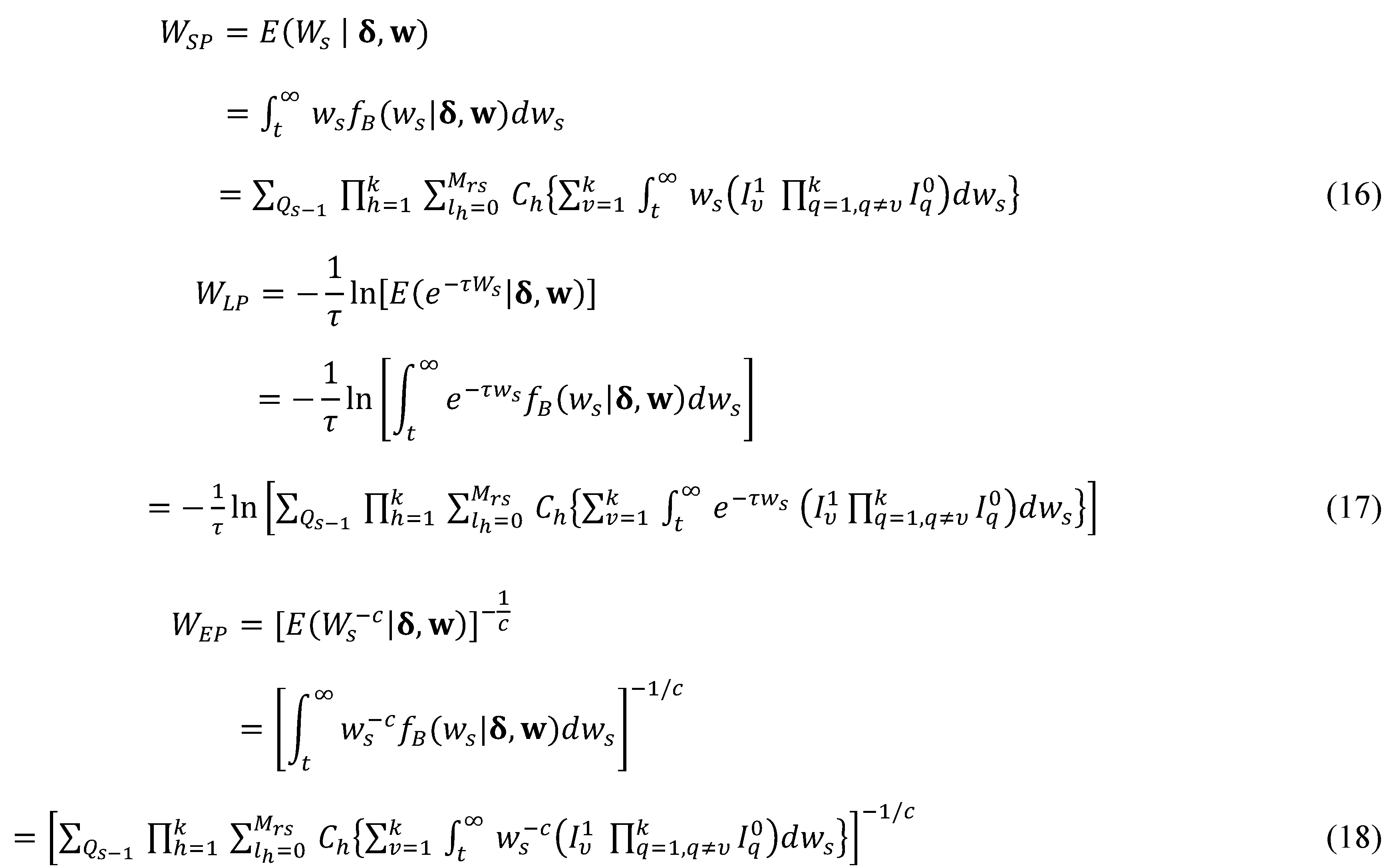1. Introduction and motivations
Generalized Bayes is a Bayesian study based on a learning rate parameter (
) as a power of the likelihood function. The traditional Bayes framework is obtained for
, and we demonstrate the effect of the learning rate parameter on the prediction results. That is, if the prior distribution of the parameter
is
then the generalized Bayes posterior distribution for
is
For more details on the generalized Bayes method and the choice of the value of the rate parameter we refer readers to
[1-
11]. An exact inference method based on maximum likelihood estimates (MLEs) was developed in [
12], and its performance was compared with that of approximate, Bayesian, and bootstrap methods. The joint progressive censoring type II and the expected number of failures for two populations under the joint progressive censoring type II were introduced and studied by [
13]. In contrast, exact likelihood inference for two exponential populations under joint progressive censoring of type II was studied in [
14] and some precise results were obtained based on the maximum likelihood estimates developed by [
15]. Exact likelihood inference for two populations of two-parameter exponential distributions under type II joint censoring was studied by [
16].
One might be interested in predicting future failures using a joint type II censored sample. To accomplish this, prediction points or intervals should be determined. Bayesian prediction bounds for future observations based on certain distributions have been discussed by several authors. A study of Bayesian estimation and prediction based on a joint censored sample of type II from two exponential populations was presented by [
17]. Prediction (various classical and Bayesian point predictors) for future failures in the Weibull distribution under hybrid censoring was studied by [
18]. A Bayesian prediction based on generalized order statistics with multiple censoring of type II was developed by [
19].
The main objective of this study is to predict future failures based on a joint type-II censoring scheme for k-exponential populations when censoring is performed on k-samples in a combined manner. Suppose that products from different lines are produced in the same factory, and independent samples of size are selected from these lines and simultaneously placed in a lifetime experiment. To reduce the cost and time of the experiment, the experimenter may decide to stop the lifetime test when a certain number () of failures occurs. The nature of the problem and the distributions used in our study are presented below.
Suppose are -samples, where are the lifetimes of samples of product line and are assumed to be independent and identically distributed (iid) random variables from a population with a a probability density function (pdf) and a cumulative distribution function (cdf) .
Furthermore, let
be the total sample size and let
be the total number of observed failures. Let
denotes the order statistics of
N random variables,
. Under the joint type- II censoring scheme for the
-samples, the observable data then consist of
, where
, with
being a pre-fixed integer and
associated to
is defined by
Letting
denote the number of
-failures in
and
, then the joint density function of
is given by
where
, is the survival functions of
population and
.
The joint density function of
is given by
where,
with .
The conditional density function of
given
, is given by
where,
When the
populations are exponential with pdf and cdf respectively,
Then the likelihood function in (3) becomes
where,
and
.
Substituting (6) into (5), we obtain the conditional density function of
, given
where,
,
and
.
Some special cases of the conditional density function are described as follows:
Case 1:
Suppose that is the number of samples satisfy and only one sample satisfies for equivalently but ; .
In Case 1, the conditional density function of
given
is given by
where,
,
,
.
Case 2:
Suppose that is the number of samples satisfy for and is the number of samples satisfy for , equivalently but .
Under case 2, let’s just consider the
samples, where
, then the conditional density function of
given
is given by
where, , and .
The remainder of this article is organized as follows.
Section 2 presents the generalized Bayesian and Bayesian prediction points and intervals using squared error, Linex, and general entropy loss functions in the point predictor. A numerical study of the results from
Section 2 is presented in
Section 3. Finally, we conclude the paper in
Section 4.
2. Generalized Bayes prediction
In this section, we introduce the concept of generalized Bayesian prediction, which is an investigation of Bayesian prediction under the influence of a learning rate parameter . To apply the concept of generalized Bayesian prediction to a prediction study, we give a brief description of generalized Bayesian prediction based on a learning rate parameter . A scheme for predicting a sample based on joint censoring of type II samples from k exponential distributions is presented. The main goal is to obtain the point predictors and prediction intervals given at the end of this section.
2.1. Generalized Bayes
The parameters
are assumed to be unknown, we may consider the conjugate prior distributions of
as independent gamma prior distributions, i.e.
. Hence, the joint prior distribution of
is given by
where
and
denotes the complete gamma function.
Combining (7) and (10) after raising (7) to the power
, the posterior joint density function of
is then
where,
.
Since is a conjugate prior, where then it follows that the posterior density function of is .
2.2. One sample prediction
A sample prediction scheme for the case of joint censoring of samples from two exponential distributions was studied in [
17] and then three cases for the future failures were derived, where in the first case the future predicted failure surly belongs to
failures if
, second case, the future predicted failure surly belongs to
failures if
, third case, it is unknown to which sample the future predicted failure belongs. Here, we generalize the results reported in [
17] and examine two special cases in addition to the general case.
In the general case, the size of any sample is greater than the number of observed failures; that is, for The first special case arises when all future values (predictors) belong to only one sample and the observations of the remaining samples are less than . The second special case arises when all future values (predictors) belong to some samples and all observations of the other samples are less than . The forms of all functions related to the second special case are similar to those related to the general case; therefore, we will introduce only the general case and the first special case.
For the general case, to predict
for
based on the observed data
, we use the conditional density function
. Let us define the following integral
Using (8), (12), and (13) the Bayesian predictive density function of
, given
, is given by
Under case 1, the Bayesian predictive density function of
, given
, is given by
where,
,
.
2.3. Bayesian point predictors
For the point predictor, we considered three types of loss functions:
(i). The squared error loss function (SE), which is classified as a symmetric function, is given by
where
is an estimate of
(ii). The Linex loss function, which is asymmetric is given by
(iii). The generalization of the entropy (GE) loss function is
It is worth noting that the Bayes estimates under the GE loss function coincide with those under the SE loss function when However, when the Bayes estimates under GE become those under the weighted squared error loss function and the precautionary loss function, respectively.
Now, the Bayesian point predictors , under different loss functions (SE, Linex, GE) can be obtained using the predictive density function (14), which are denoted respectively by and given as follows:
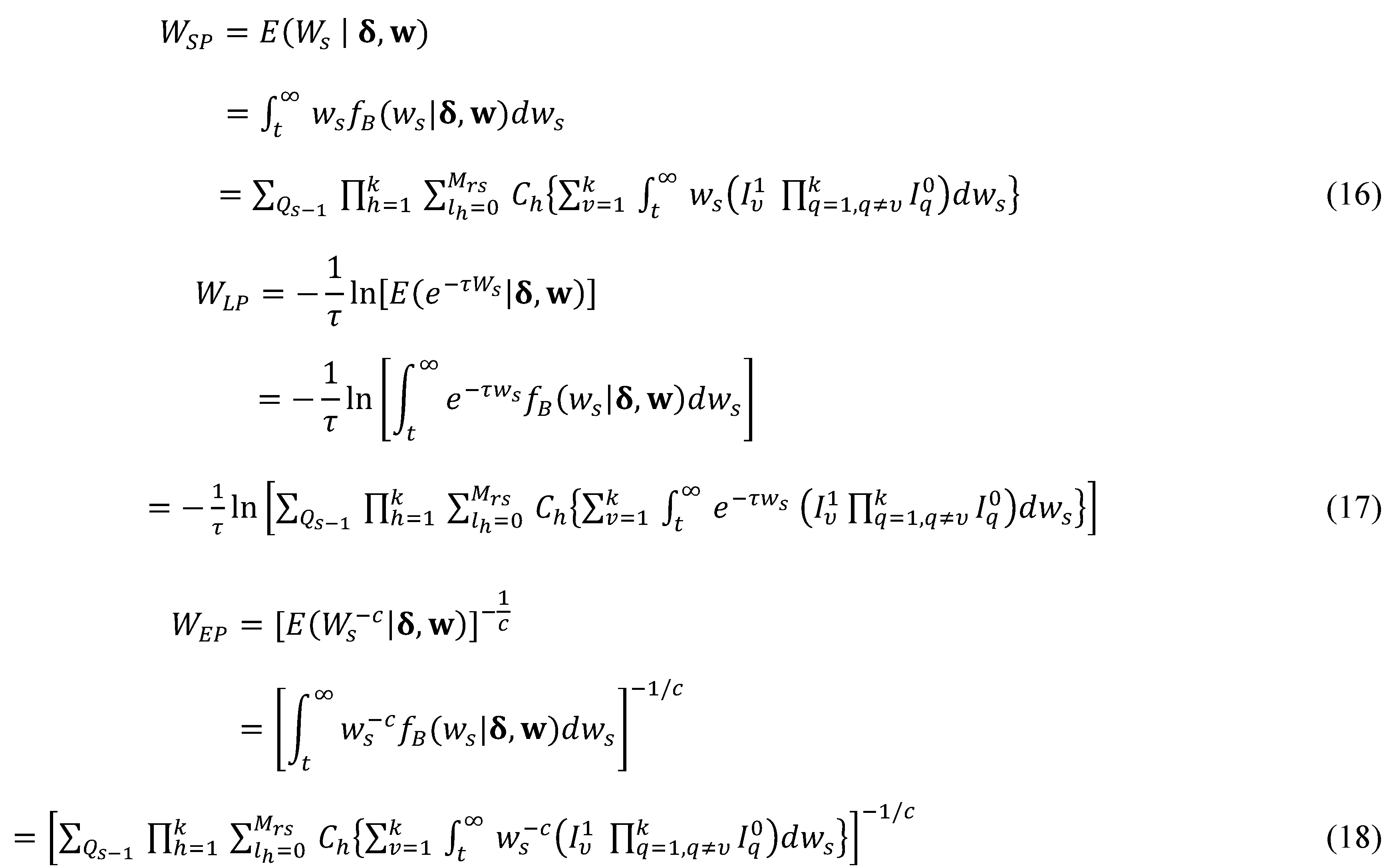
Under case 1,
are respectively given by
Numerical integration is required to obtain the predictors .
2.4. Prediction Interval
The predictive survival function of
is given by
Numerical integration is required to obtain the predictive distribution function in Equation (22). In Case 1, the predictive survival function of
is given by
The Bayesian predictive bounds of a two-sided equi-tailed
interval for
, can be obtained by solving the following two equations,
3. Numerical study
In this section, the results of the Monte Carlo simulation study are conducted to evaluate the performance of the prediction study derived in the previous section, and an example is presented to illustrate the prediction methods discussed here.
3.1. Simulation study
We considered three samples from three populations with for choices and . In case 1, we choose the exponential parameters as (2, 1, 0.1) based on the hyperparameters represented by , where .
In the general case we choose the exponential parameters as (2, 2.5, 3 ) based on the hyperparameters .
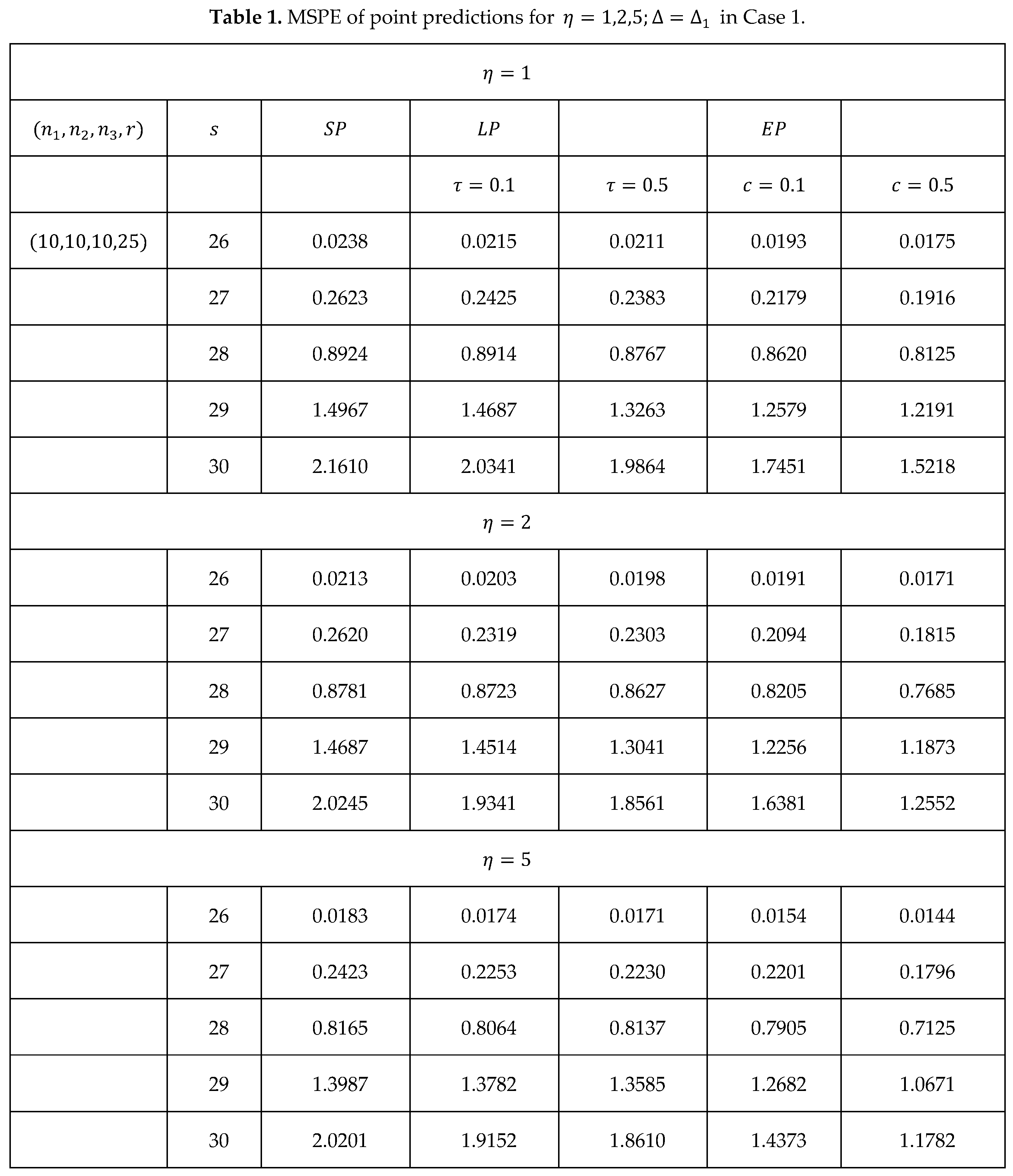
For the generalized Bayesian study, three values are chosen for the learning rate parameter and 10,000 repetitions are used for the Monte Carlo simulations. For , under case 1 we use (19), (20) and (21), to calculate the mean squared prediction errors (MSPEs) of the point predictors () for , where ; and the results are presented in Table 1.
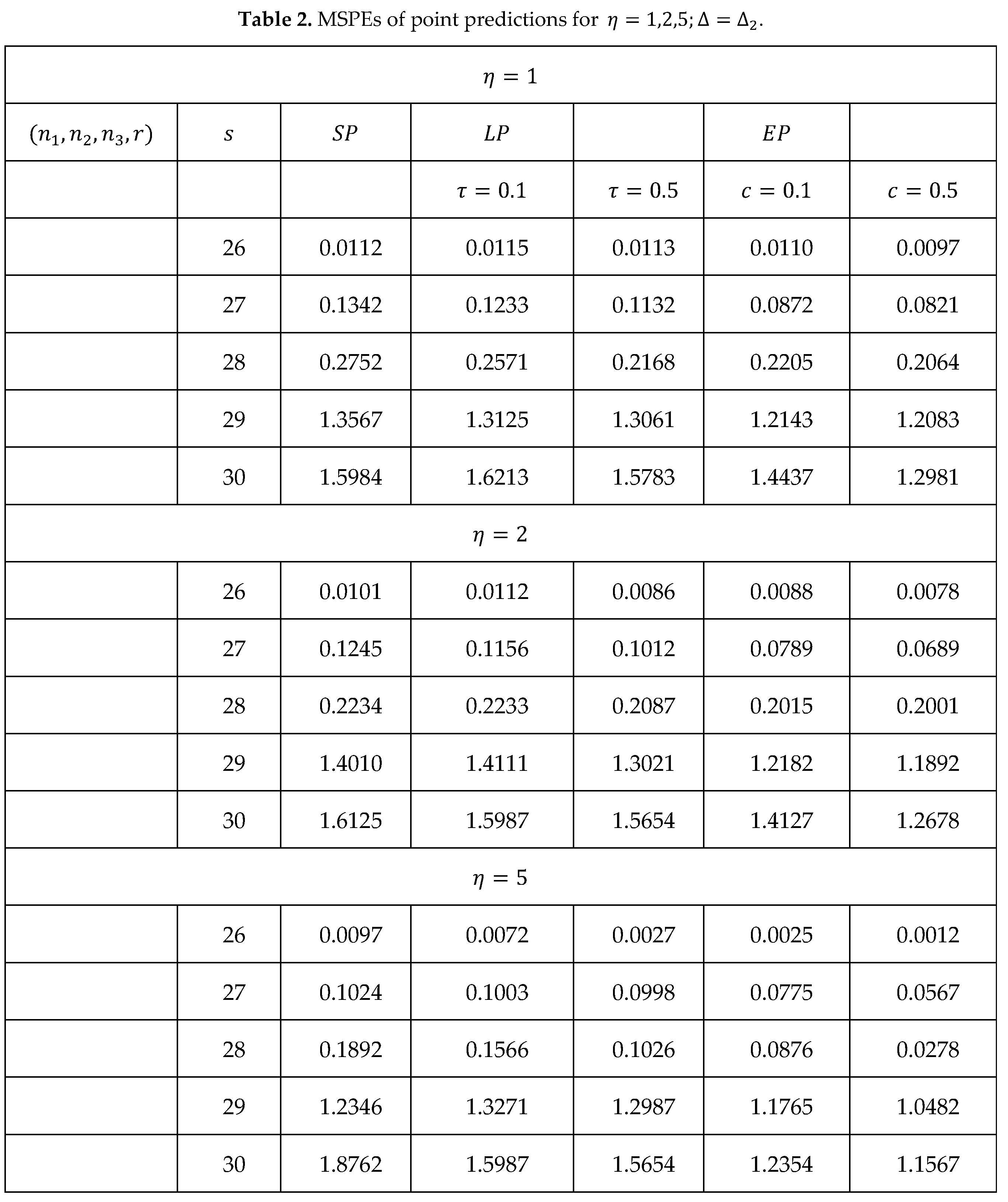
The results of the MSPEs in the general case are calculated using (16), (17), and (18) and shown in Table 2.
For and the results of the prediction bounds of and , respectively are calculated using (23), (24) in Case 1 then are presented in Table 3.
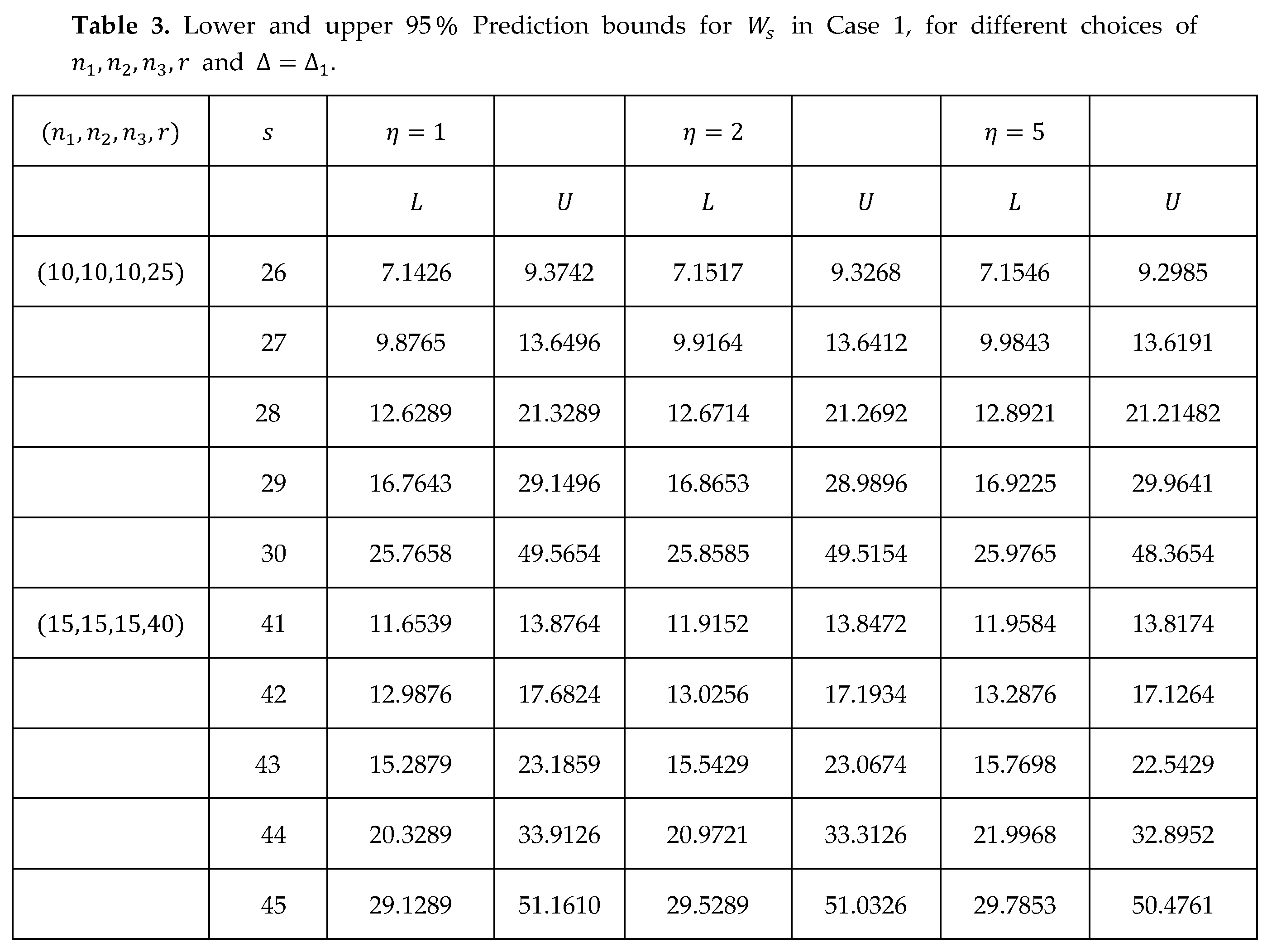
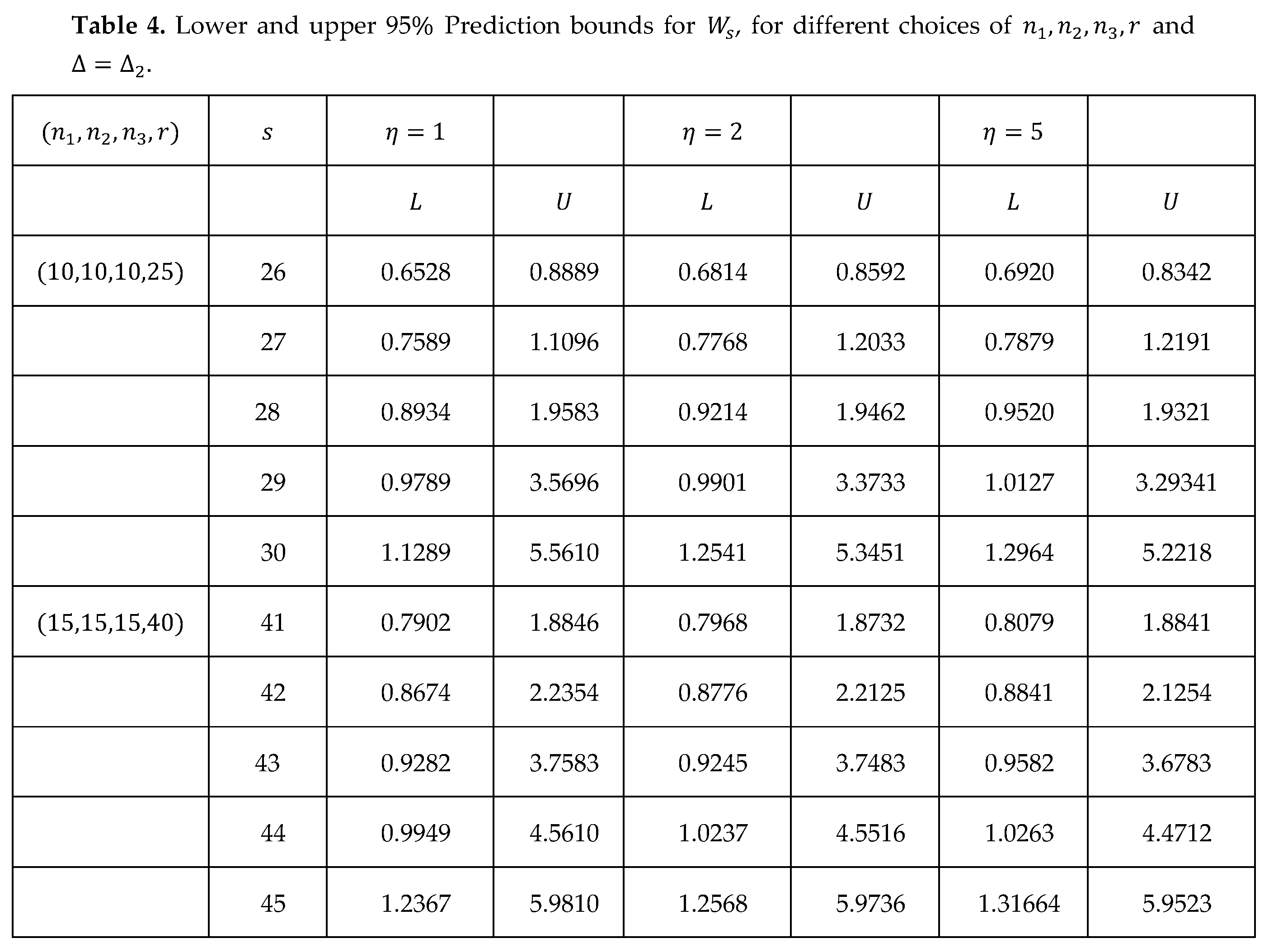
Table 4 presents the prediction bounds using (22), (24) to show the results of the general case.
3.2. Illustrative example
To illustrate the usefulness of the results developed in the previous sections, we consider three samples of size
from Nelson's data (groups 1, 4, and 5) corresponding to the breakdown of an insulating fluid subjected to a high-stress load (see [
20] p. 462), These breakdown times, referred to here as samples
, are jointly type-II censored data in the form of
obtained from these three samples with
and are shown in Table 5.

Using (16), (17), (18), (22), and (24), the MSPEs of the point predictors and prediction intervals
of ws, s = 25,..., 30 are calculated and presented in Table 6 using 𝜂 = 1, 2, 5 and Δ = Δ3 =
(1,2.6,1,2,1,3); 𝜏 = 0.1, 0.5 and c = 0.1, 0.5.