Submitted:
22 June 2023
Posted:
24 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
2.1. In Silico Comparative Analysis of Bovine (Bos taurus) and Human (Homo sapiens) Hemoglobin
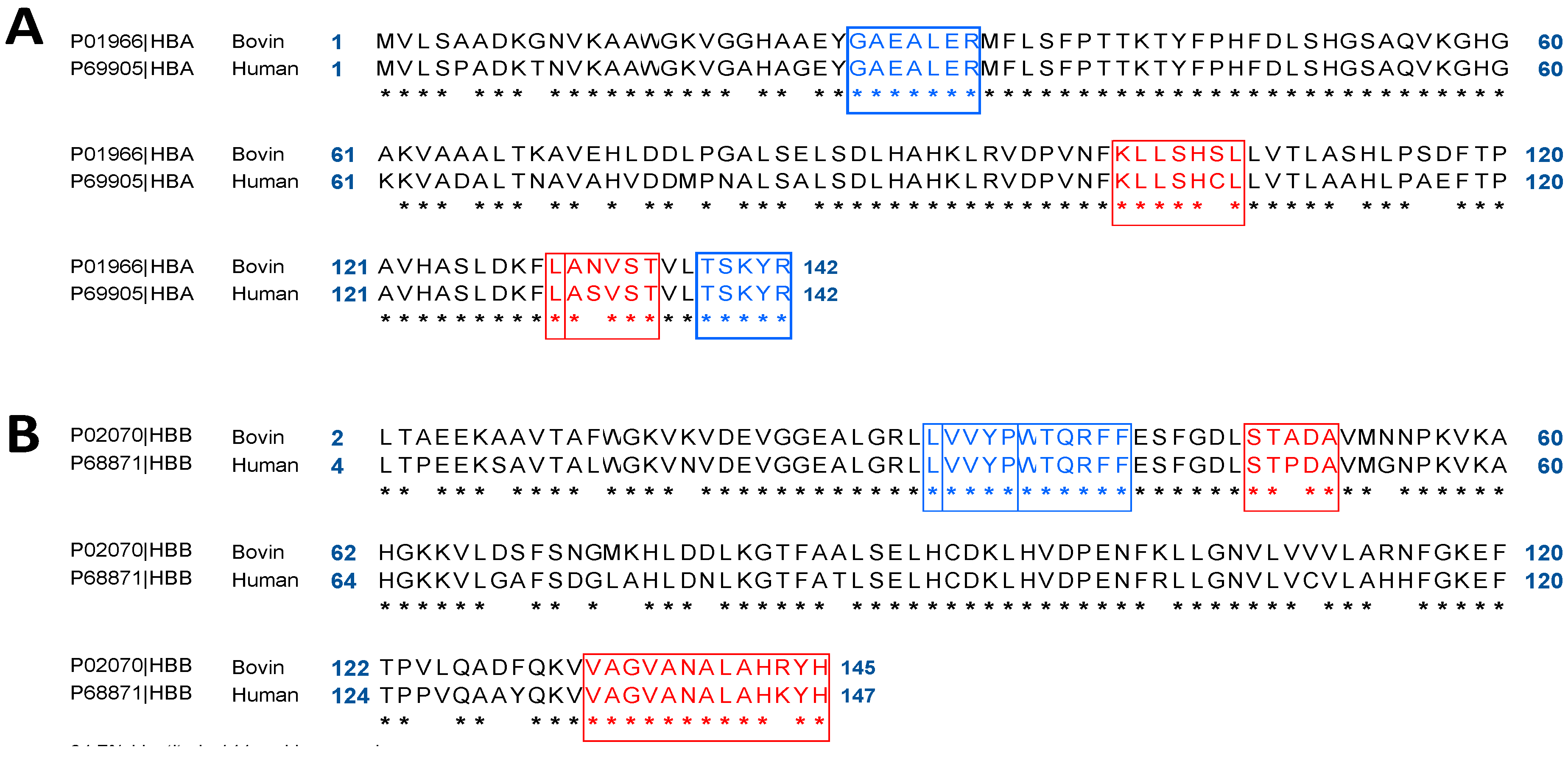
2.2. Bioinformatics Approach to Predict Peptides Derived from the Pepsic Hydrolysis of Bovine Hemoglobin Bos Taurus and Human Hemoglobin Homo sapiens
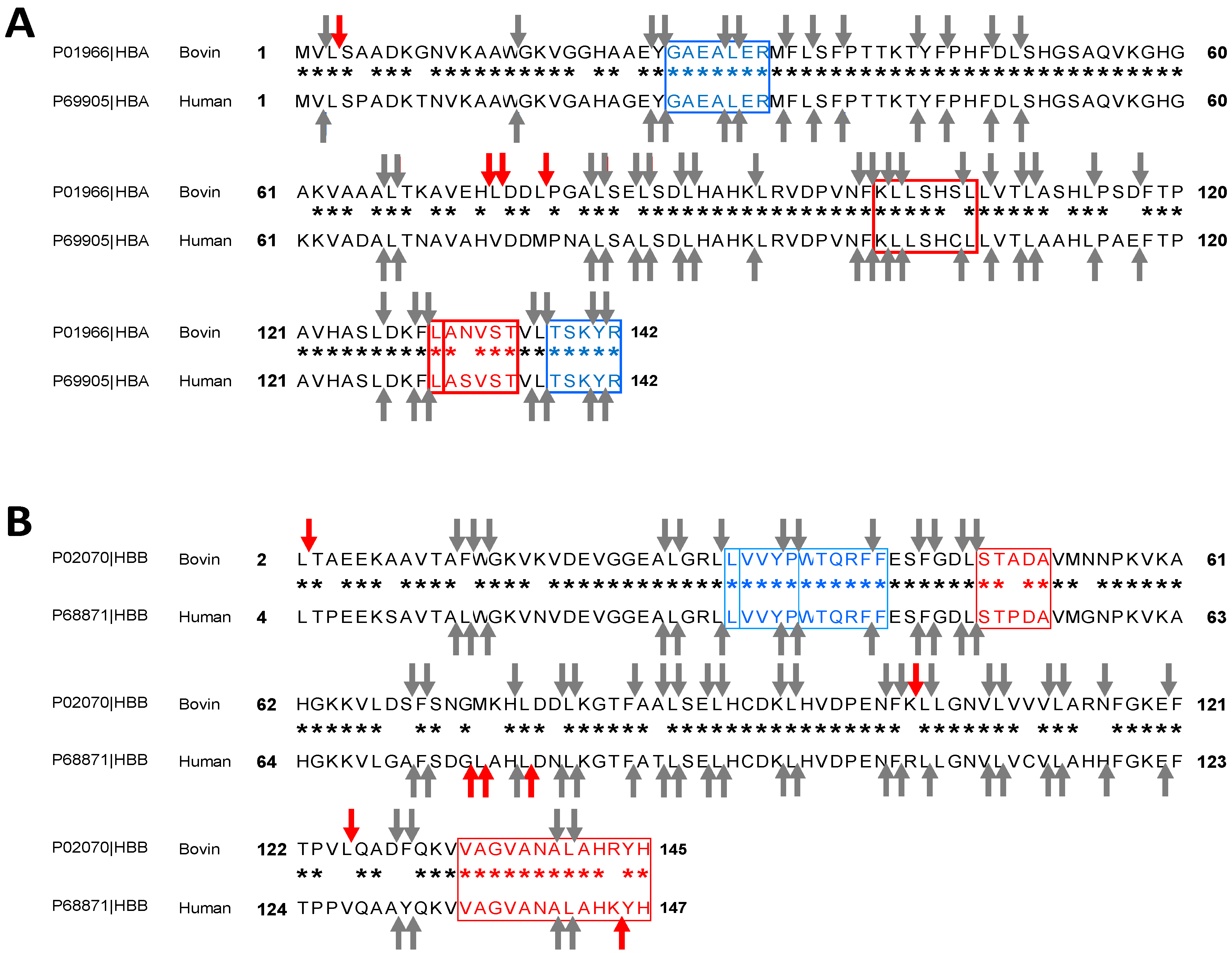
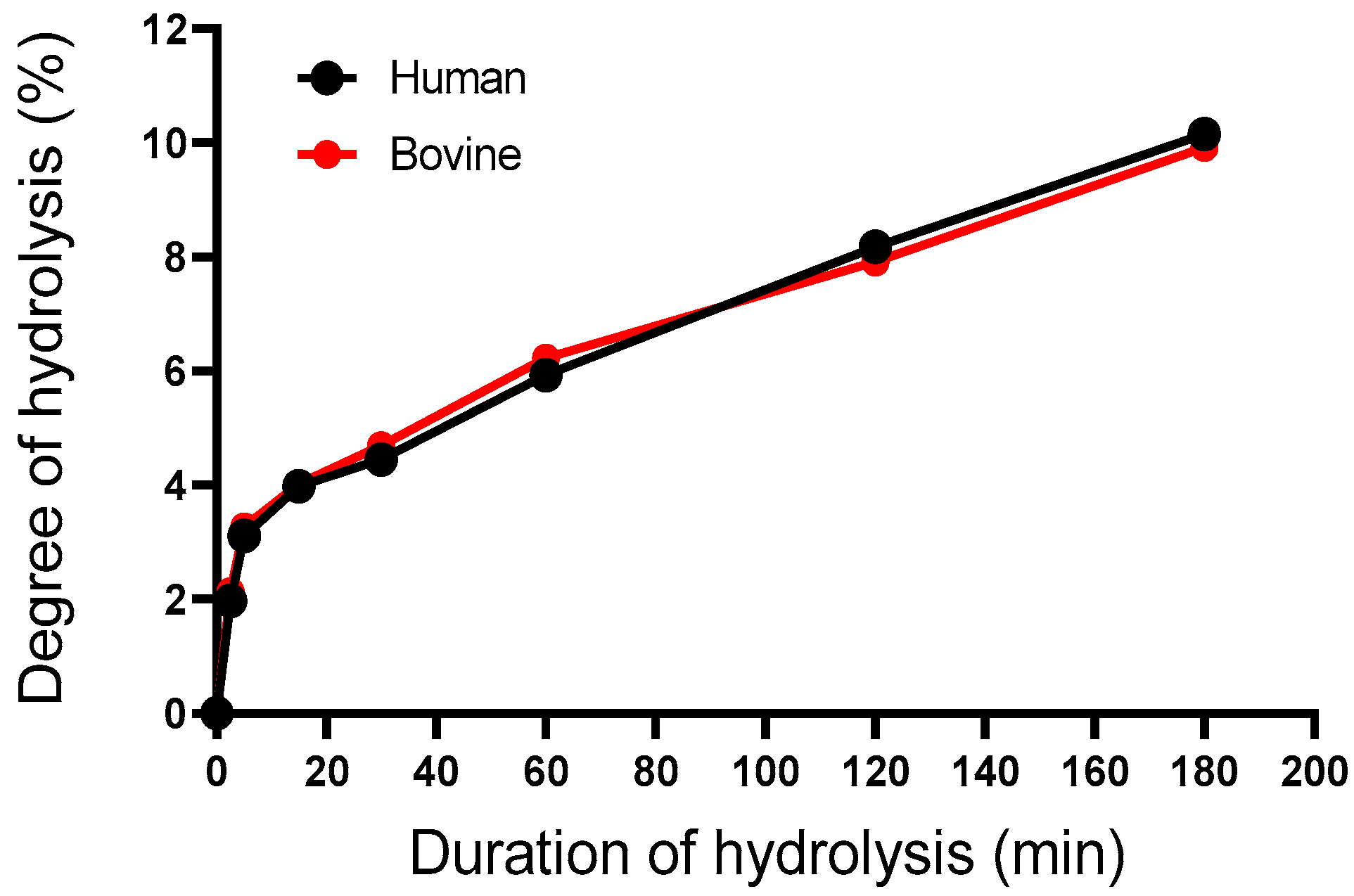
2.3. Enzymatic Kinetics and Mechanism of Action Involved
2.3.1. Determination of the Degree of Hydrolysis
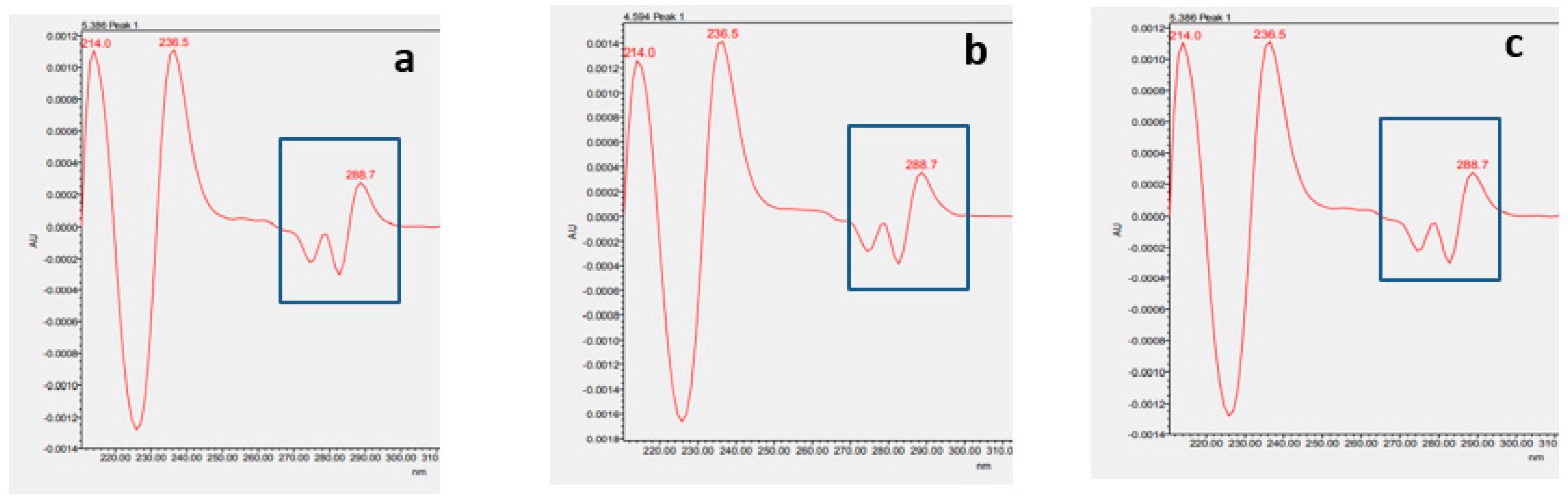
2.3.2. Identification of α137-141 by Waters Software
2.3.3. Reaction Mechanism of the Enzymatic Hydrolysis of Human Hemoglobin by Pepsin
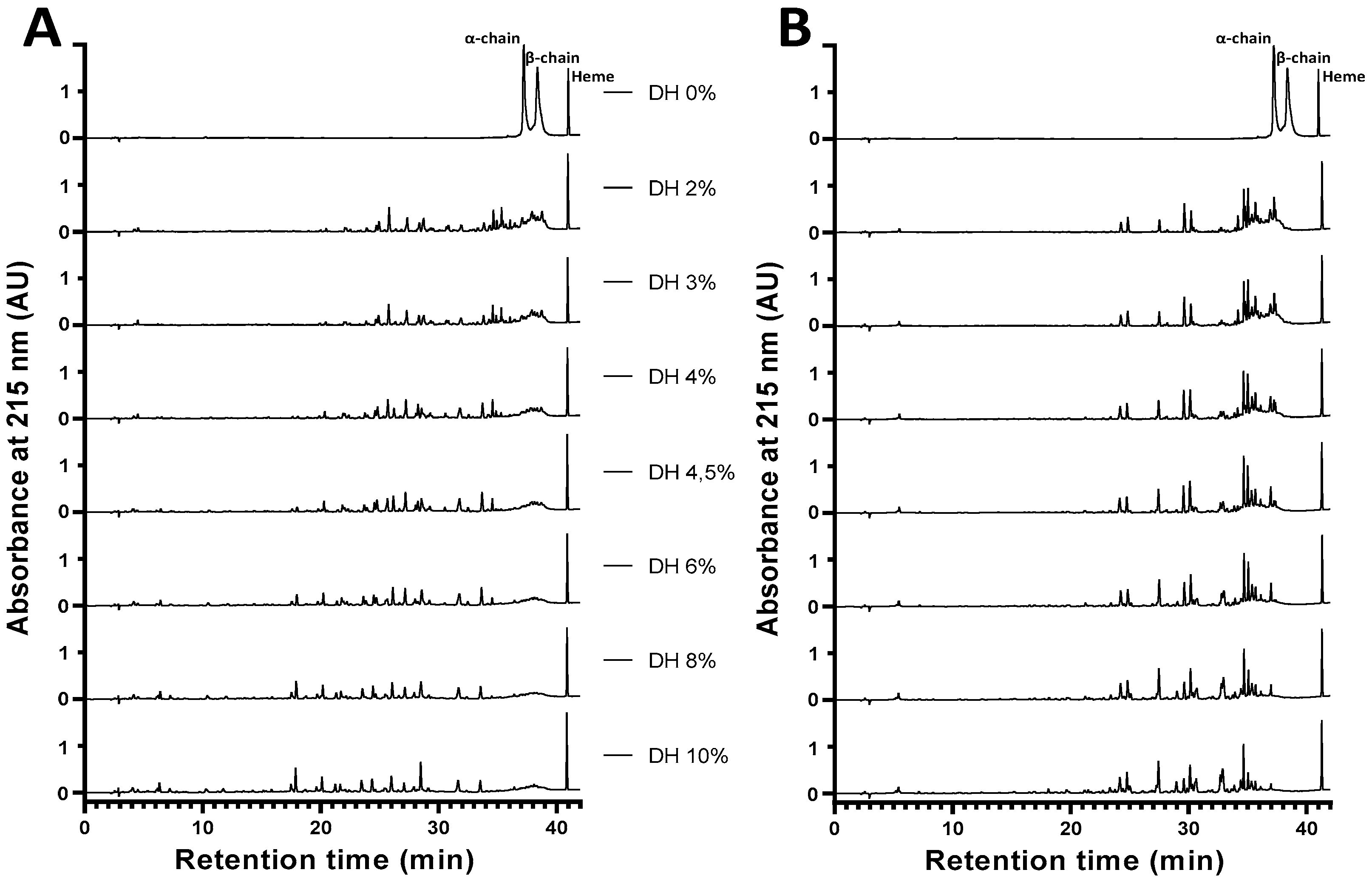
2.4. Obtaining the Active Peptide α137-141 during the Peptide Hydrolysis of Human and Bovine Hemoglobins
2.4.1. Effect of Increasing the Initial Concentration of Bovine Hemoglobin on the Enzymatic Obtention of Active Peptide α137-141
2.4.2. Quantification of the Production of the Active Peptide α137-141
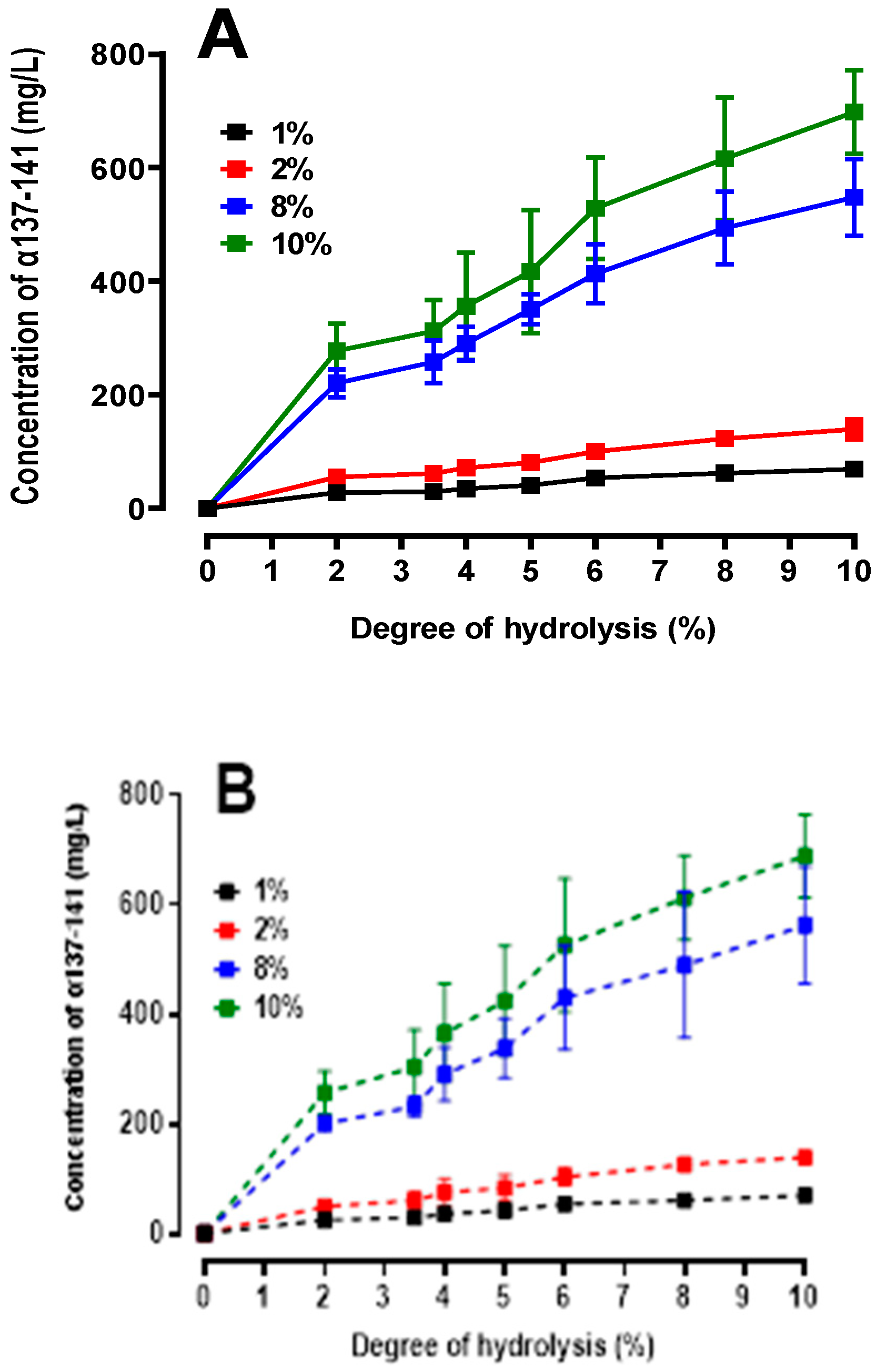
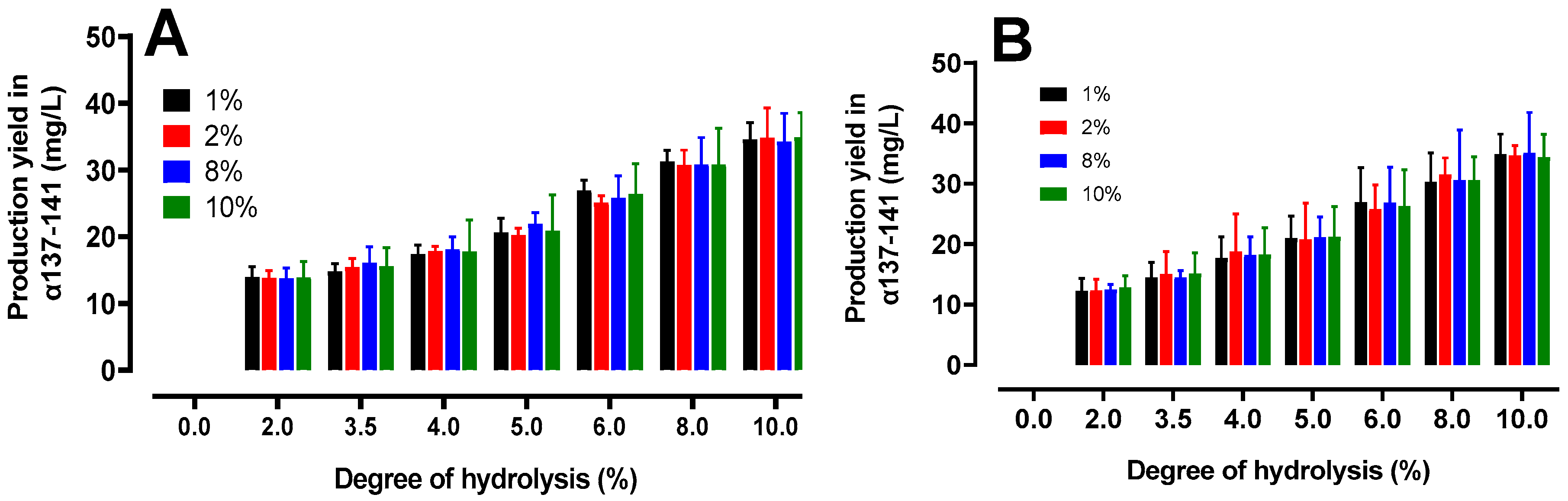
2.5. Peptidomics Approach to Characterizing the Peptide Populations.
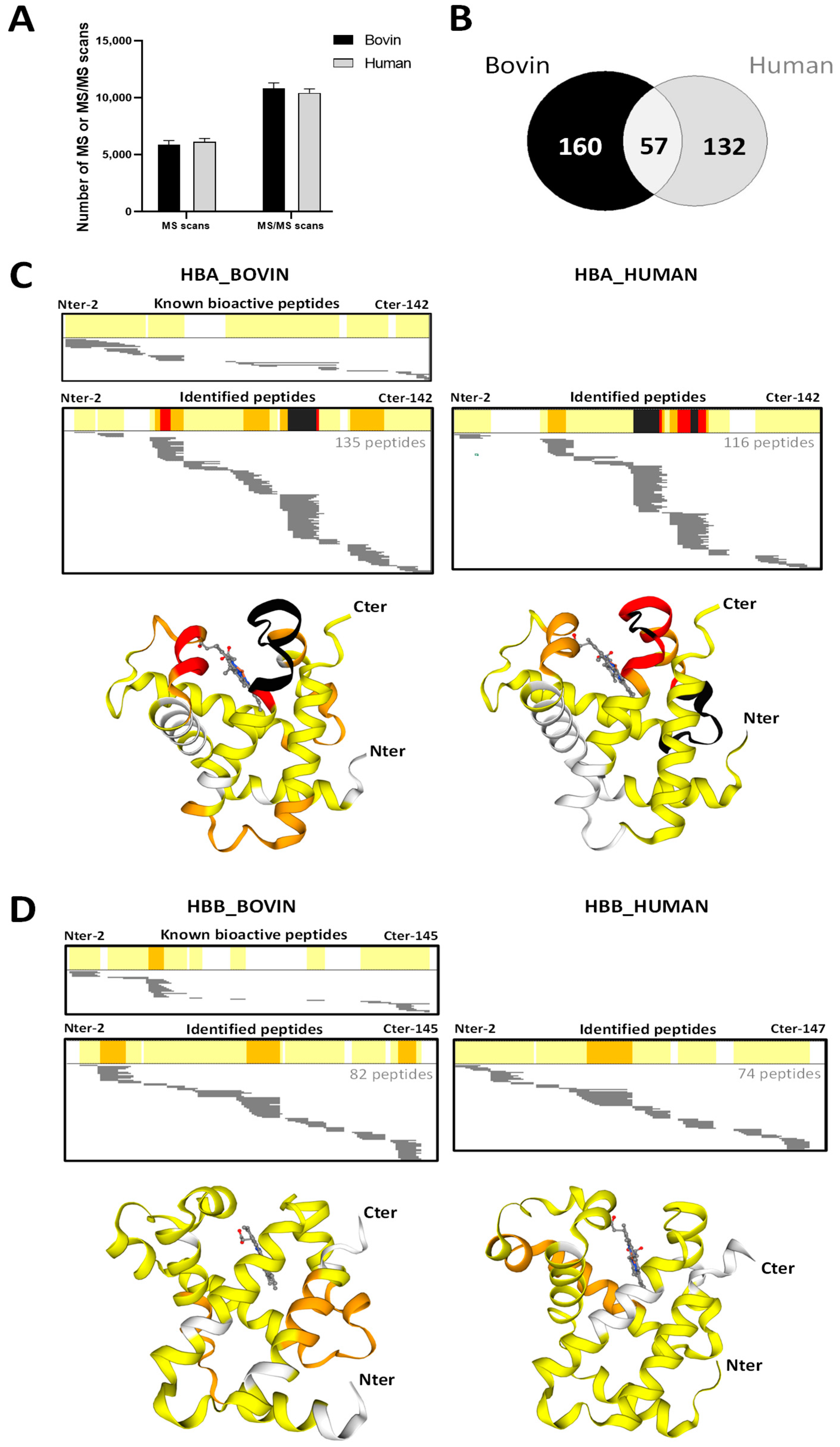
2.6. Focus on Bioactive Peptides from Bovine and Human Hemoglobin Hydrolysis
| Biological Activity | Position | Sequence | Monoisotopic Molecular (Da) | Bovine hemoglobin | Human hemoglobin |
|---|---|---|---|---|---|
| Antimicrobial | α34-46 | LSFPTTKTYFPHF | 1584.787 | + | - |
| α33-46 | FLSFPTTKTYFPHF | 1731.855 | + | - | |
| α37-46 | PTTKTYFPHF | 1237.602 | + | + | |
| α36-45 | FPTTKTYFPH | 1237.602 | + | + | |
| α137-141 | TSKYR | 653.339 | + | + | |
| α133-141 | STVLTSKYR | 1053.571 | + | + | |
| α99-105 | KLLSHSL | 796.470 | + | KLLSHCL | |
| α100-105 | LLSHSL | 668.375 | + | - | |
| α99-106 | KLLSHSLL | 909.554 | + | KLLSHCLL | |
| β140-145 | LAHRYH | 795.403 | + | LAHKYH | |
| Hematopoietic | α76-82 | LPGALSE | 685.354 | + | MPNALSA |
| β115-122 | RNFGKEFT | 997.487 | + | HHFGKEFT | |
| Opioid | α137-141 | TSKYR | 653.339 | + | + |
| β32-40 | VVYPWTQRF | 1194.608 | + | + | |
| β31-40 | LVVYPWTQRF | 1307.692 | + | + | |
| β31-37 | LVVYPWT | 876.464 | + | ||
| Analgesic and Potentiator of bradykinin | α129-134 | LANVST | 603.312 | + | LASVST |
| β 129-134 | QKVVAG | 600.349 | + | - | |
| dipeptidyl-peptidase Inhibitor | α130-134 | ANVST | 490.228 | + | ASVST |
| β6-10 | KAAVT | 488.285 | + | KSAVT | |
| ACE inhibition | β129-135 | KVVAGVA | 642.395 | + | + |
| Antihypertensive | α99-105 | KLLSHSL | 796.470 | + | KLLSHCL |
| Antioxidant | α137-141 | TSKYR | 653.339 | + | + |
| Bacterial growth stimulator | β48-52 | STADA | 463.180 | + | STPDA |
| anticancer | β33–39 | VVYPWTQ | 891.438 | + | + |
| Position | Sequence | Monoisotopic Molecular (Da) | Bovine hemoglobin | Human hemoglobin | Ref |
| The first family is situated on the N-terminal end of the α-chain, with the active portion found between residues 1 and 23. | |||||
| α1-40 | VLSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTTK | 4249 | - | + | b |
| α1-33 | VLSPADKTNVKAAWGKVGAHAGEYGAEALERMF | 3474 | - | + | b |
| α1-32 | VLSPADKTNVKAAWGKVGAHAGEYGAEALERM VLSAADKGNVKAAWGKVGGHAAEYGAEALERM (bovine) | 3327 3257 |
+ + |
+ - |
a |
| α1 -31 | VLSPADKTNVKAAWGKVGAHAGEYGAEALER | 3196 | - | + | b |
| α 1-29 | VLSPADKTNVKAAWGKVGAHAGEYGAEAL VLSAADKGNVKAAWGKVGGHAAEYGAEAL (bovine) |
2911 2841 |
- + |
+ - |
b a |
| α1-28 | VLSAADKGNVKAAWGKVGGHAAEYGAEA | 2728 | + | - | a |
| α1-27 | VLSAADKGNVKAAWGKVGGHAAEYGAE | 2656 | + | - | a |
| α1-23 | VLSAADKGNVKAAWGKVGGHAAE | 2237 | + | + | a |
| α1 -20 | VLSPADKTNVKAAWGKVGAH | 2949 | - | + | b |
| α18-44 | VGAHAGEYGAEALERMFLSFPTTKTYF | 2994 | - | + | b |
| A second family of peptides located between residues 32 and 98, with an active sequence between residues 36 and 46. | |||||
| α32-41 | FLSFPTTKTY | 1204 | - | + | c |
| α33-46 | FLSFPTTKTYFPHF | 1731 | + | + | a/e |
| α34-46 | LSFPTTKTYFPHF | 1585 | + | + | a/e |
| α36-45 | FPTTKTYFPH | 1238 | + | + | a/e |
| α37-46 | PTTKTYFPHF | 1238 | + | + | a/e |
| α33- 98 | FLSFPTTKTYFPHFDLSHGSAQVKGHGAKVAAALTKAVEHLDDLPGALSELSDLHAHKLRVDPVNF | 7151 | + | - | a |
| α33-97 | FLSFPTTKTYFPHFDLSHGSAQVKGHGAKVAAALTKAVEHLDDLPGALSELSDLHAHKLRVDPVN | 7004 | + | - | a |
| α34-98 | LSFPTTKTYFPHFDLSHGSAQVKGHGAKVAAALTKAVEHLDDLPGALSELSDLHAHKLRVDPVNF | 7004 | + | - | a |
| α36-97 | SFPTTKTYFPHFDLSHGSAQVKGHGAKVAAALTKAVEHLDDLPGALSELSDLHAHKLRVDPVN | 6744 | + | - | a |
| α37-98 | PTTKTYFPHFDLSHGSAQVKGHGAKVAAALTKAVEHLDDLPGALSELSDLHAHKLRVDPVNF | 6657 | + | - | a |
| α 33-83 | FLSFPTTKTYFPHFDLSHGSAQVKGHGAKVAAALTKAVEHLDDLPGALSEL | 5422 | + | - | a |
| α34-83 | LSFPTTKTYFPHFDLSHGSAQVKGHGAKVAAALTKAVEHLDDLPGALSEL | 5274 | + | - | a |
| α33-66 | FLSFPTTKTYFPHFDLSHGSAQVKGHGAKVAAAL | 3632 | + | - | a |
| α34-66 | LSFPTTKTYFPHFDLSHGSAQVKGHGAKVAAAL | 3484 | + | - | a |
| α35-56 | SFPTTKTYFPHFDLSHGSAQVK | 2495 | - | + | b |
| α35-80 | SFPTTKTYFPHFDLSHGSAQVKGHGKKVADALTNAVAHVDDMPNAL | 4922 | - | + | b |
| α35-77 | SFPTTKTYFPHFDLSHGSAQVKGHGKKVADALTNAVAHVDMP | 4624 | - | + | b |
| Position | Sequence | Monoisotopic Molecular (Da) | Bovine hemoglobin | Human hemoglobin | Ref |
| The third family is located on the c-terminal side of the α-chain. | |||||
| α110-131 | AAHLPAEFTPAVHASLDKFLAS | 2293 | + | - | a |
| α107-141 | VTLASHLPSDFTPAVHASLDKFLANVSTVLTSKYR | 3788 | + | - | a |
| α107-136 | VTLASHLPSDFTPAVHASLDKFLANVSTVL | 3152 | + | - | a |
| α107-133 | VTLASHLPSDFTPAVHASLDKFLANVS | 2838 | + | - | a |
| α133-141 | STVLTSKYR | 1055 | + | + | a/e |
| α137-141 | TSKYR | 654 | + | + | a/e |
| α99-105 | KLLSHSL (bovine) KLLSHCL |
796 813 |
+ - |
- + |
a e |
| α100-105 | LLSHSL | 668 | + | - | a |
| α99-106 | KLLSHSLL KLLSHCLL |
910 926 |
+ - |
- + |
a e |
| The last family of peptides is located in the c terminal region of the β chain of hemoglobin | |||||
| β1-30 | MLTAEEKAAVTAFWGKVKVDEVGGEALGRL (bovine) MVH LTPEEKSA VTALWGKVNVDEVGGEALG |
3176 3137 |
+ | + | a |
| β1-55 | MVHLTPEEKSAVTALWGKVNVDEVGGEALGRLLVVYPWTQRFFESFGDLSTPDAV | 6063 | - | + | d |
| β56–146 | MGNPKVKAHGKKVLGAFSDGLAHLDNLKGTFATLSELHCDKLHVDPENFRLLGNVLVCVLAHHFGKEFTPPVQAAYQKVVAGVANALAHKY | 9815 | - | + | d |
| β116-146 | LLGNVLVCVLAHHFGKEFTPPVQAAYQKVVAGVANALAHKY | 4375 | - | + | d |
| β56–72 | MGNPKVKAHGKKVLGAF | 1782 | - | + | d |
| β43-83 | RFFESFGDLSTPDAVMGNPKVKAHGKKVLGAFSDGLAHLDNLK | 4616 | - | + | b |
| β111-146 | LVCVLAHHFGKEFTPPVQAAYQKVVAGVANALAHKY | 3878 | - | + | b |
| β115-146 | LAHHFGKEFTPPVQAAYQKVVAGVANALAHKY | 3463 | - | + | b |
| β114-145 | ARNFGKEFTPVLQADFQKVVAGVANALAHRYH | 3556 | + | - | a |
| β121-145 | FTPVLQADFQKVVAGVANALAHRYH | 2753 | - | + | b |
| β126-145 | QADFQKVVAGVANALAHRYH | 2196 | + | - | b |
| β1-13 | MLTAEEKAAVTAF | 1381 | + | - | a |
| β140-145 | LAHRYH (bovine) LAHKYH |
795 767 |
+ - |
- + |
a/e e |
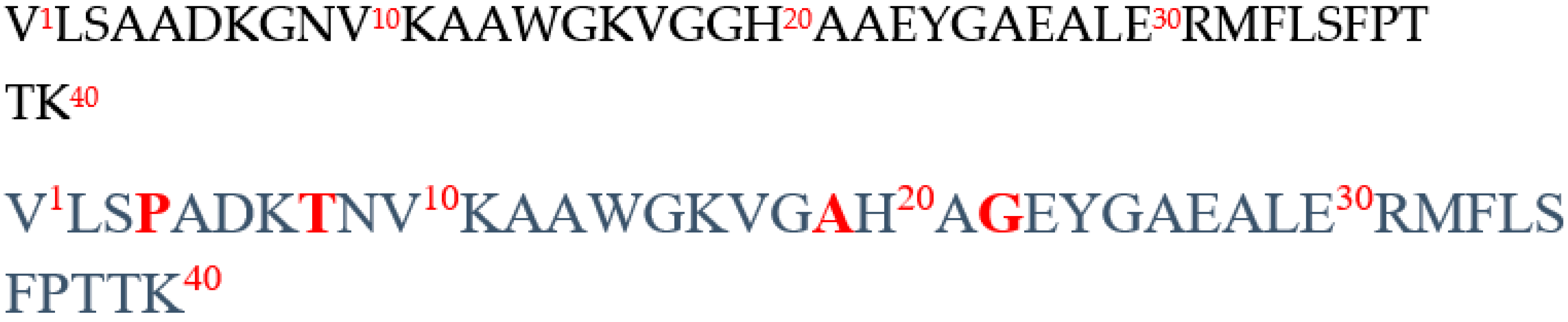
4. Materials and Methods
2.1. Comparison of Peptide Sequences by Bioinformatics Approach
2.2. Prediction of Clipping Sites by Bioinformatics Approach
2.3. Materials: Reagents, Solvents and Standards Used
- -
- - Purified bovine hemoglobin powder (H2625), dark brown and purified human hemoglobin (H7379), dark red, were obtained from Sigma-Aldrich. Hemoglobins were stored at 4°C before use.
- -
- - Pepsin, a lyophilized powder sourced from porcine gastric mucosa, purchased from Sigma-Aldrich (P6887). Its activity was measured at 3250 AU/mg protein using a protocol established by the supplier. To maintain the stability of the pepsin, it was stored at a -20°C.
2.4. Preparation of Hydrolysates
2.4.1. Preparation of the Stock Solution
2.4.2. Hydrolysis Process
2.5. Determination of the Degree of Bovine/Human Hemoglobin Hydrolysis
2.6. Analysis of the Peptide Hydrolysate by RP-UPLC and RP-HPLC
2.6.1. Hardware, Software and Protocol Used
Analysis by RP-UPLC
2.6.2. Identification of the α137-141 Peptide
2.6.3. Quantification of α137-141 in Hydrolysates
2.7. Peptidomics Approach
2.8. Two-Dimensional (2D) and Tri-Dimensional (3D) Heatmaps
2.9. RP-UPLC Statistical Analysis
5. Conclusion
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gatnau, R.; Polo, J.; Robert, E. Plasma protein antimicrobial substitution at negligible risk, Feed Manufacturing in the Mediterranean Region. Improving Safety: From Feed to Food 2001, 54, 141–150. [Google Scholar]
- Mora, L.; Reig, M.; Toldrá, F. Bioactive peptides generated from meat industry by-products. Food Research International. 2014, 65, 344–349. [Google Scholar] [CrossRef]
- Gómez-Juárez, C.; Castellanos, R.; Ponce-Noyola, T.; Calderón, V.; Figueroa, J. Protein recovery from slaughterhouse wastes. Bioresource Technology 1999, 70, 129–133. [Google Scholar] [CrossRef]
- Bah, C.S.; Bekhit, A.E.-D.A.; Carne, A.; McConnell, M.A. Slaughterhouse blood: an emerging source of bioactive compounds. Comprehensive Reviews in Food Science and Food Safety 2013, 12, 314–331. [Google Scholar] [CrossRef]
- Lafarga, T.; Hayes, M. Bioactive peptides from meat muscle and by-products: generation, functionality and application as functional ingredients. Meat Science 2014, 98, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Toldrá, F.; Aristoy, M.-C.; Mora, L.; Reig, M. Innovations in value-addition of edible meat by-products. Meat Science 2012, 92, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.-H.P.; Ofori, J.A. Blood-derived products for human consumption. Revelation and Science 2011, 1. [Google Scholar]
- Sasakawa, S. Studies on Hemoglobin VI. Amino Acid Compositions of the Fractionated Bovine Globin α and β. The Journal of Biochemistry 1961, 50, 345–351. [Google Scholar] [CrossRef]
- Lignot, B.; Froidevaux, R.; Nedjar-Arroume, N.; Guillochon, D. Solvent effect on kinetics of appearance of neokyotorphin, VV-haemorphin-4 and a bradykinin-potentiating peptide in the course of peptic hydrolysis of bovine haemoglobin. Biotechnology and Applied Biochemistry 1999, 30, 201–207. [Google Scholar]
- Zhao, Q.; Molina, P.; Piot, J.M. Peptic peptide mapping by HPLC, on line with photodiode array detection, of a hemoglobin hydrolysate produced at pilot-plant scale from an ultrafiltration process. Journal of Liquid Chromatography & Related Technologies 1997, 20, 1717–1739. [Google Scholar]
- Vercaigne-Marko, D.; Kosciarz, E.; Nedjar-Arroume, N.; Guillochon, D. Improvement of Staphylococcus aureus-V8-protease hydrolysis of bovine haemoglobin by its adsorption on to a solid phase in the presence of SDS: peptide mapping and obtention of two haemopoietic peptides. Biotechnology and Applied Biochemistry 2000, 31, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Adje, E.Y. Hydrolyse ménagée de l’hémoglobine bovine par la pepsine porcine en mélanges hydroalcooliques et obtention d’une nouvelle famille de peptides antimicrobiens, Lille 1, 2010.
- Daoud, R.; Dubois, V.; Bors-Dodita, L.; Nedjar-Arroume, N.; Krier, F.; Chihib, N.-E.; Mary, P.; Kouach, M.; Briand, G.; Guillochon, D. New antibacterial peptide derived from bovine hemoglobin. Peptides 2005, 26, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Nedjar-Arroume, N.; Dubois-Delval, V.; Adje, E.Y.; Traisnel, J.; Krier, F.; Mary, P.; Kouach, M.; Briand, G.; Guillochon, D. Bovine hemoglobin: an attractive source of antibacterial peptides. Peptides 2008, 29, 969–977. [Google Scholar] [CrossRef]
- Choisnard, L.; Froidevaux, R.; Nedjar-Arroume, N.; Lignot, B.; Vercaigne-Marko, D.; Krier, F.; Dhulster, P.; Guillochon, D. Kinetic study of the appearance of an anti-bacterial peptide in the course of bovine haemoglobin peptic hydrolysis. Biotechnology and Applied Biochemistry 2002, 36, 187–194. [Google Scholar] [CrossRef]
- Przybylski, R.; Firdaous, L.; Châtaigné, G.; Dhulster, P.; Nedjar, N. Production of an antimicrobial peptide derived from slaughterhouse by-product and its potential application on meat as preservative. Food Chemistry 2016, 211, 306–313. [Google Scholar] [CrossRef]
- de Melo, J.G.; de Sousa Araújo, T.A.; de Almeida e Castro, V.T.N.; de Vasconcelos Cabral, D.L.; Rodrigues, M.D.D.; Nascimento, S.C.D.; de Amorim, E.L.C.; De Albuquerque, U.P. Antiproliferative activity, antioxidant capacity and tannin content in plants of semi-arid northeastern Brazil. Molecules 2010, 15, 8534–8542. [Google Scholar] [CrossRef]
- Sakanaka, S.; Tachibana, Y.; Okada, Y. Preparation and antioxidant properties of extracts of Japanese persimmon leaf tea (kakinoha-cha). Food Chemistry 2005, 89, 569–575. [Google Scholar] [CrossRef]
- Chen, L.; Xiong, Z.; Sun, L.; Yang, J.; Jin, Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Research 2012, 40, D641–D645. [Google Scholar] [CrossRef]
- Saadi, S.; Saari, N.; Anwar, F.; Hamid, A.A.; Ghazali, H.M. Recent advances in food biopeptides: Production, biological functionalities and therapeutic applications. Biotechnology Advances 2015, 33, 80–116. [Google Scholar] [CrossRef]
- Linderstrom-Lang, K. Les phases initiales de la dégradation des protéines par les enzymes. Bulletin de La Société de Chimie Biologique 1953, 100–116. [Google Scholar]
- Takagi, H.; Shiomi, H.; Fukui, K.; Hayashi, K.; Kiso, Y.; Kitagawa, K. Isolation of a novel analgesic pentapeptide, neo-kyotorphin, from bovine brain. Life Sciences 1982, 31, 1733–1736. [Google Scholar] [CrossRef]
- Catiau, L.; Traisnel, J.; Delval-Dubois, V.; Chihib, N.-E.; Guillochon, D.; Nedjar-Arroume, N. Minimal antimicrobial peptidic sequence from hemoglobin alpha-chain: KYR. Peptides 2011, 32, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Dunn, B.M. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chemical Reviews 2002, 102, 4431–4458. [Google Scholar] [CrossRef]
- Church, F.C.; Swaisgood, H.E.; Porter, D.H.; Catignani, G.L. Spectrophotometric Assay Using o-Phthaldialdehyde for Determination of Proteolysis in Milk and Isolated Milk Proteins. Journal of Dairy Science 1983, 66, 1219–1227. [Google Scholar] [CrossRef]
- Church, F.C.; Swaisgood, H.E.; Porter, D.H.; Catignani, G.L. Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. Journal of Dairy Science 1983, 66, 1219–1227. [Google Scholar] [CrossRef]
- Spellman, D.; McEvoy, E.; O’cuinn, G.; FitzGerald, R.J. Proteinase and exopeptidase hydrolysis of whey protein: Comparison of the TNBS, OPA and pH stat methods for quantification of degree of hydrolysis. International Dairy Journal 2003, 13, 447–453. [Google Scholar] [CrossRef]
- Church, F.C.; Porter, D.H.; Catignani, G.L.; Swaisgood, H.E. An o-phthalaldehyde spectrophotometric assay for proteinases. Analytical Biochemistry 1985, 146, 343–348. [Google Scholar] [CrossRef]
- Zhao, Q.; Sannier, F.; Piot, J.M. Kinetics of appearance of four hemorphins from bovine hemoglobin peptic hydrolysates by HPLC coupled with photodiode array detection. Biochimica et Biophysica Acta (BBA)-Protein Structure and Molecular Enzymology 1996, 1295, 73–80. [Google Scholar] [CrossRef]
- Zhao, Q.; Sannier, F.; Ricart, G.; Piot, J.M. A rapid detection and identification of hemorphins released from bovine hemoglobin enzymatic hydrolysis by use of HPLC coupled with photodiode array detector. Journal of Liquid Chromatography & Related Technologies 1995, 18, 93–103. [Google Scholar]
- Dubois, V.; Nedjar-Arroume, N.; Guillochon, D. Influence of pH on the appearance of active peptides in the course of peptic hydrolysis of bovine haemoglobin. Preparative Biochemistry and Biotechnology 2005, 35, 85–102. [Google Scholar] [CrossRef]
- Ivanov, V.; Andrei, T.; Karelin, A.; Mikhaleva, I.; Vaskovsky, B.V.; Sviryaev, V.L.; Nazimov, I.V. Isolation, structure and properties of endogenous peptides. Russian Journal of Bioorganic Chemistry 1992, 1271–1311. [Google Scholar]
- Piot, J.-M.; Zhao, Q.; Guillochon, D.; Ricart, G.; Thomas, D. Isolation and characterization of two opioid peptides from a bovine hemoglobin peptic hydrolysate. Biochemical and Biophysical Research Communications 1992, 189, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Abou-Diab, M.; Thibodeau, J.; Deracinois, B.; Flahaut, C.; Fliss, I.; Dhulster, P.; Nedjar, N.; Bazinet, L. Bovine Hemoglobin Enzymatic Hydrolysis by a New Ecoefficient Process—Part I: Feasibility of Electrodialysis with Bipolar Membrane and Production of Neokyotorphin (α137-141). Membranes 2020, 10, 257. [Google Scholar] [CrossRef] [PubMed]
- Caron, J.; Cudennec, B.; Domenger, D.; Belguesmia, Y.; Flahaut, C.; Kouach, M.; Lesage, J.; Goossens, J.-F.; Dhulster, P.; Ravallec, R. Simulated GI digestion of dietary protein: Release of new bioactive peptides involved in gut hormone secretion. Food Research International 2016, 89, 382–390. [Google Scholar] [CrossRef]
- Kagawa, K.; Matsutaka, H.; Fukuhama, C.; Watanabe, Y.; Fujino, H. Globin digest, acidic protease hydrolysate, inhibits dietary hypertriglyceridemia and Val-Val-Tyr-Pro, one of its constituents, possesses most superior effect. Life Sciences 1996, 58, 1745–1755. [Google Scholar] [CrossRef]
- Sousa, R., Jr.; Lopes, G.P.; Tardioli, P.W.; Giordano, R.L.C.; Almeida, P.I.F.; Giordano, R.C. Kinetic model for whey protein hydrolysis by alcalase multipoint-immobilized on agarose gel particles. Brazilian Journal of Chemical Engineering 2004, 21, 147–153. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, M.; Zhang, J.; Zhao, D.; Sun, J. Peptidomics insights into the interplay between the pre-digestion effect of mixed starters and the digestive pattern of sausage proteins. Food Research International 2022, 162, 111963. [Google Scholar] [CrossRef]
- Albuquerque, W.; Ghezellou, P.; Lee, K.-Z.; Schneider, Q.; Gross, P.; Kessel, T.; Omokungbe, B.; Spengler, B.; Vilcinskas, A.; Zorn, H. Peptidomics as a Tool to Assess the Cleavage of Wine Haze Proteins by Peptidases from Drosophila suzukii Larvae. Biomolecules 2023, 13, 451. [Google Scholar] [CrossRef]
- Barkhudaryan, N.; Oberthuer, W.; Lottspeich, F.; Galoyan, A. Structure of hypothalamic coronaro-constrictory peptide factors. Neurochemical Research 1992, 17, 1217–1221. [Google Scholar] [CrossRef]
- Blishchenko, E.Y.; Mernenko, O.A.; Mirkina, I.I.; Satpaev, D.K.; Ivanov, V.S.; Tchikin, L.D.; Ostrovsky, A.G.; Karelin, A.A.; Ivanov, V.T. Tumor cell cytolysis mediated by valorphin, an opioid-like fragment of hemoglobin β-chain. Peptides 1997, 18, 79–85. [Google Scholar] [CrossRef]
- Mak, P.; Wójcik, K.; Wicherek, Ł.; Suder, P.; Dubin, A. Antibacterial hemoglobin peptides in human menstrual blood. Peptides 2004, 25, 1839–1847. [Google Scholar] [CrossRef] [PubMed]
- Parish, C.A.; Jiang, H.; Tokiwa, Y.; Berova, N.; Nakanishi, K.; McCabe, D.; Zuckerman, W.; Xia, M.M.; Gabay, J.E. Broad-spectrum antimicrobial activity of hemoglobin. Bioorganic & Medicinal Chemistry 2001, 9, 377–382. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





