelli, S.; Desideri, A.; Sadler, P.J.; Deflon, V.M. Palladium(ii) complexes with thiosemicarbazones derived from pyrene as topoisomerase IB inhibitors. Dalton Trans 2019, 48, 16509-16517. [CrossRef]
- Heng, M.P.; Sinniah, S.K.; Teoh, W.Y.; Sim, K.S.; Ng, S.W.; Cheah, Y.K.; Tan, K.W. Synthesis of a DNA-targeting nickel (II) complex with testosterone thiosemicarbazone which exhibits selective cytotoxicity towards human prostate cancer cells (LNCaP). Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2015, 150, 360-372. [CrossRef]
- Qi, J.; Zheng, Y.; Li, B.; Ai, Y.; Chen, M.; Zheng, X. Pyridoxal hydrochloride thiosemicarbazones with copper ions inhibit cell division via Topo-I and Topo-IIa. J. Inorg. Biochem. 2022, 232, 111816. [CrossRef]
- Bisceglie, F.; Musiari, A.; Pinelli, S.; Alinovi, R.; Menozzi, I.; Polverini, E.; Tarasconi, P.; Tavone, M.; Pelosi, G. Quinoline-2-carboxaldehyde thiosemicarbazones and their Cu(II) and Ni(II) complexes as topoisomerase IIα inhibitors. J. Inorg. Biochem. 2015, 152, 10-19. [CrossRef]
- Miller III, M.C.; Stineman, C.N.; Vance, J.R.; West, D.X.; Hall, I.H. Multiple mechanisms for cytotoxicity induced by copper(II) complexes of 2-acetylpyrazine-N-substituted thiosemicarbazones. Appl. Organomet. Chem. 1999, 13, 9-19. [CrossRef]
- Sandhaus, S.; Taylor, R.; Edwards, T.; Huddleston, A.; Wooten, Y.; Venkatraman, R.; Weber, R.T.; Gonzalez-Sarrias, A.; Martin, P.M.; Cagle, P.; et al. A novel copper(II) complex identified as a potent drug against colorectal and breast cancer cells and as a poison inhibitor for human topoisomerase IIalpha. Inorg. Chem. Commun. 2016, 64, 45-49. [CrossRef]
- Bacher, F.; Enyedy, E.; Nagy, N.V.; Rockenbauer, A.; Bognar, G.M.; Trondl, R.; Novak, M.S.; Klapproth, E.; Kiss, T.; Arion, V.B. Copper(II) complexes with highly water-soluble L- and D-proline-thiosemicarbazone conjugates as potential inhibitors of Topoisomerase IIalpha. Inorg. Chem. 2013, 52, 8895-8908. [CrossRef]
- Bisceglie, F.; Pinelli, S.; Alinovi, R.; Goldoni, M.; Mutti, A.; Camerini, A.; Piola, L.; Tarasconi, P.; Pelosi, G. Cinnamaldehyde and cuminaldehyde thiosemicarbazones and their copper(II) and nickel(II) complexes: a study to understand their biological activity. J. Inorg. Biochem. 2014, 140, 111-125. [CrossRef]
- Lindsey, R.H.; Bender, R.P.; Osheroff, N. Stimulation of topoisomerase II-mediated DNA cleavage by benzene metabolites. Chem. Biol. Interact. 2005, 153-154, 197-205. [CrossRef]
- Lindsey, R.H., Jr.; Bender, R.P.; Osheroff, N. Effects of benzene metabolites on DNA cleavage mediated by human topoisomerase IIα: 1,4-hydroquinone is a topoisomerase II poison. Chem. Res. Toxicol. 2005, 18, 761-770. [CrossRef]
- Lindsey, R.H., Jr.; Bromberg, K.D.; Felix, C.A.; Osheroff, N. 1,4-Benzoquinone is a topoisomerase II poison. Biochemistry 2004, 43, 7563-7574. [CrossRef]
- Dutta, R.; Inouye, M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci 2000, 25, 24-28. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





