1. Introduction
1,2,3,4,6 Pentagalloyl glucose (PGG), as well as its parent compound, tannic acid (decagalloyl glucose), were first introduced in the early 1970s as efficient mordants (i.e., fixation agents that bind to tissue components and enhance further binding of metal ions) during tissue preparation for transmission electron microscopy(TEM)[
1,
2]. Notably, Kajikawa et al. showed that elastin could be visualized at the TEM level using a tannic acid method[
3], inferring that tannic acid has a high affinity towards elastin. Since then, many papers have been published utilizing this method of highlighting elastic fibers for TEM[
3,
4,
5]. While searching for alternative fixatives for the preparation of bioprosthetic heart valves, our group showed that tannic acid binds to pure elastin and vascular elastic fibers, protecting elastin from degradation by elastase[
6]. In a subsequent paper, we showed that tannic-acid-mediated elastin stabilization also protects elastin from calcification in vivo[
7]. In 2006, we determined that the polyphenolic groups are responsible for the stabilization properties and that PGG, resulting from tannic acid hydrolysis, is equally effective as an elastin stabilizer and more stable than tannic acid[
8]. Since then, we continued to utilize PGG as a non-cytotoxic agent to stabilize collagen and elastin scaffolds for tissue engineering purposes[
9,
10,
11,
12]. PGG has found potential applications in cardiovascular medicine exhibiting excellent safety profiles[
13,
14]. In 2007, using periadventitial treatment of rodent AAA, our group published the first in vivo evidence that delivery of PGG to aneurysmal aorta inhibits elastin degradation and attenuates aneurysmal expansion[
15]. Lindholt et al. then showed that intraluminal infusion of PGG could impair AAA development in an elastase-induced aneurysm swine model[
16], and Schack et al. confirmed the beneficial effect of intraluminal PGG in rodents[
17]. In 2020, Simionescu et al. reported that PGG diffuses rapidly through arterial tissues and that a single 3-minute intraluminal delivery of PGG to the aneurysmal aorta was sufficient to reduce AAA expansion in a swine model[
18]. The use of PGG, delivered endovascularly, to slow the growth of AAA has now moved to first-in-human (FIH) clinical trials (
https://clinicaltrials.gov/ct2/show/NCT05133492).
Despite extensive preclinical experimentation, the molecular mechanisms of the action of PGG on the aneurysmal wall still need to be better understood. The studies described herein were undertaken to identify molecular interactions between PGG and individual components of the aneurysmal aorta, including extracellular matrix components, cytokines, and the intra-luminal thrombus. A short description of these target elements follows.
1.1. Extracellular Matrix (ECM) in the Normal Aorta
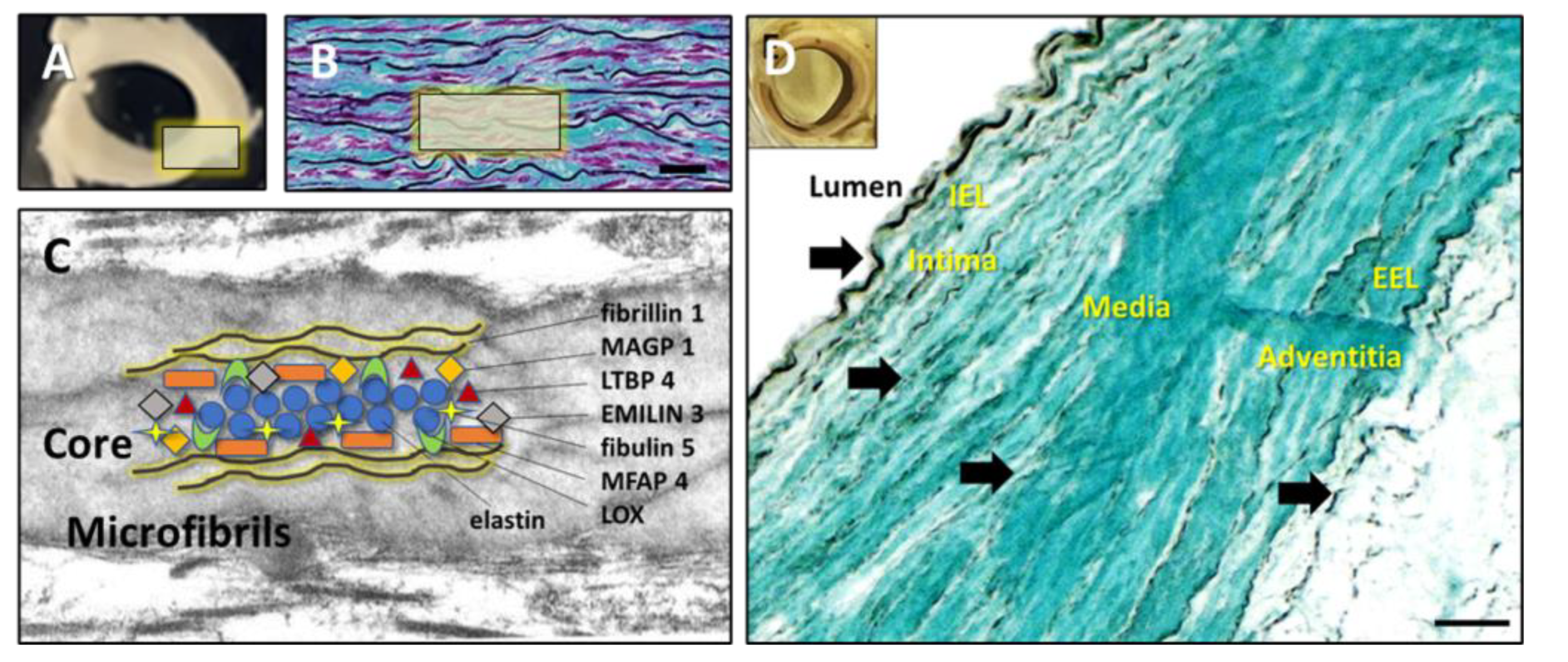
Large arteries comprise about 50% of elastic fibers, with the remainder comprising collagconsisting and cells (
Figure 1). Zooming into the ultrastructural and molecular composition, elastic fibers are complex structures composed of a dense core of amorphous, crosslinked elastin (ELN) and multiple elastin-associated microfibrillar (EAMF) proteins (
Figure 1). The basis of this complexity resides in the multi-step process of elastic fiber formation, which starts with the extra-cellular secretion of the tropoelastin monomers, which coacervate into spherical aggregates on fibrillin templates that are stabilized by proteoglycans and fibulins. For a more detailed review of vascular extracellular matrix organization see the excellent, reviews by Heinz, A, et al. [
19,
20]. The elastin is cross-linked by the action of lysyl oxidase (LOX) which remains wi,thin the fiber[
21]. The structure matures by acquiring other EAMF proteins such as microfibril-associated glycoprotein (MAGP), latent-transforming growth factor beta-binding protein (LTBP), microfibril-associated glycoprotein (MFAP) and elastin-microfbril-interface-located-protein (EMILIN) (
Figure 1) [
21,
22,
23]. When PGG is infused intraluminally into the abdominal aorta for 3 minutes or more, it diffuses rapidly through the tissue and binds to the internal elastic lamina[
18]. With saturating concentrations, PGG binds the medial elastic fibers, external elastic lamina, and continues to diffuse swiftly through the vessel wall reaching the adventitial fibers (
Figure 1). Due to the complex nature of the ECM in the aorta, we decided to investigate the molecular interactions of PGG with each of these components.
1.2. Aneurysmal aorta components
AAA is a chronic disorder characterized by the weakening and progressive dilatation of the aorta with an increasing risk of rupture. The pathogenesis of AAA is associated with cell death, localized inflammation, leucocyte infiltration, and degradation of the aortic matrix elastin and collagen fibers by matrix metalloproteinases (MMPs). Many cytokines are present in AAA throughout its development and progression[
24,
25]. Among these, interleukin 1 (IL-1), IL-6[
26], IL-8, IL-10, TNF-alpha, CCL-5, MCP 1 were found to be associated with experimentally induced AAA, and in human AAA tissue extracts[
27]. MMPs have been implicated in the pathogenesis of AAA for decades[
28], specifically focusing on MMP 2, 9 and 12. Inhibition of MMPs has been an obvious target in experimental animal models of AAA[
29]; however, in a recent clinical trial, aneurysm growth was not inhibited by systemic administration of doxycycline, an MMP inhibitor although interestingly use of systemic doxycycline did inhibit local MMP activity in the AAA wall[
30].
1.3. Intraluminal Thrombus in AAA
About 70-80% of AAA patients have intraluminal thrombus (ILT), which generally is eccentric and does not impede blood flow[
31,
32]. Typically, the ILT is around 8-9 cm long, has a volume of about 90 ml, and is positioned in the anterior portion of the AAA sac[
33,
34]. The ILT has been considered a bystander to AAA. Recent research, however, highlighted the role of ILT in AAA development and in the progression of endoleaks after endovascular repair of abdominal aneurysms (EVAR)[
31,
33]. Moreover, drug diffusion through the ILT becomes important when agents targeting the aneurysmal wall are delivered intraluminally. The transport of macromolecules through ILT is likely occurring from the lumen towards the abluminal side through a system of channels or canaliculi[
34]. Most channels are between 5-36 µm
2 in area, large enough for macromolecules to diffuse through[
34]. Toluidine Blue diffusion measurements through human ILT revealed a 0.91+/-0.54 mm
4/N permeability, three orders of magnitude higher than that of the aortic wall or that of articular cartilage[
34]. To our knowledge, research has yet to be conducted to assess the diffusivity of PGG through ILT and the molecular interactions of PGG with EAMF, MMPs, and cytokines involved in AAA.
Our working hypothesis is that PGG can bind to some of the main components of the vascular wall, including elastin, EAMF, proteoglycans, and collagen, thus accounting for the observed tissue stabilization effect of PGG. We also hypothesize that PGG could bind to MMPs and cytokines in the aneurysmal aorta as a secondary means to impact on AAA development and progression. Finally, we sought to understand whether patient ILT poses a barrier to PGG diffusion at physiologic pressures. To understand these interactions, we performed three complementary studies.
First, we exposed purified human aortic elastin to a solution containing PGG for 3 minutes, a duration sufficient for tissue saturation based on bench testing and preclinical studies[
18]. We then evaluated the effects of PGG on elastin structure by LC-MS/MS and peptide fingerprinting after elastase digestion compared to appropriate controls.
In the second experiment, we used molecular docking software to predict the most probable binding sites and calculate the affinity of PGG to the elastin protein molecule, to naturally occurring EAMF proteins, collagens, and proteoglycans, as well as to other major elements found in the AAA environment such as MMPs and cytokines.
Finally, we evaluated PGG binding to fibrin by molecular simulations and performing PGG diffusion studies in pure fibrin gels and human ILT samples.
2. Materials and Methods
Purified human aortic elastin (SH476) and human neutrophil elastase (HNE) (SE563) were obtained from Elastin Products Company. A formulation of 3.0 mg/mL PGG concentration in buffer and contrast material (iopamidol) consistent with that used in a recent human FIH clinical trial as well as vehicle controls (buffer, contrast material, no PGG) was provided by Nectero Medical Inc., Tempe, AZ. The PGG solution was neutralized with sodium bicarbonate (6.6 ml of a 4.2% solution added to 100 ml PGG or vehicle solution) before use. All other materials and chemicals were of the highest purity available.
2.1. Treatment of pure elastin with PGG and sample preparation for peptide fingerprinting
Human aortic elastin samples, at 2 mg each, were randomly divided into three Groups (n=3 per group). Group 1 was treated with the vehicle control buffer with contrast material, Group 2 was treated with a vehicle control buffer without contrast material, and Group 3 was treated with buffered, neutralized 3.0 mg/mL PGG in vehicle with contrast material. Dry elastin samples were transferred to 2 ml tubes, and 1 ml solution was added to the elastin samples; the tubes were vortexed twice for 5 seconds and placed in a water bath at 37oC for 3 minutes. The samples were centrifuged at 12000 rpm for 5 minutes, and the supernatant was aspirated with a transfer pipette. The samples were rinsed twice with 1 ml saline, vortexed, centrifuged again, and the supernatant aspirated. For enzymatic digestion, human neutrophil elastase (HNE) was prepared at a concentration of 43U/ml in PBS with 5 mM CaCl2. One (1) ml HNE solution was added to each elastin sample which were then stirred with a Teflon-coated micro magnetic stirrer at 37oC for 20 hours. To finalize digestion, more enzyme (0.5 ml of the same HNE solution) was added to each sample followed by at 37 oC for another 20 hours. Samples were centrifuged at 12000 rpm for 5 minutes and filtered through a 10k MW filter (Centricon Ultracel YM 10, 0.5 ml). The insoluble fibers, undigested elastin, and elastase remained on the filter while the filtrate contained the digested elastin peptides. Peptides were quantified on a Nanodrop machine at 205 nm and all samples were stored at -20 oC. Most samples contained around 1.6-1.8 mg peptides/ml (data not shown). Samples were transported frozen to the MUAL lab on the Clemson campus for analysis.
2.2. Peptide fingerprinting
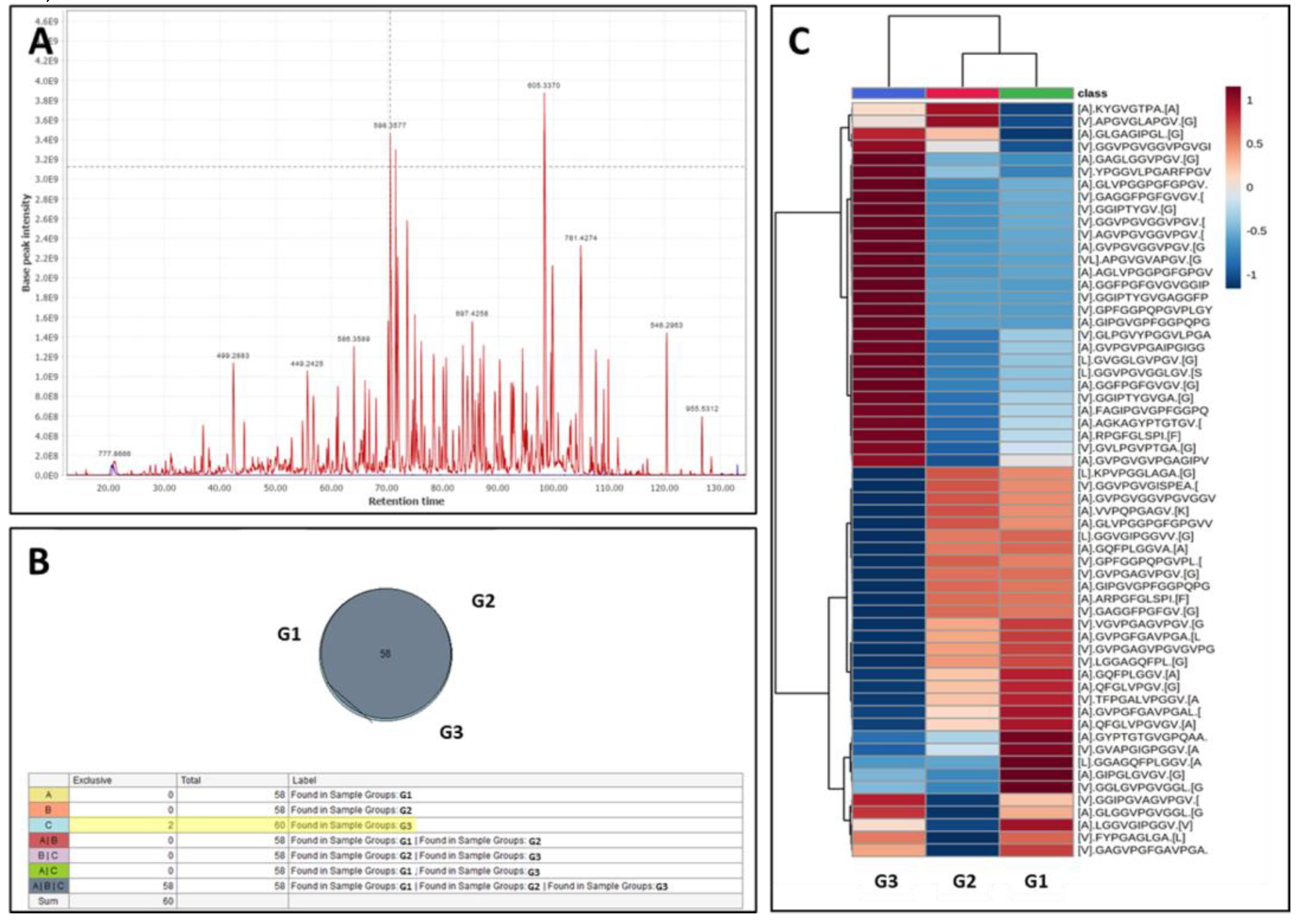
Elastase-digested samples were prepared as described above and analyzed using tandem liquid chromatography mass spectrometry (LC-MS/MS). For better separation and resolution of peptide fragments, samples were first run through a C-18 spin column (Pierce 89870). Samples were then dried and reconstituted in 0.1 % formic acid. Retention time calibrants were added (Pierce 88321) and samples were analyzed on a nano-LC-MS/MS system for peptide profiling using a 120 min gradient. Solvent A was 0.1% Formic Acid, and Solvent B was 80% Acetonitrile + 0.1% Formic Acid. The column was PepMapRSLC C18 (2um, 100A, 75um, 50 cm). Injections were 1 µL with duplicate injections per sample. Blanks were injected between each sample. Peptide fingerprinting yielded accurate mass and retention time of elastin peptides, and relative abundance was calculated based on the areas of each peak using the canonical human elastin (ELN) amino acid sequence (Entrez GeneID2006, UniProt P15502). Data were processed using Proteome Discoverer 2.4 and Skyline software. A comparative analysis between the three groups was performed using the heat map function and volcano plots, looking at the number of peptides and the abundance of peptides in each group.
2.3. Molecular Docking
Docking is an in-silico method used to predict the binding modes of a ligand to a target. This is a standard drug discovery pipeline for FDA-approved drugs, enzyme inhibitors, and others. The ligand is typically a small molecule; the target is a protein, an enzyme or its catalytic site, DNA sequences, etc. Molecular docking in-silico can help reduce in vitro screening efforts. The main steps in docking include 1) preparing the ligand, 2) preparing the target, 3) defining the 3D reaction volume (box) within the target, 4) activating the molecular simulation software, and 5) analyzing results. There are multiple software packages available, with AutodockVina being the most popular. Most of these require additional software packages and Unix coding and a requirement for advanced knowledge. Recently, however, web interfaces incorporating all the needed software into easy-to-use platforms have been developed. We utilized SeamDock, developed by the University of Paris, which integrates all required software into a user-friendly cloud platform[
35].
The PGG structure was downloaded from Pubchem (CID 65238) in SDF format. The target proteins were obtained as pdb files from the RCSB PDB data bank (RCSB.org), UniProt (uniport.org) or AlphaFold (alphafold.ebi.ac.uk)[
36].
Table 1 summarizes the target proteins and their ID codes used in this docking study. To perform the docking, the SeamDock program was launched (
https://bioserv.rpbs.univ-paris-diderot.fr/services/SeamDock/), the ligand (PGG) sdf structure and the target protein (receptor) pdb file were uploaded into the appropriate sites, and, after the software had prepared the two reactants, the interaction box coordinates were set to cover the catalytic sites for enzymes, or the entire protein molecule for most proteins, except elastin. Since elastin protein is a very large structure (786 amino acids), we performed 6 separate docking simulations using different ligand-target interaction boxes, thus systematically covering the entire molecule. The docking parameters were set at Vina software, mode 2, energy range 12, exhaustiveness 8, and the docking program was launched. After the docking process has finalized (10-20 minutes per protein), affinity values or calculated binding energy were displayed as (-) kcal/mol. If binding energy appeared as a positive value, it meant that binding of PGG to the target was energy-consuming, pointing to low affinity. Conversely, if binding energy was negative, PGG bound spontaneously to the target without energy consumption, releasing energy while binding. Therefore, the lower the binding energy of the docked PGG, the stronger the interactions with the target protein. In our studies, all PGG-target protein interactions had negative values (see Results). In addition, the software displayed the number and location of hydrophobic interactions and hydrogen bonds for each target-bound PGG simulation.
2.4. PGG binding patterns analysis in fresh aorta
To mimic AAA, fresh, cannulated porcine abdominal aorta segments were treated endoluminally with a mixture of elastase (10 U/mL) and collagenase (50 U/mL) in 100 mM Tris-HCl buffer and 1 mM CaCl2 in saline (pH 8) by infusion through the lumen, followed by capping the 2 Luer adapters. After incubation for 5 minutes, the arteries were rinsed in saline, and then treated by endoluminal infusion with a clinical formulation containing neutralized 3.0 mg/mL PGG solution for 3 minutes, to mimic the clinical application, followed by a saline rinse, as previously described.[
37] Tissue samples were then stained
en bloc with FeCl
3 for 5 minutes, rinsed in saline, and photographed. Cryosections at 5 µm thickness were counterstained with Light Green and imaged. Tissue-bound PGG appeared as black deposits on a green background on histology.
2.5. Diffusion of PGG through pure fibrin gels and intra-luminal thrombus (ILT)
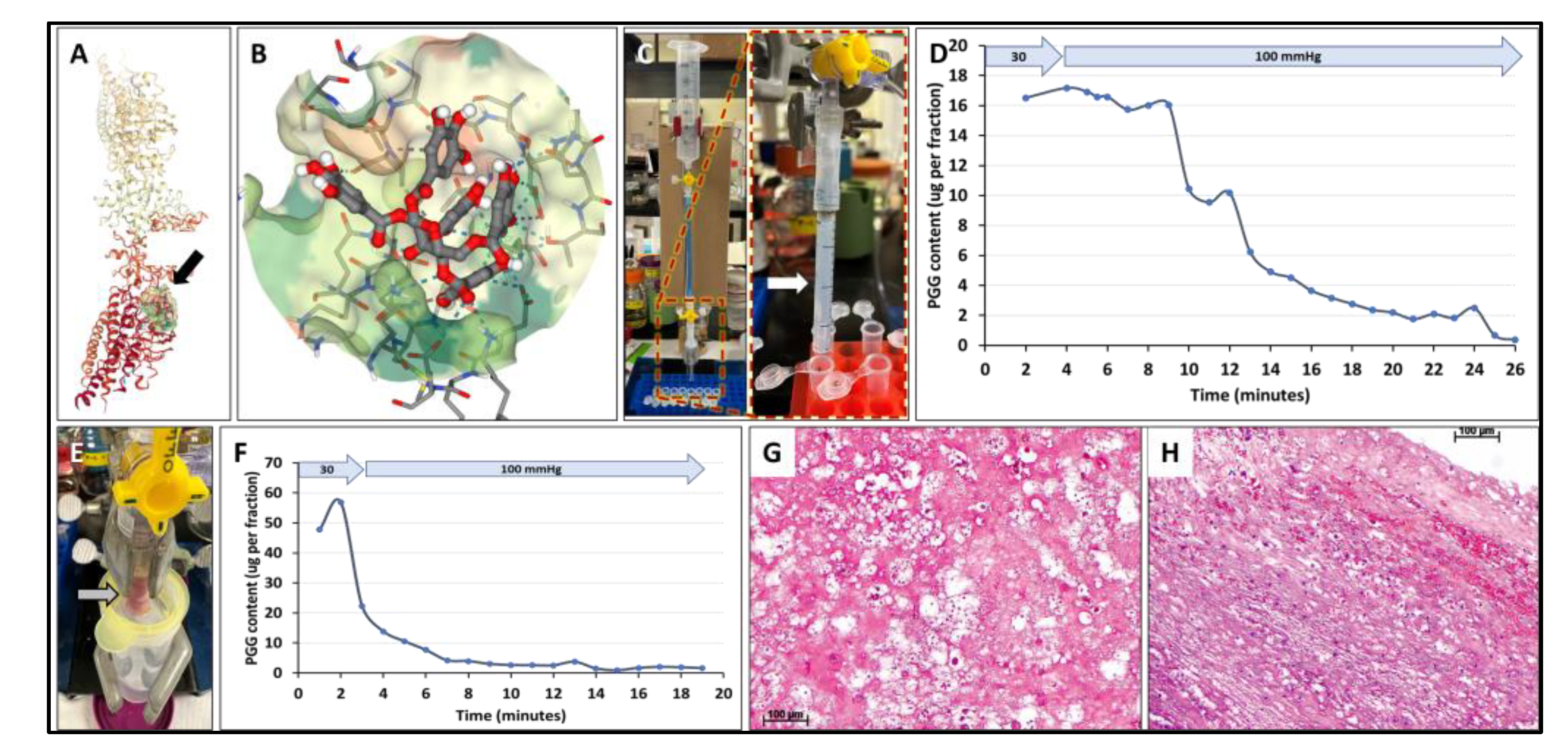
The experimental setup consisted of 5 mL borosilicate glass pipettes (VWR #93000-696), cut to 7 cm length using a glass tubing cutter, connected to a 3-way stopcock via a barbed Luer adaptor and a segment of 2 cm long silicone tubing. A 60 ml syringe without the plunger served as a reservoir filled with saline. This was connected to the 3-way stopcock via a 25 cm long vertical tube (for the 30-mmHg experiment) and a 120 cm long vertical tube (for the 100-mmHg application) (
Figure 6).
The fibrin gels were prepared by mixing bovine plasma fibrinogen and bovine plasma thrombin at 4mg/mL and 0.8U/mL final concentrations, respectively, in 20 mM HEPES buffer, 150 mM NaCl, 5 mM CaCl
2. The mixture was drawn into the cut glass pipettes, the tube ends were sealed with parafilm, placed vertically in a stand, and allowed to clot at 37
oC for 30 minutes. The parafilm was removed from the glass tubes just before utilization. In pilot studies, migration of red food dye (FD&C Red #40, 50 µL) through the 5 cm long gel column was observed visually by taking time-stamped photographs every 3-4 minutes. The dye diffused at a rate of around 1.25 mm/min at 30 mmHg and at 10 mm/min at 100 mmHg. The 50 µL volume applied was calculated from a typical situation where 25 mL of PGG is applied to an AAA of 4.5 cm diameter, covered 70% with a thrombus. PGG solution (50 µl) spiked with red dye exhibited similar diffusion characteristics (not shown). To test PGG diffusion through pure fibrin gels, 50 µl of PGG solution was loaded on top of the fibrin gel, pressure was applied at 30 mmHg for 3 minutes, during which 2 fractions of 100 µL each were collected, then pressure was increased to 100 mmHg and 100 µL fractions collected every minute for up to 25 minutes. The 3-minute, 30-mmHg application mimics the FIH clinical scenario where pressure inside the AAA was reduced to approximately 30-mmHg during infusion of PGG intraluminally for 3 minutes, followed by exposure of the aneurysmal sac to normal blood pressure of approximately 100 mmHg. PGG content was assayed in each fraction using the Folin-Ciocalteu reagent as described earlier [
15,
37].
De-identified leftover ILT tissue was obtained from a consenting AAA patient during an open surgical repair procedure performed at PRISMA Health Upstate, Vascular Surgery, Greenville, SC. The freshly collected ILT tissue was made available as part of a research agreement between Clemson University and the PRISMA Health Biorepository tissue bank. The patient signed a written informed consent to participate in the PRISMA Health Biorepository. The Biorepository is an IRB approved human tissue bank which provides deidentified human tissue specimens for a wide variety of research nationally and internationally. Patients are consented to participate in the Biorepository separately from their procedural consent. As a participant, the patient was informed that the tissue being collected was routinely disposed of unless participating in the Biorepository. The tissue was of no value to the patient, its collection did not pose any additional medical or privacy risks, and that by participating in the Biorepository, the tissue would be used for further research. This was all reviewed and confirmed before the tissue was obtained.
The Biorepository obtained the tissue samples from the OR, de-identified the sample, ensured the consent process was complete and then provided us in a blinded fashion, access to specimens. ILT was collected into sterile saline and after deidentification was immediately transported to the Clemson lab. The authors of this study did not have access to information that could identify individual participants during or after data collection. Tissue collection for this study followed the recommendations outlined in the FDA OMB#0910-0582 “Guidance On Informed Consent For In Vitro Diagnostic Device Studies Using Leftover Human Specimens That Are Not Individually Identifiable”.
ILT core samples, obtained within 2 hours of collection, were used for PGG diffusion studies using a modified perfusion setup (
Figure 6). PGG solution was applied on top of the thrombus, the system was connected to the perfusion setup and 30 mmHg applied for 3 minutes to mimic the clinical PGG application, followed by 100 mmHg physiologic pressure. Elution fractions were collected manually at the distal end of the ILT and analyzed for PGG content (
Figure 6). ILT samples were also fixed in formalin, processed for histology, and stained by H&E for overall morphology of the thrombus.
3. Results
3.1. Peptide fingerprinting
In the first study, we treated pure human aortic elastin with PGG and analyzed the type and abundance of elastin peptides. Results were compared to elastin samples treated with the vehicle control (buffer and contrast material) and the vehicle without contrast material (to assess the effects of contrast material). Overall, the digestion of elastin treated with vehicle generated 58 peptides, covering about 50% of the elastin sequence. By comparison, PGG-treated elastin generated 60 peptides, of which 2 new peptides had appeared in addition to the 58 peptides characteristic of our digestion conditions (
Figure 2.1B). These peptides were: [A].GIPGVGPFGGPQPGVPLGYPI.[K] localized at amino acids 188-208 and [V].GPFGGPQPGVPLGYPI.[K] at amino acids 192-208 in the elastin sequence (
Figure 3D). When comparing the abundance of elastin peptides in the three groups, G1 and G2 controls were more similar, suggesting that contrast material does not inhibit elastase and only slightly changes peptide generation after elastase. G3 samples (PGG-treated elastin) were distinct from G1 and G2 controls, as evidenced by the heat map representation (
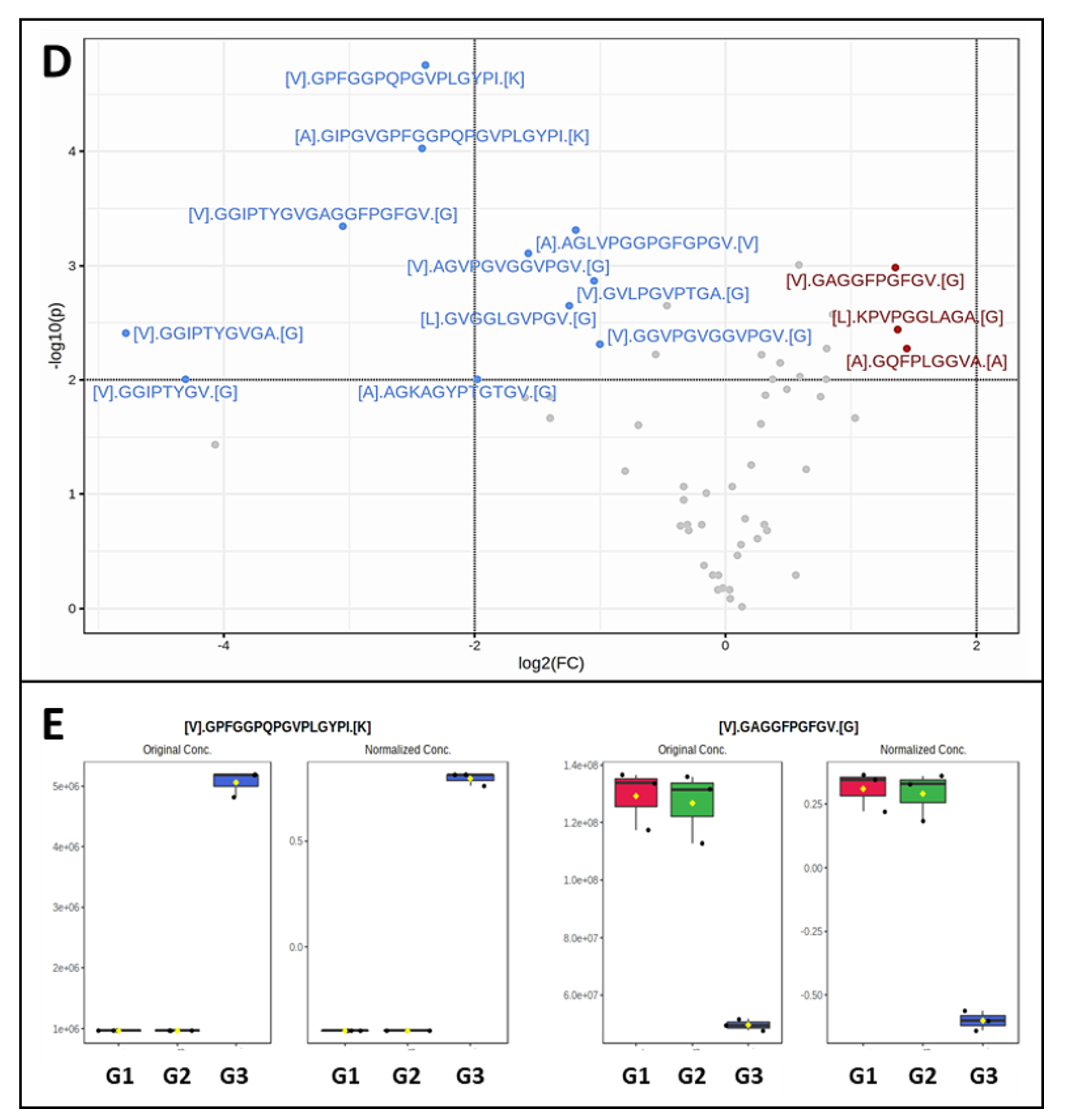
Figure 2.1C), where G3 samples exhibited significantly altered abundance. About half of the peptides obtained from PGG-treated elastin were generated at higher abundance and half at lower abundance. Comparing individual peptides using volcano plots, 3 peptides appeared in significantly lower abundance while 11 peptides appeared as significantly higher abundance (
Figure 2.2D, E) in the PGG-treated elastin. The three peptides in lower abundance were [L].KPVPGGLAGA.[G] (63-74), [V].GAGGFPGFGV.[G] (398-410) and [A].GQFPLGGVA.[A] (750-761). These peptides were also mapped to the amino acid composition of the canonical human elastin (
Figure 3D).
Figure 2.
Peptide fingerprinting of human aortic elastin. A) Representative chromatogram of elastin peptides obtained by elastase digestion. B) Comparison of elastin peptides generated by HNE elastase in the buffer control group (G1), vehicle control group (G2) and PGG-treated group (G3). Fifty-eight peptides were common to the three groups, with only 2 peptides appearing as specific to G3. C) Heat map of the abundance of elastin peptides across G3 (PGG), G2 (vehicle control), and G1 (control) groups. Dark red, highest abundance, light blue, lowest abundance.
Figure 2.
Peptide fingerprinting of human aortic elastin. A) Representative chromatogram of elastin peptides obtained by elastase digestion. B) Comparison of elastin peptides generated by HNE elastase in the buffer control group (G1), vehicle control group (G2) and PGG-treated group (G3). Fifty-eight peptides were common to the three groups, with only 2 peptides appearing as specific to G3. C) Heat map of the abundance of elastin peptides across G3 (PGG), G2 (vehicle control), and G1 (control) groups. Dark red, highest abundance, light blue, lowest abundance.
Figure 2.
Cont. D) Volcano plot (FC>2; P<0.01) comparing specific elastin peptides in G3 (PGG treated elastin) vs. G2 (vehicle control). Red points – lower concentrations of peptides in G3, blue points – higher concentration of peptides in G3. E) Two representative examples of alterations in individual peptide abundance after PGG treatment. Left, higher abundance of peptide [V].GPFGGPQPGVPLGYPI.[K] in PGG-treated elastin (G3), right, lower abundance of [V].GAGGFPGFGV.[G] elastin peptide after PGG treatment (G3).
Figure 2.
Cont. D) Volcano plot (FC>2; P<0.01) comparing specific elastin peptides in G3 (PGG treated elastin) vs. G2 (vehicle control). Red points – lower concentrations of peptides in G3, blue points – higher concentration of peptides in G3. E) Two representative examples of alterations in individual peptide abundance after PGG treatment. Left, higher abundance of peptide [V].GPFGGPQPGVPLGYPI.[K] in PGG-treated elastin (G3), right, lower abundance of [V].GAGGFPGFGV.[G] elastin peptide after PGG treatment (G3).
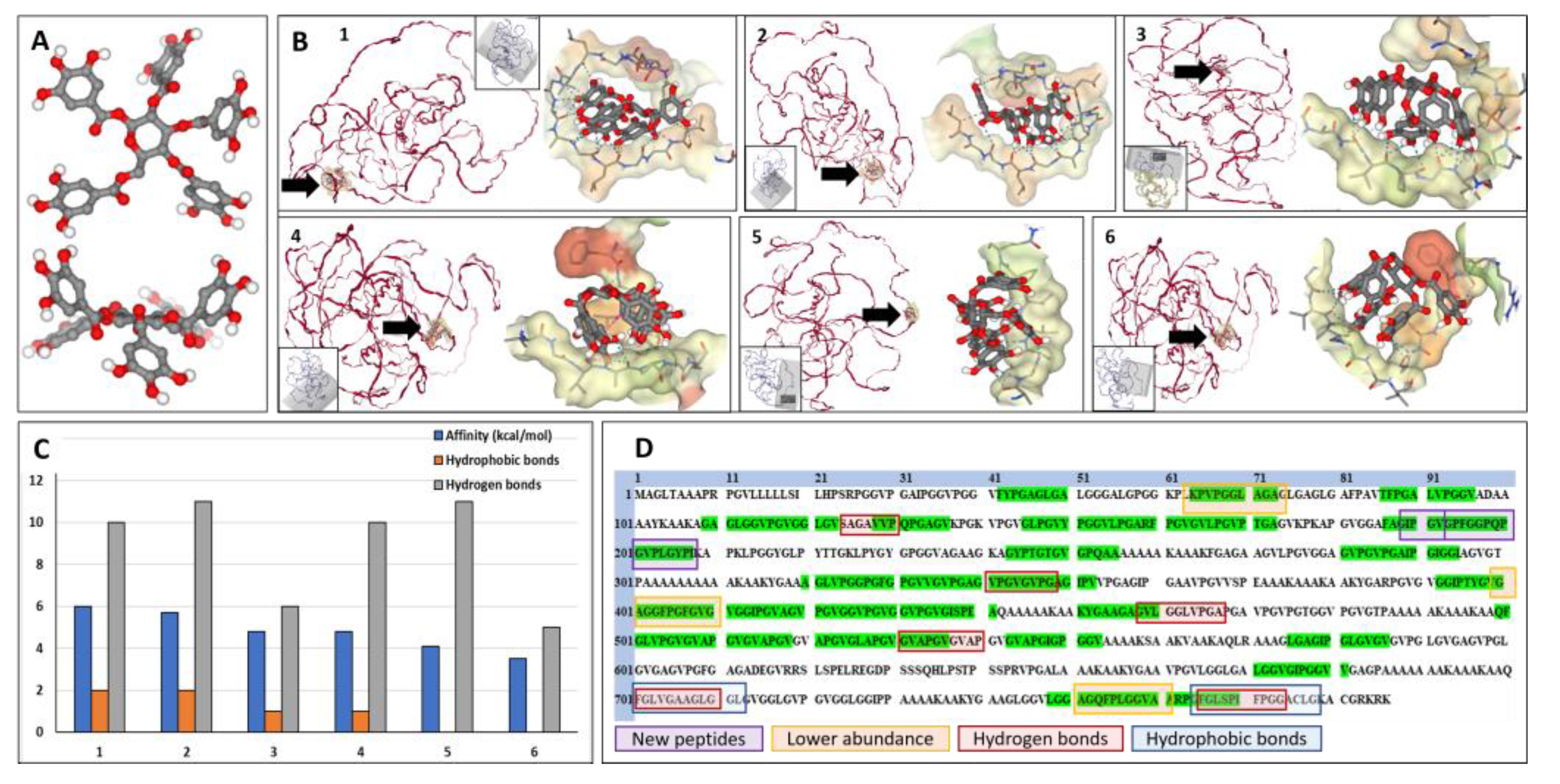
3.2. Molecular docking of PGG onto elastin
To better understand the interactions between PGG and elastin, we performed docking simulations using SeamDock as a web interface and the canonical human elastin structure. In water, PGG appears as a relatively planar molecule with the five galloyl residues pointing away from the center of the molecule and away from each other, similar to a “fan blade” (
Figure 3A). Elastin appears as a random coil structure with very few alpha-helices in its structure. Molecular docking of PGG onto elastin in different areas of the molecule (
Figure 3B), covering the entire elastin structure, showed that PGG binds preferentially to 6-7 areas with different affinities, as shown by the binding energy ranging from -3.5 to -6 kcal/mol and sustained by 0 to 2 hydrophobic bonds and 5 to 11 hydrogen bonds. These bonds were also mapped to the amino acid composition of the canonical human elastin (
Figure 3D). Hydrophobic bonds between PGG and elastin appeared mostly towards the C-terminal of the elastin molecule (amino acids 700-712 and 764-778). Hydrogen bonds between PGG and elastin were localized in the same C-terminal area (701-710, 766-744) and throughout the elastin molecule (124-130, 341-348, 458-467, 531-540). Analysis of the docking images showed that PGG could acquire multiple flexible configurations to fit within the binding “pocket,” either spread out with all galloyl residues establishing bonds in multiple directions or a “clenched fist” configuration, with 3-4 galloyl residues concentrated in one single plane.
Figure 3D depicts the salient features of PGG binding to human elastin resulting from overlapping the peptide fingerprinting data with the docking simulations.
Figure 3.
Docking of PGG onto elastin and localization of molecular interactions. A) 3-dimensional structure of the PGG molecule, top view, and side view. Red balls depict the oxygen moieties. B) Docking results from the study on PGG interactions with elastin (ELN). Interactions were mapped in 6 different ELN protein areas (labeled 1 to 6) by changing the interaction box coordinates (gray box in each insert), covering the entire protein structure. In each figure (1 to 6), the ELN protein is shown in red on the left, and a black arrow highlights the PGG binding site; on the right, a close-up of the PGG molecule interacting with the elastin chain. Note that PGG can acquire multiple flexible configurations. C) Docking results show affinity values (binding energy) of PGG binding to the different areas in the ELN protein, expressed as (-) kcal/mol (blue bars), the number of hydrophobic bonds (orange bars), and hydrogen bonds (gray bars). D) Aminoacid structure of the canonical human ELN protein and mapping of salient PGG interactions. Green highlights depict the elastase-generated peptide sequences as determined by LC/MS. Purple squares depict the 2 new elastase-generated peptides that appeared only after PGG treatment, and the orange squares, the 3 peptides that exhibited lower abundance after PGG treatment of elastin. Overlapping the peptide fingerprinting data with the docking data reveals 6 areas of hydrogen bonding between PGG and elastin (red squares) and 2 main areas of hydrophobic bonding between PGG and ELN (blue squares).
Figure 3.
Docking of PGG onto elastin and localization of molecular interactions. A) 3-dimensional structure of the PGG molecule, top view, and side view. Red balls depict the oxygen moieties. B) Docking results from the study on PGG interactions with elastin (ELN). Interactions were mapped in 6 different ELN protein areas (labeled 1 to 6) by changing the interaction box coordinates (gray box in each insert), covering the entire protein structure. In each figure (1 to 6), the ELN protein is shown in red on the left, and a black arrow highlights the PGG binding site; on the right, a close-up of the PGG molecule interacting with the elastin chain. Note that PGG can acquire multiple flexible configurations. C) Docking results show affinity values (binding energy) of PGG binding to the different areas in the ELN protein, expressed as (-) kcal/mol (blue bars), the number of hydrophobic bonds (orange bars), and hydrogen bonds (gray bars). D) Aminoacid structure of the canonical human ELN protein and mapping of salient PGG interactions. Green highlights depict the elastase-generated peptide sequences as determined by LC/MS. Purple squares depict the 2 new elastase-generated peptides that appeared only after PGG treatment, and the orange squares, the 3 peptides that exhibited lower abundance after PGG treatment of elastin. Overlapping the peptide fingerprinting data with the docking data reveals 6 areas of hydrogen bonding between PGG and elastin (red squares) and 2 main areas of hydrophobic bonding between PGG and ELN (blue squares).
3.3. Molecular docking of PGG onto other normal vascular extracellular matrix proteins
To complement the data on elastin, we evaluated the potential of PGG to bind to the major fibrillar components of the elastic fiber and collagens using docking simulations.
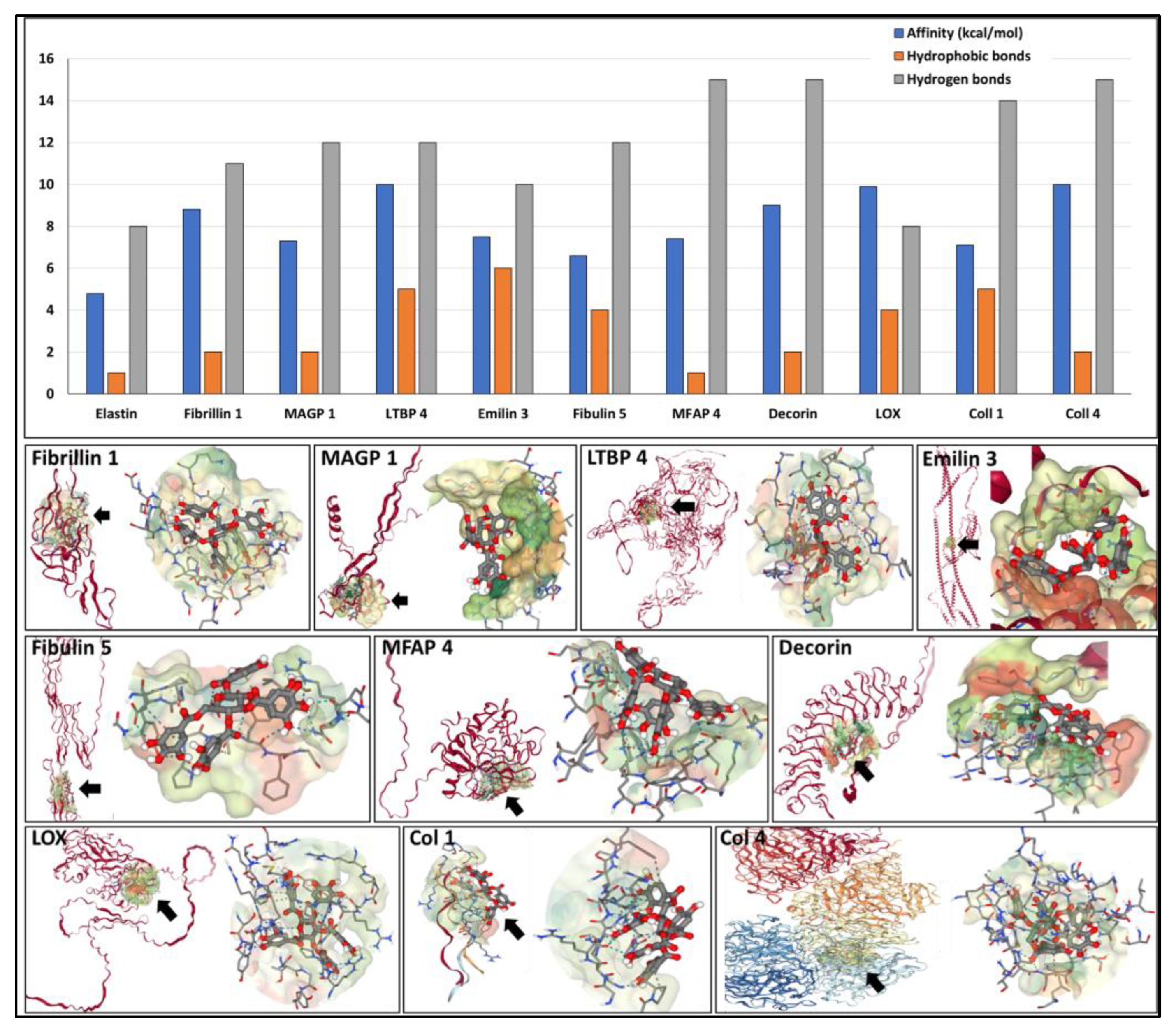
Table 1 provides a summary of the proteins analyzed and docking parameters. PGG exhibited a high affinity towards all proteins tested, including fibrillin 1 (binding energy of -9 kcal/mol), which makes up most of the elastin-associated microfibrillar “sheath” covering elastic fibers. A very high affinity of PGG was found towards MFAP 4 (-15 kcal/mol), followed by fibulin 5, LTBP 4, and MAGP 1 (-12 kcal/mol), followed by EMILIN 3 (-10 kcal/mol) and LOX (-8 kcal/mol). These high affinities were sustained by multiple hydrophobic bonds and hydrogen bonds (
Figure 4). PGG was also predicted to bind strongly to the collagen type I triple-helical repetitive sequence and to the collagen type IV NC1 domain (-7 and -10 kcal/mol, respectively) and to the decorin proteoglycan core protein (-9 kcal/mol), which is closely associated with collagen fibers. These simulations show that PGG has the ability to bind to single protein chains (intramolecular) or to “pockets” created by multiple chains (intermolecular).
Figure 4.
(top) Affinity of PGG to the different components of the aortic wall depicted as binding energy values expressed as (-) kcal/mol (blue bars), the number of hydrophobic bonds (orange bars), and hydrogen bonds (gray bars) established between PGG and the proteins. (bottom) Docking of PGG onto the major components of the aortic wall. The target protein (ribbon format) is shown in red on the left in each figure. A black arrow points to the most favorable calculated PGG binding site; on the right, a close-up of the PGG molecule interacting with the elastin chain within the “pocket.” Note that PGG can acquire multiple flexible configurations.
Figure 4.
(top) Affinity of PGG to the different components of the aortic wall depicted as binding energy values expressed as (-) kcal/mol (blue bars), the number of hydrophobic bonds (orange bars), and hydrogen bonds (gray bars) established between PGG and the proteins. (bottom) Docking of PGG onto the major components of the aortic wall. The target protein (ribbon format) is shown in red on the left in each figure. A black arrow points to the most favorable calculated PGG binding site; on the right, a close-up of the PGG molecule interacting with the elastin chain within the “pocket.” Note that PGG can acquire multiple flexible configurations.
3.4. Molecular docking of PGG onto proteins present in the aneurysmal aorta
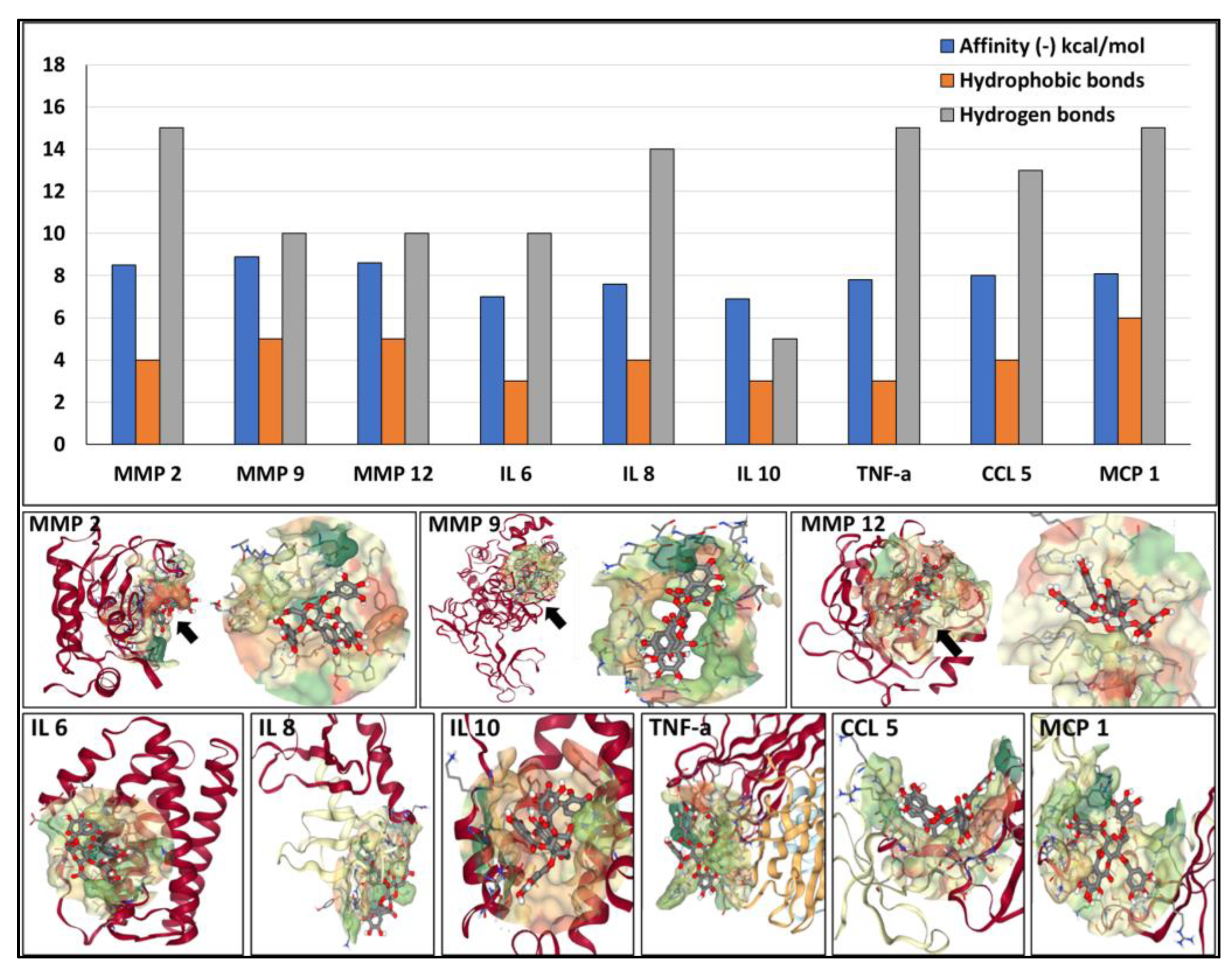
Next, we investigated the potential of PGG to bind to several components associated with the development and progression of AAA, namely active MMPs and cytokines, using docking simulations. PGG was predicted to bind with high affinity to the catalytic site of MMP 2, MMP 9, and MMP 12 (-8.2, -8.5 and -8.3 kcal/mol, respectively), sustained by multiple hydrophobic bonds and hydrogen bonds (
Figure 5). PGG also exhibited significant affinity towards IL 6, IL 8, IL 10, TNF-a, CCL 5, and MCP 1 (-7 to -8 kcal/mol) with the ability to bind to single protein chains or to pockets created by multiple chains (
Figure 5).
Figure 5.
Affinity of PGG to MMPs and cytokines involved with AAA development and progression. (Top) Binding energy values are expressed as (-) kcal/mol, blue bars, the number of hydrophobic bonds (orange bars), and hydrogen bonds (gray bars) established between PGG and the proteins. (Bottom) Docking of PGG onto the MMPs and cytokines.
Figure 5.
Affinity of PGG to MMPs and cytokines involved with AAA development and progression. (Top) Binding energy values are expressed as (-) kcal/mol, blue bars, the number of hydrophobic bonds (orange bars), and hydrogen bonds (gray bars) established between PGG and the proteins. (Bottom) Docking of PGG onto the MMPs and cytokines.
3.5. Diffusion of PGG through fibrin gels and intra-luminal thrombus
SeamDock simulation of PGG binding to fibrin was performed as described above. PGG was predicted to bind to the D-dimer of crosslinked fibrin with high affinity (-8.5 kcal/mol), involving 3 hydrophobic bonds and 15 hydrogen bonds. Despite this potential for binding PGG was shown to diffuse relatively rapidly through 5 cm of pure fibrin gel columns at a perfusion pressure of 30 mmHg for 3 minutes followed by 100 mmHg. Transit occurred within 20 - 30 minutes with almost complete recovery in fractions collected in the outflow of the fibrin gel column (
Figure 6D). Subsequent experiments utilizing a diffusion chamber assessed whether the presence of patient intra-luminal thrombus (ILT) poses a barrier to PGG diffusion at physiologic pressures. When PGG was applied to a 4 cm long column of human intra-luminal thrombus it was found to diffuse rapidly through the thrombus under 30 mmHg, followed by rapid elution from the ILT by saline at 100 mmHg (
Figure 6F). Quantification of PGG in the ILT after the diffusion study showed that about 17-20% of applied PGG remained in the thrombus (data not shown). Histology analysis of the ILT sample showed the presence of multiple pores of various diameters (10-100 µm) (
Figure 6G and 6H).
Figure 6.
Interactions of PGG with fibrin and intraluminal thrombus. A) Docking simulation of PGG binding to fibrin (black arrow). B) Binding of PGG to a pocket in the fibrin molecule. C) Hydrostatic pressure setup for evaluating PGG diffusion through fibrin gels. Fibrin gels were cast in a small glass tube attached to a funnel placed at various heights to simulate 30 mmHg and 100 mmHg. Right, detailed image of the fibrin gel (white arrow). D) PGG diffusion through fibrin gels at 30 mmHg followed by 100 mmHg pressures. PGG content was assayed in each eluting fraction collected at the outflow. E) Hydrostatic pressure setup for evaluating PGG diffusion through intraluminal thrombus (arrow). F) PGG diffusion through intraluminal thrombus at 30 mmHg followed by 100 mmHg pressures. PGG content was assayed in each eluting fraction collected at the outflow. G-H) histological evaluation of the ILT. Hematoxylin and Eosin stain, bar, 100 µm.
Figure 6.
Interactions of PGG with fibrin and intraluminal thrombus. A) Docking simulation of PGG binding to fibrin (black arrow). B) Binding of PGG to a pocket in the fibrin molecule. C) Hydrostatic pressure setup for evaluating PGG diffusion through fibrin gels. Fibrin gels were cast in a small glass tube attached to a funnel placed at various heights to simulate 30 mmHg and 100 mmHg. Right, detailed image of the fibrin gel (white arrow). D) PGG diffusion through fibrin gels at 30 mmHg followed by 100 mmHg pressures. PGG content was assayed in each eluting fraction collected at the outflow. E) Hydrostatic pressure setup for evaluating PGG diffusion through intraluminal thrombus (arrow). F) PGG diffusion through intraluminal thrombus at 30 mmHg followed by 100 mmHg pressures. PGG content was assayed in each eluting fraction collected at the outflow. G-H) histological evaluation of the ILT. Hematoxylin and Eosin stain, bar, 100 µm.
4. Discussion
In our studies, PGG was predicted to bind to all normal vascular matrix components tested, such as elastin, elastin-associated microfibrils, collagens and proteoglycan core proteins. The binding affinities were in the same range as those reported before for other proteins. Binding of PGG to a variety of targets has been investigated before by molecular docking including using SeamDock[
38]. For example, molecular docking of PGG to two prostaglandin receptors was performed to understand its gastroprotective effects[
39]. PGG was docked to bcl-2 family of anti-apoptotic targets (Bcl-2, BCL-XL, Caspase 3, and Caspase 9) with binding energies of -8.6,-7,-7.5 and 4.4 kcal/mol respectively[
40]. Also, PGG was shown by molecular docking to interact with VEGF signaling molecules VEGF-A,VEGFR-2, PKC, RAF, MEK, ERK and AKT with binding affinity of -7.9,-8.3,-8.6, -3.7,10.1,-9 and -10.8 kcal/mol[
41]. Recently, PGG was found to bind to spike-receptor binding domain of SARS-CoV-2 by molecular docking, with a -8 kcal/mol binding energy[
42]. Furthermore, in our work, PGG was projected to bind to the active site of three major MMPs and to several cytokines, essential elements associated with the development and progression of AAA.
PGG binding to pure human elastin is expected to occur at several sites with high affinity. PGG binding to elastin altered its cleavage pattern by elastase, suggesting a mechanism of elastin stabilization, alluded to in earlier work[
13,
17,
18,
43,
44,
45,
46,
47,
48,
49,
50,
51]. Based on peptide fingerprinting and the molecular docking experiments, the C-terminal area of the elastin molecule around the amino acids 700-770 appears to be a region of high affinity binding for PGG, with significant effects on elastase digestion patterns.
PGG binding to matrix proteins typically spans 8-9 amino acids, sometimes involving intramolecular interactions with a single linear protein chain or intermolecular bonds within “pockets” created by multiple neighboring chains. With alpha-helical proteins such as collagen, PGG tends to bind on the outside of the structures in a “clenched fist” configuration, surrounding or “wrapping” the fibrils like a glove. PGG binding to matrix proteins occurs mainly through multiple hydrophobic and hydrogen bonds. Hydrophobic bonds are typically established with Val and Leu in elastin and with Glu, Phe, Pro, Gln and Arg in collagen, while hydrogen bonds are formed with Leu, Ala, and Gly in elastin and Gly, Gln, Arg, Pro in collagen. PGG established hydrophobic bonds with other proteins with Thr, Pro, Val, Tyr, Leu, Phe, His, Ala, Ile, and Trp.
In this study, PGG was also predicted to bind to catalytic sites of MMPs with high affinity, suggesting that PGG may act as a direct MMP inhibitor, as was suggested earlier[
15,
52,
53]. PGG was also predicted to react with some of the cytokines involved in AAA, potentially inactivating them. This would result in reducing the inflammatory burden typical of AAA, possibly reducing inflammatory cell recruitment, and potentially facilitating tissue healing. Overall, treatment of AAA tissues with PGG could potentially stabilize the existing matrix components and reduce their enzymatic degradation. As the peptide fragments generated during enzymatic degradation (matrix degradation products or “matrikines”) are typically pro-inflammatory, protection from further degradation, in turn, could further diminish inflammation. These actions might be responsible for the positive effects of PGG delivery to the aneurysmal aorta in rodents and swine. In these studies, delivery of PGG intra-luminally or peri-adventitially reduced aneurysmal expansion over time, sometimes leading to healing and reversal of the disease.
In our work, PGG was predicted to bind to crosslinked fibrin with high affinity but to diffuse rapidly through pure fibrin gels and human ILT under physiologic pressures, potentially because of the highly porous nature of the fibrin network. In our testing conditions, about 80% of PGG diffused freely through ILT, suggesting that ILT does not pose a barrier to PGG diffusion at physiologic pressures.
This study has several limitations: 1) One caveat of docking is that only one ligand molecule can be simulated simultaneously; multiple PGG molecules would likely bind to large molecules such as elastin or collagen. 3) The list of targets was not exhaustive but representative of our hypothesis. More studies are needed to cover the entire range of matrix components, enzymes, and multiple inflammatory cytokines to understand better the effects of PGG on AAA development and progression. It is also important to be able to validate results of molecular docking simulation with evidence from in vivo binding studies. 4) In the aneurysmal wall, there are probably multiple forms of degraded matrix proteins (elastin, EAMFs, collagen, etc.), cell debris, and infiltrated cells. We need to find out how PGG interacts with these components as well as with normal intact proteins. 5) we only had access to one ILT specimen; since ILT was reported to be heterogenous in nature and composition, more samples are needed to reach definitive conclusions.
5. Conclusions
PGG binds to the amorphous elastin core and the elastin-associated proteins, potentially protecting the entire macromolecular elastic fibril structure from degradation. Similarly, PGG binds to the collagen protein per se and to the proteoglycans that decorate and protect the fibers from degradation. PGG also binds to MMPs, and cytokines known to be present in AAA, potentially exerting anti-inflammatory activities. These observations could potentially explain the observed tissue stabilization effect of PGG against the action of MMPs, and the “healing” effects of PGG on AAA development and progression. Finally, the patient ILT does not pose a barrier to PGG diffusion at physiologic pressures, opening opportunities for a deeper understanding of the clinical potential of PGG as a non-surgical treatment of AAA.
Author Contributions
Conceptualization, D.S., W.C., Al.S., and Ag.S.; methodology, N.T., E.L., C.C., J.C.C.G., A.C., C.O.; formal analysis, K.N.; writing—original draft preparation, D.S.; writing—review and editing, W.C., A.S. All authors have read and agreed to the published version of the manuscript.
Funding
The present study was funded by Nectero Medical, Inc (to N.T., E.L, W.C, A.S., K.N.), the Harriet and the Jerry Dempsey Endowment (to D.S.)
Ethical and Informed Consent Statement
De-identified leftover ILT tissue was obtained from a consenting AAA patient during an open surgical repair procedure performed at PRISMA Health Upstate, Vascular Surgery, Greenville, SC. The freshly collected ILT tissue was made available as part of a research agreement between Clemson University and the PRISMA Health Biorepository tissue bank. The patient signed a written informed consent to participate in the PRISMA Health Biorepository. The Biorepository is an IRB approved human tissue bank which provides deidentified human tissue specimens for a wide variety of research nationally and internationally. Patients are consented to participate in the Biorepository separately from their procedural consent. As a participant, the patient was informed that the tissue being collected was routinely disposed of unless participating in the Biorepository. The tissue was of no value to the patient, its collection did not pose any additional medical or privacy risks, and that by participating in the Biorepository, the tissue would be used for further research. This was all reviewed and confirmed before the tissue was obtained. The Biorepository obtained the tissue samples from the OR, de-identified the sample, ensured the consent process was complete and then provided us in a blinded fashion, access to specimens. ILT was collected into sterile saline and after deidentification was immediately transported to the Clemson lab. The authors of this study did not have access to information that could identify individual participants during or after data collection. Tissue collection for this study followed the recommendations outlined in the FDA OMB#0910-0582 “Guidance On Informed Consent For In Vitro Diagnostic Device Studies Using Leftover Human Specimens That Are Not Individually Identifiable”.
Data Availability Statement
All relevant data are available within the manuscript.
Conflicts of Interest
Nectero Medical, Inc. funded most of this project; however, all performance data analysis and interpretation were performed by the authors, independent of input or interpretation from Nectero Medical, Inc. Dr Simionescu Dan is a co-inventor of Clemson University technology using 1,2,3,4,6-pentagalloylglucose for the treatment of abdominal aortic aneurysms (U.S. Patent and Trademark Office no. 8,435,553). Nectero Medical, Inc. licensed the PGG technology from Clemson University. C.C., A.C., C.O., A.S., have no conflicts of interest.
References
- Simionescu, N.; Simionescu, M. Galloylglucoses of low molecular weight as mordant in electron microscopy. II. The moiety and functional groups possibly involved in the mordanting effect. J Cell Biol 1976, 70, 622–633. [Google Scholar] [CrossRef] [PubMed]
- Simionescu, N.; Simionescu, M. Galloylglucoses of low molecular weight as mordant in electron microscopy. I. Procedure, and evidence for mordanting effect. J Cell Biol 1976, 70, 608–621. [Google Scholar] [CrossRef]
- Kajikawa, K.; Yamaguchi, T.; Katsuda, S.; Miwa, A. An improved electron stain for elastic fibers using tannic acid. J Electron Microsc (Tokyo) 1975, 24, 287–289. [Google Scholar]
- Cotta-Pereira, G.; Rodrigo, F.G.; David-Ferreira, J.F. The use of tannic acid-glutaraldehyde in the study of elastic and elastic-related fibers. Stain Technol 1976, 51, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, T.; Hamada, T. Elastotic material and elastic fibers in aged skin: an ultrastructural study with conventional and tannic acid stain. Acta Derm Venereol 1981, 61, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Isenburg, J.C.; Simionescu, D.T.; Vyavahare, N.R. Elastin stabilization in cardiovascular implants: improved resistance to enzymatic degradation by treatment with tannic acid. Biomaterials 2004, 25, 3293–3302. [Google Scholar] [CrossRef] [PubMed]
- Isenburg, J.C.; Simionescu, D.T.; Vyavahare, N.R. Tannic acid treatment enhances biostability and reduces calcification of glutaraldehyde fixed aortic wall. Biomaterials 2005, 26, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Isenburg, J.C.; Karamchandani, N.V.; Simionescu, D.T.; Vyavahare, N.R. Structural requirements for stabilization of vascular elastin by polyphenolic tannins. Biomaterials 2006, 27, 3645–3651. [Google Scholar] [CrossRef]
- Tedder, M.E.; Simionescu, A.; Chen, J.; Liao, J.; Simionescu, D.T. Assembly and testing of stem cell-seeded layered collagen constructs for heart valve tissue engineering. Tissue Eng Part A 2011, 17, 25–36. [Google Scholar] [CrossRef]
- Tedder, M.E.; Liao, J.; Weed, B.; Stabler, C.; Zhang, H.; Simionescu, A.; Simionescu, D.T. Stabilized collagen scaffolds for heart valve tissue engineering. Tissue Eng Part A 2009, 15, 1257–1268. [Google Scholar] [CrossRef]
- Chuang, T.H.; Stabler, C.; Simionescu, A.; Simionescu, D.T. Polyphenol-stabilized tubular elastin scaffolds for tissue engineered vascular grafts. Tissue Eng Part A 2009, 15, 2837–2851. [Google Scholar] [CrossRef] [PubMed]
- Sierad, L.N.; Simionescu, A.; Albers, C.; Chen, J.; Maivelett, J.; Tedder, M.E.; Liao, J.; Simionescu, D.T. Design and Testing of a Pulsatile Conditioning System for Dynamic Endothelialization of Polyphenol-Stabilized Tissue Engineered Heart Valves. Cardiovasc Eng Technol 2010, 1, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, S.S.; Simionescu, D.T.; Goergen, C.J.; Hoyt, K.; Sirsi, S.; Finol, E.A. Pentagalloyl Glucose and Its Functional Role in Vascular Health: Biomechanics and Drug-Delivery Characteristics. Ann Biomed Eng 2019, 47, 39–59. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, S.S.; Piskin, S.; Pillalamarri, N.R.; Romero, G.; Escobar, G.P.; Sprague, E.; Finol, E.A. Biomechanical Restoration Potential of Pentagalloyl Glucose after Arterial Extracellular Matrix Degeneration. Bioengineering (Basel) 2019, 6. [Google Scholar] [CrossRef]
- Isenburg, J.C.; Simionescu, D.T.; Starcher, B.C.; Vyavahare, N.R. Elastin stabilization for treatment of abdominal aortic aneurysms. Circulation 2007, 115, 1729–1737. [Google Scholar] [CrossRef] [PubMed]
- Kloster, B.O.; Lund, L.; Lindholt, J.S. Inhibition of early AAA formation by aortic intraluminal pentagalloyl glucose (PGG) infusion in a novel porcine AAA model. Ann Med Surg (Lond) 2016, 7, 65–70. [Google Scholar] [CrossRef]
- Schack, A.S.; Stubbe, J.; Steffensen, L.B.; Mahmoud, H.; Laursen, M.S.; Lindholt, J.S. Intraluminal infusion of Penta-Galloyl Glucose reduces abdominal aortic aneurysm development in the elastase rat model. PLoS One 2020, 15, e0234409. [Google Scholar] [CrossRef]
- Simionescu, D.; Casco, M.; Turner, J.; Rierson, N.; Yue, J.; Ning, K. Chemical stabilization of the extracellular matrix attenuates growth of experimentally induced abdominal aorta aneurysms in a large animal model. JVS Vasc Sci 2020, 1, 69–80. [Google Scholar] [CrossRef]
- Heinz, A. Elastases and elastokines: elastin degradation and its significance in health and disease. Crit Rev Biochem Mol Biol 2020, 55, 252–273. [Google Scholar] [CrossRef]
- Heinz, A.; Schrader, C.U.; Baud, S.; Keeley, F.W.; Mithieux, S.M.; Weiss, A.S.; Neubert, R.H.; Schmelzer, C.E. Molecular-level characterization of elastin-like constructs and human aortic elastin. Matrix Biol 2014, 38, 12–21. [Google Scholar] [CrossRef]
- Hedtke, T.; Schrader, C.U.; Heinz, A.; Hoehenwarter, W.; Brinckmann, J.; Groth, T.; Schmelzer, C.E.H. A comprehensive map of human elastin cross-linking during elastogenesis. FEBS J 2019, 286, 3594–3610. [Google Scholar] [CrossRef] [PubMed]
- Kielty, C.M.; Shuttleworth, C.A. Microfibrillar elements of the dermal matrix. Microsc Res Tech 1997, 38, 413–427. [Google Scholar] [CrossRef]
- Mecham, R.P. Elastin in lung development and disease pathogenesis. Matrix Biol 2018, 73, 6–20. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, D.; Sun, B.; Zhang, X.; Li, F.; Liu, Z.; Zheng, Y. Discovery of crucial cytokines associated with abdominal aortic aneurysm formation by protein array analysis. Exp Biol Med (Maywood) 2019, 244, 1648–1657. [Google Scholar] [CrossRef] [PubMed]
- Puchenkova, O.A.; Soldatov, V.O.; Belykh, A.E.; Bushueva, O.; Piavchenko, G.A.; Venediktov, A.A.; Shakhpazyan, N.K.; Deykin, A.V.; Korokin, M.V.; Pokrovskiy, M.V. Cytokines in Abdominal Aortic Aneurysm: Master Regulators With Clinical Application. Biomark Insights 2022, 17, 11772719221095676. [Google Scholar] [CrossRef]
- Nishihara, M.; Aoki, H.; Ohno, S.; Furusho, A.; Hirakata, S.; Nishida, N.; Ito, S.; Hayashi, M.; Imaizumi, T.; Fukumoto, Y. The role of IL-6 in pathogenesis of abdominal aortic aneurysm in mice. PLoS One 2017, 12, e0185923. [Google Scholar] [CrossRef]
- Golledge, J. Is there a new target in the renin-angiotensin system for aortic aneurysm therapy? Arterioscler Thromb Vasc Biol 2013, 33, 1456–1457. [Google Scholar] [CrossRef]
- Rabkin, S.W. The Role Matrix Metalloproteinases in the Production of Aortic Aneurysm. Prog Mol Biol Transl Sci 2017, 147, 239–265. [Google Scholar] [CrossRef]
- Nosoudi, N.; Nahar-Gohad, P.; Sinha, A.; Chowdhury, A.; Gerard, P.; Carsten, C.G.; Gray, B.H.; Vyavahare, N.R. Prevention of abdominal aortic aneurysm progression by targeted inhibition of matrix metalloproteinase activity with batimastat-loaded nanoparticles. Circ Res 2015, 117, e80–e89. [Google Scholar] [CrossRef]
- Baxter, B.T.; Matsumura, J.; Curci, J.A.; McBride, R.; Larson, L.; Blackwelder, W.; Lam, D.; Wijesinha, M.; Terrin, M.; Investigators, N.T.C. Effect of Doxycycline on Aneurysm Growth Among Patients With Small Infrarenal Abdominal Aortic Aneurysms: A Randomized Clinical Trial. JAMA 2020, 323, 2029–2038. [Google Scholar] [CrossRef]
- Piechota-Polanczyk, A.; Jozkowicz, A.; Nowak, W.; Eilenberg, W.; Neumayer, C.; Malinski, T.; Huk, I.; Brostjan, C. The Abdominal Aortic Aneurysm and Intraluminal Thrombus: Current Concepts of Development and Treatment. Front Cardiovasc Med 2015, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- Hans, S.S.; Jareunpoon, O.; Balasubramaniam, M.; Zelenock, G.B. Size and location of thrombus in intact and ruptured abdominal aortic aneurysms. J Vasc Surg 2005, 41, 584–588. [Google Scholar] [CrossRef] [PubMed]
- Whaley, Z.L.; Cassimjee, I.; Novak, Z.; Rowland, D.; Lapolla, P.; Chandrashekar, A.; Pearce, B.J.; Beck, A.W.; Handa, A.; Lee, R.; et al. The Spatial Morphology of Intraluminal Thrombus Influences Type II Endoleak after Endovascular Repair of Abdominal Aortic Aneurysms. Ann Vasc Surg 2020, 66, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Adolph, R.; Vorp, D.A.; Steed, D.L.; Webster, M.W.; Kameneva, M.V.; Watkins, S.C. Cellular content and permeability of intraluminal thrombus in abdominal aortic aneurysm. J Vasc Surg 1997, 25, 916–926. [Google Scholar] [CrossRef] [PubMed]
- Murail, S.; de Vries, S.J.; Rey, J.; Moroy, G.; Tuffery, P. SeamDock: An Interactive and Collaborative Online Docking Resource to Assist Small Compound Molecular Docking. Front Mol Biosci 2021, 8, 716466. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Simionescu, D. , Casco, M, Turner, J, Rierson, N., Yue, J., Ning, N. Chemical stabilization of the extracellular matrix attenuates growth of experimentally induced abdominal aorta aneurysms in a large animal model. Journal of Vascualr Surgery - Vascular Science 2020, 1, 69–80. [Google Scholar]
- Chen, Z.; Li, Y.; Chen, E.; Hall, D.L.; Darke, P.L.; Culberson, C.; Shafer, J.A.; Kuo, L.C. Crystal structure at 1.9-A resolution of human immunodeficiency virus (HIV) II protease complexed with L-735,524, an orally bioavailable inhibitor of the HIV proteases. J Biol Chem 1994, 269, 26344–26348. [Google Scholar] [CrossRef]
- Mahmoud, M.F.; Nabil, M.; Hasan, R.A.; El-Shazly, A.M.; El-Ansari, M.A.; Sobeh, M. Pentagalloyl Glucose, a Major Compound in Mango Seed Kernel, Exhibits Distinct Gastroprotective Effects in Indomethacin-Induced Gastropathy in Rats via Modulating the NO/eNOS/iNOS Signaling Pathway. Front Pharmacol 2022, 13, 800986. [Google Scholar] [CrossRef]
- Dharmalingam, K.; Dharmalingam, V.; Durairaj, S.; Sharma, P.; Jayaraman, S.; Choudhary, S. Molecular docking analysis of penta galloyl glucose with the bcl-2 family of anti-apoptotic targets. Bioinformation 2021, 17, 861–865. [Google Scholar] [CrossRef]
- Dharmalingam, K.; Dharmalingam, V.; Durairaj, S.; Sharma, P.; Jayaraman, S. Molecular docking analysis of penta-galloyl-glucose with VEGF signaling molecules. Bioinformation 2021, 17, 924–927. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.H.; Yang, L.J.; Hamdoun, S.; Chung, S.K.; Lam, C.W.; Zhang, K.X.; Guo, X.; Xia, C.; Law, B.Y.K.; Wong, V.K.W. 1,2,3,4,6-Pentagalloyl Glucose, a RBD-ACE2 Binding Inhibitor to Prevent SARS-CoV-2 Infection. Front Pharmacol 2021, 12, 634176. [Google Scholar] [CrossRef] [PubMed]
- Nosoudi, N.; Chowdhury, A.; Siclari, S.; Parasaram, V.; Karamched, S.; Vyavahare, N. Systemic Delivery of Nanoparticles Loaded with Pentagalloyl Glucose Protects Elastic Lamina and Prevents Abdominal Aortic Aneurysm in Rats. J Cardiovasc Transl Res 2016, 9, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Dhital, S.; Vyavahare, N.R. Nanoparticle-based targeted delivery of pentagalloyl glucose reverses elastase-induced abdominal aortic aneurysm and restores aorta to the healthy state in mice. PLoS One 2020, 15, e0227165. [Google Scholar] [CrossRef]
- Arnold, F.; Muzzio, N.; Patnaik, S.S.; Finol, E.A.; Romero, G. Pentagalloyl Glucose-Laden Poly(lactide-co-glycolide) Nanoparticles for the Biomechanical Extracellular Matrix Stabilization of an In Vitro Abdominal Aortic Aneurysm Model. ACS Appl Mater Interfaces 2021, 13, 25771–25782. [Google Scholar] [CrossRef]
- Dhital, S.; Rice, C.D.; Vyavahare, N.R. Reversal of elastase-induced abdominal aortic aneurysm following the delivery of nanoparticle-based pentagalloyl glucose (PGG) is associated with reduced inflammatory and immune markers. Eur J Pharmacol 2021, 910, 174487. [Google Scholar] [CrossRef]
- Thirugnanasambandam, M.; Simionescu, D.T.; Escobar, P.G.; Sprague, E.; Goins, B.; Clarke, G.D.; Han, H.C.; Amezcua, K.L.; Adeyinka, O.R.; Goergen, C.J.; et al. The Effect of Pentagalloyl Glucose on the Wall Mechanics and Inflammatory Activity of Rat Abdominal Aortic Aneurysms. J Biomech Eng 2018, 140, 0845021–0845029. [Google Scholar] [CrossRef]
- Kennamer, A.; Sierad, L.; Pascal, R.; Rierson, N.; Albers, C.; Harpa, M.; Cotoi, O.; Harceaga, L.; Olah, P.; Terezia, P.; et al. Bioreactor Conditioning of Valve Scaffolds Seeded Internally with Adult Stem Cells. Tissue Eng Regen Med 2016, 13, 507–515. [Google Scholar] [CrossRef]
- Deborde, C.; Simionescu, D.T.; Wright, C.; Liao, J.; Sierad, L.N.; Simionescu, A. Stabilized Collagen and Elastin-Based Scaffolds for Mitral Valve Tissue Engineering. Tissue Eng Part A 2016, 22, 1241–1251. [Google Scholar] [CrossRef]
- Sierad, L.N.; Shaw, E.L.; Bina, A.; Brazile, B.; Rierson, N.; Patnaik, S.S.; Kennamer, A.; Odum, R.; Cotoi, O.; Terezia, P.; et al. Functional Heart Valve Scaffolds Obtained by Complete Decellularization of Porcine Aortic Roots in a Novel Differential Pressure Gradient Perfusion System. Tissue Eng Part C Methods 2015, 21, 1284–1296. [Google Scholar] [CrossRef]
- Pennel, T.; Fercana, G.; Bezuidenhout, D.; Simionescu, A.; Chuang, T.H.; Zilla, P.; Simionescu, D. The performance of cross-linked acellular arterial scaffolds as vascular grafts; pre-clinical testing in direct and isolation loop circulatory models. Biomaterials 2014, 35, 6311–6322. [Google Scholar] [CrossRef] [PubMed]
- Parasaram, V.; Wang, X.; Krisanarungson, P.; Vyavahare, N. Targeted delivery of pentagalloyl glucose inhibits matrix metalloproteinase activity and preserves elastin in emphysematous lungs. Respir Res 2021, 22, 249. [Google Scholar] [CrossRef] [PubMed]
- Parasaram, V.; Nosoudi, N.; Chowdhury, A.; Vyavahare, N. Pentagalloyl glucose increases elastin deposition, decreases reactive oxygen species and matrix metalloproteinase activity in pulmonary fibroblasts under inflammatory conditions. Biochem Biophys Res Commun 2018, 499, 24–29. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).