Submitted:
02 June 2023
Posted:
05 June 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
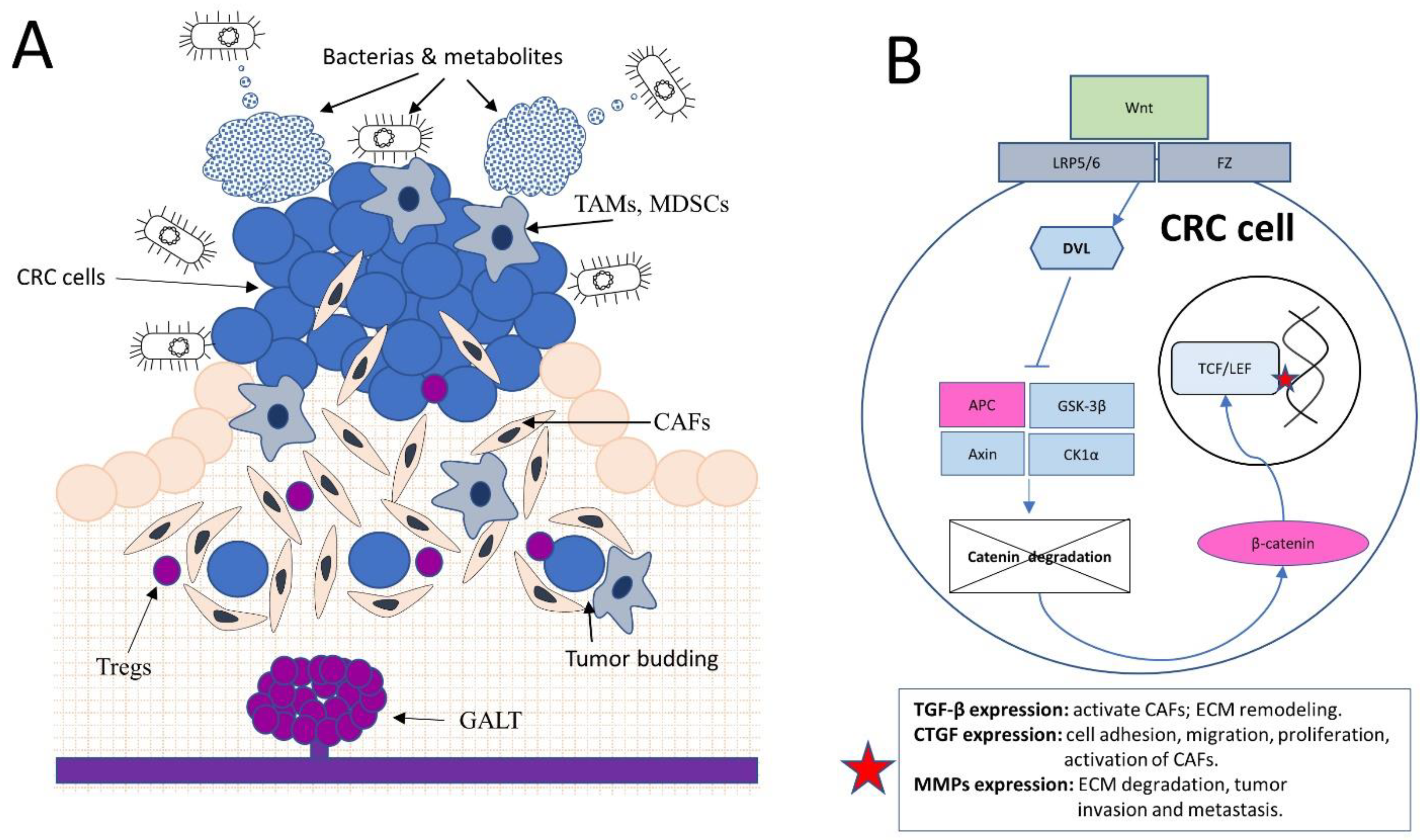
2. The Gut Microbiome and Colorectal Cancer: Dysbiosis, Tumor Stroma Modulation, and Emerging Therapeutic Strategies
3. Gut-Associated Lymphoid Tissue (GALT) and its Influence on Colorectal Tumor Stroma Development and Stability
4. The Role of the Wnt Signaling Pathway in Colorectal Cancer Tumor Stroma Development and Maintenance
- Porcupine inhibitors: Porcupine is an enzyme required for the palmitoylation and secretion of Wnt ligands. Inhibition of Porcupine prevents the secretion of Wnt ligands, thereby blocking Wnt signaling activation [76]. Several Porcupine inhibitors, such as LGK974 and ETC-159, are currently being evaluated in clinical trials for the treatment of Wnt-driven cancers, including CRC [77,78].
- Frizzled receptor antagonists: Frizzled receptors are cell surface receptors that bind to Wnt ligands and activate Wnt signaling [77]. Targeting Frizzled receptors with antagonistic antibodies or small molecules can prevent Wnt ligand binding and subsequent pathway activation. OMP-18R5 (vantictumab) and OMP-54F28 (ipafricept) are two examples of Frizzled receptor antagonists under clinical investigation [79,80].
- Tankyrase inhibitors: Tankyrases are enzymes that regulate the stability of the scaffold protein AXIN, a component of the β-catenin destruction complex. Tankyrase inhibitors stabilize AXIN, promoting the degradation of β-catenin and thus inhibiting Wnt signaling [81,82]. G007-LK and NVP-TNKS656 are examples of Tankyrase inhibitors in preclinical development [83].
- β-catenin inhibitors: Directly targeting β-catenin can inhibit its interaction with TCF/LEF transcription factors, preventing the activation of Wnt target genes [82]. Small molecules, such as PRI-724 and BC2059, have been developed to target the β-catenin/TCF interaction and are currently in clinical trials [84,85].
5. Tumor Budding and Its Influence on the Tumor Stroma in Colorectal Cancer
- EMT Inhibitors: Targeting the EMT process can inhibit tumor budding and potentially prevent invasion and metastasis. Inhibition of EMT-driving pathways, such as TGF-β, Wnt, and Notch, can restore epithelial properties and reduce the invasive potential of tumor cells [93,94,95]. Small molecules, monoclonal antibodies, and other therapeutic agents targeting these pathways are under investigation.
- Targeting CAFs: CAFs play a critical role in supporting tumor budding by secreting factors that promote EMT, invasion, and angiogenesis. Inhibiting CAF activation, proliferation, or function may disrupt the tumor-stroma crosstalk and reduce the supportive role of CAFs in tumor budding. Several strategies, including targeting CAF-derived factors, such as fibroblast activation protein (FAP) [96] or transforming growth factor-beta (TGF-β), are being explored [97].
- Inhibition of MMPs: MMPs are enzymes that degrade the ECM, facilitating tumor invasion and metastasis [98]. Budding tumor cells produce MMPs to invade the stroma and promote tumor progression. Inhibiting MMP activity may prevent the degradation of the ECM and reduce tumor budding-associated invasion and metastasis. Several MMP inhibitors have been developed, and some are currently being tested in preclinical and clinical studies for their potential in CRC treatment [99,100].
- Targeting Immune Cells: The immune cells within the tumor stroma, such as tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells (MDSCs), contribute to the supportive tumor microenvironment and facilitate tumor budding. Modulating the function of these immune cells or reprogramming them to adopt an anti-tumor phenotype may help inhibit tumor budding and improve CRC outcomes. Immunotherapies, such as immune checkpoint inhibitors and adoptive cell transfer, are being investigated for their potential to target these immune cells [101,102].
- Targeting Tumor-Stroma Interactions: The reciprocal feedback loop between tumor budding cells and the stroma is crucial for CRC progression. Disrupting the signaling pathways involved in this crosstalk, such as the chemokine ligand-receptor axis (e.g., CXCL12-CXCR4), may inhibit tumor budding and its associated invasion and metastasis. Therapeutic agents targeting these signaling pathways are under investigation [103,104].
- Therapeutic Approaches Targeting Tumor-Initiating Cells: Tumor-initiating cells (TICs) or cancer stem cells are a subpopulation of tumor cells that possess self-renewal and differentiation capabilities, driving tumor heterogeneity and resistance to therapy. Tumor budding has been linked to the presence of TICs, which are capable of initiating new tumor growth at the invasive front. Targeting TICs using specific surface markers, such as CD133, CD44, and Lgr5, or cellular processes like self-renewal and differentiation, can help inhibit TICs and tumor budding [105,106,107]. Targeting metabolic reprogramming in TICs, such as glucose metabolism, glutamine metabolism, or fatty acid synthesis, may selectively affect TICs and impair their survival and function [108,109].
6. Conserved Oncogenic Signatures in Colorectal Cancer Stroma
7. Culmination of Colorectal Cancer Tumor-Stroma Interactions in Metastasis: The Seed and Soil Hypothesis
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int J Cancer 2021. https://doi.org/10.1002/ijc.33588. [CrossRef]
- Ladabaum, U.; Dominitz, J.A.; Kahi, C.; Schoen, R.E. Strategies for Colorectal Cancer Screening. Gastroenterology 2020, 158, 418-432. https://doi.org/10.1053/j.gastro.2019.06.043. [CrossRef]
- Biller, L.H.; Schrag, D. Diagnosis and Treatment of Metastatic Colorectal Cancer: A Review. JAMA 2021, 325, 669-685. https://doi.org/10.1001/jama.2021.0106. [CrossRef]
- Song, M.; Chan, A.T.; Sun, J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology 2020, 158, 322-340. https://doi.org/10.1053/j.gastro.2019.06.048. [CrossRef]
- Rubinstein, M.R.; Baik, J.E.; Lagana, S.M.; Han, R.P.; Raab, W.J.; Sahoo, D.; Dalerba, P.; Wang, T.C.; Han, Y.W. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/beta-catenin modulator Annexin A1. EMBO Rep 2019, 20. https://doi.org/10.15252/embr.201847638. [CrossRef]
- Brennan, C.A.; Clay, S.L.; Lavoie, S.L.; Bae, S.; Lang, J.K.; Fonseca-Pereira, D.; Rosinski, K.G.; Ou, N.; Glickman, J.N.; Garrett, W.S. Fusobacterium nucleatum drives a pro-inflammatory intestinal microenvironment through metabolite receptor-dependent modulation of IL-17 expression. Gut Microbes 2021, 13, 1987780. https://doi.org/10.1080/19490976.2021.1987780. [CrossRef]
- Sui, H.; Zhang, L.; Gu, K.; Chai, N.; Ji, Q.; Zhou, L.; Wang, Y.; Ren, J.; Yang, L.; Zhang, B.; et al. YYFZBJS ameliorates colorectal cancer progression in Apc(Min/+) mice by remodeling gut microbiota and inhibiting regulatory T-cell generation. Cell Commun Signal 2020, 18, 113. https://doi.org/10.1186/s12964-020-00596-9. [CrossRef]
- Knippel, R.J.; Drewes, J.L.; Sears, C.L. The Cancer Microbiome: Recent Highlights and Knowledge Gaps. Cancer Discov 2021, 11, 2378-2395. https://doi.org/10.1158/2159-8290.CD-21-0324. [CrossRef]
- Chattopadhyay, I.; Dhar, R.; Pethusamy, K.; Seethy, A.; Srivastava, T.; Sah, R.; Sharma, J.; Karmakar, S. Exploring the Role of Gut Microbiome in Colon Cancer. Appl Biochem Biotechnol 2021, 193, 1780-1799. https://doi.org/10.1007/s12010-021-03498-9. [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443-1448. https://doi.org/10.1126/science.aal5240. [CrossRef]
- Engevik, M.A.; Danhof, H.A.; Ruan, W.; Engevik, A.C.; Chang-Graham, A.L.; Engevik, K.A.; Shi, Z.; Zhao, Y.; Brand, C.K.; Krystofiak, E.S.; et al. Fusobacterium nucleatum Secretes Outer Membrane Vesicles and Promotes Intestinal Inflammation. mBio 2021, 12. https://doi.org/10.1128/mBio.02706-20. [CrossRef]
- Casasanta, M.A.; Yoo, C.C.; Udayasuryan, B.; Sanders, B.E.; Umana, A.; Zhang, Y.; Peng, H.; Duncan, A.J.; Wang, Y.; Li, L.; et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal 2020, 13. https://doi.org/10.1126/scisignal.aba9157. [CrossRef]
- Hills, R.D., Jr.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019, 11. https://doi.org/10.3390/nu11071613. [CrossRef]
- Cheng, W.T.; Kantilal, H.K.; Davamani, F. The Mechanism of Bacteroides fragilis Toxin Contributes to Colon Cancer Formation. Malays J Med Sci 2020, 27, 9-21. https://doi.org/10.21315/mjms2020.27.4.2. [CrossRef]
- Lee, C.W.; Chen, H.J.; Chien, Y.H.; Hsia, S.M.; Chen, J.H.; Shih, C.K. Synbiotic Combination of Djulis (Chenopodium formosanum) and Lactobacillus acidophilus Inhibits Colon Carcinogenesis in Rats. Nutrients 2019, 12. https://doi.org/10.3390/nu12010103. [CrossRef]
- Faghfoori, Z.; Pourghassem Gargari, B.; Saber, A.; Seyyedi, M.; Fazelian, S.; Khosroushahi, A.Y. Prophylactic effects of secretion metabolites of dairy lactobacilli through downregulation of ErbB-2 and ErbB-3 genes on colon cancer cells. Eur J Cancer Prev 2020, 29, 201-209. https://doi.org/10.1097/CEJ.0000000000000393. [CrossRef]
- Williamson, A.J.; Jacobson, R.; van Praagh, J.B.; Gaines, S.; Koo, H.Y.; Lee, B.; Chan, W.C.; Weichselbaum, R.; Alverdy, J.C.; Zaborina, O.; et al. Enterococcus faecalis promotes a migratory and invasive phenotype in colon cancer cells. Neoplasia 2022, 27, 100787. https://doi.org/10.1016/j.neo.2022.100787. [CrossRef]
- Banna, G.L.; Torino, F.; Marletta, F.; Santagati, M.; Salemi, R.; Cannarozzo, E.; Falzone, L.; Ferrau, F.; Libra, M. Lactobacillus rhamnosus GG: An Overview to Explore the Rationale of Its Use in Cancer. Front Pharmacol 2017, 8, 603. https://doi.org/10.3389/fphar.2017.00603. [CrossRef]
- Nouri, R.; Hasani, A.; Shirazi, K.M.; Alivand, M.R.; Sepehri, B.; Sotoodeh, S.; Hemmati, F.; Rezaee, M.A. Escherichia coli and Colorectal Cancer: Unfolding the Enigmatic Relationship. Curr Pharm Biotechnol 2022, 23, 1257-1268. https://doi.org/10.2174/1389201022666210910094827. [CrossRef]
- Dikeocha, I.J.; Al-Kabsi, A.M.; Chiu, H.T.; Alshawsh, M.A. Faecalibacterium prausnitzii Ameliorates Colorectal Tumorigenesis and Suppresses Proliferation of HCT116 Colorectal Cancer Cells. Biomedicines 2022, 10. https://doi.org/10.3390/biomedicines10051128. [CrossRef]
- Long, X.; Wong, C.C.; Tong, L.; Chu, E.S.H.; Ho Szeto, C.; Go, M.Y.Y.; Coker, O.O.; Chan, A.W.H.; Chan, F.K.L.; Sung, J.J.Y.; et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol 2019, 4, 2319-2330. https://doi.org/10.1038/s41564-019-0541-3. [CrossRef]
- Pasquereau-Kotula, E.; Martins, M.; Aymeric, L.; Dramsi, S. Significance of Streptococcus gallolyticus subsp. gallolyticus Association With Colorectal Cancer. Front Microbiol 2018, 9, 614. https://doi.org/10.3389/fmicb.2018.00614. [CrossRef]
- Slezak, M.; Smolar, M.; Drobna Saniova, B.; Hosala, M.; Miklusica, J. Clostridium septicum foot gangrene associated with colorectal cancer. Neuro Endocrinol Lett 2022, 43, 57-64.
- Ebert, M.N.; Klinder, A.; Peters, W.H.; Schaferhenrich, A.; Sendt, W.; Scheele, J.; Pool-Zobel, B.L. Expression of glutathione S-transferases (GSTs) in human colon cells and inducibility of GSTM2 by butyrate. Carcinogenesis 2003, 24, 1637-1644. https://doi.org/10.1093/carcin/bgg122. [CrossRef]
- Stoeva, M.K.; Garcia-So, J.; Justice, N.; Myers, J.; Tyagi, S.; Nemchek, M.; McMurdie, P.J.; Kolterman, O.; Eid, J. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes 2021, 13, 1-28. https://doi.org/10.1080/19490976.2021.1907272. [CrossRef]
- Park, C.H.; Eun, C.S.; Han, D.S. Intestinal microbiota, chronic inflammation, and colorectal cancer. Intest Res 2018, 16, 338-345. https://doi.org/10.5217/ir.2018.16.3.338. [CrossRef]
- Ryu, T.Y.; Kim, K.; Han, T.S.; Lee, M.O.; Lee, J.; Choi, J.; Jung, K.B.; Jeong, E.J.; An, D.M.; Jung, C.R.; et al. Human gut-microbiome-derived propionate coordinates proteasomal degradation via HECTD2 upregulation to target EHMT2 in colorectal cancer. ISME J 2022, 16, 1205-1221. https://doi.org/10.1038/s41396-021-01119-1. [CrossRef]
- Liu, Y.; Zhang, S.; Zhou, W.; Hu, D.; Xu, H.; Ji, G. Secondary Bile Acids and Tumorigenesis in Colorectal Cancer. Front Oncol 2022, 12, 813745. https://doi.org/10.3389/fonc.2022.813745. [CrossRef]
- Fang, Y.; Yan, C.; Zhao, Q.; Xu, J.; Liu, Z.; Gao, J.; Zhu, H.; Dai, Z.; Wang, D.; Tang, D. The roles of microbial products in the development of colorectal cancer: a review. Bioengineered 2021, 12, 720-735. https://doi.org/10.1080/21655979.2021.1889109. [CrossRef]
- Caliceti, C.; Punzo, A.; Silla, A.; Simoni, P.; Roda, G.; Hrelia, S. New Insights into Bile Acids Related Signaling Pathways in the Onset of Colorectal Cancer. Nutrients 2022, 14. https://doi.org/10.3390/nu14142964. [CrossRef]
- Huang, C.Y.; Fang, Y.J.; Abulimiti, A.; Yang, X.; Li, L.; Liu, K.Y.; Zhang, X.; Feng, X.L.; Chen, Y.M.; Zhang, C.X. Dietary Polyamines Intake and Risk of Colorectal Cancer: A Case-Control Study. Nutrients 2020, 12. https://doi.org/10.3390/nu12113575. [CrossRef]
- Bolte, L.A.; Vich Vila, A.; Imhann, F.; Collij, V.; Gacesa, R.; Peters, V.; Wijmenga, C.; Kurilshikov, A.; Campmans-Kuijpers, M.J.E.; Fu, J.; et al. Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome. Gut 2021, 70, 1287-1298. https://doi.org/10.1136/gutjnl-2020-322670. [CrossRef]
- Yang, Y.; Li, L.; Xu, C.; Wang, Y.; Wang, Z.; Chen, M.; Jiang, Z.; Pan, J.; Yang, C.; Li, X.; et al. Cross-talk between the gut microbiota and monocyte-like macrophages mediates an inflammatory response to promote colitis-associated tumourigenesis. Gut 2020, 70, 1495-1506. https://doi.org/10.1136/gutjnl-2020-320777. [CrossRef]
- Stephens, M.; von der Weid, P.Y. Lipopolysaccharides modulate intestinal epithelial permeability and inflammation in a species-specific manner. Gut Microbes 2020, 11, 421-432. https://doi.org/10.1080/19490976.2019.1629235. [CrossRef]
- Rodrigues, D.M.; Sousa, A.J.; Hawley, S.P.; Vong, L.; Gareau, M.G.; Kumar, S.A.; Johnson-Henry, K.C.; Sherman, P.M. Matrix metalloproteinase 9 contributes to gut microbe homeostasis in a model of infectious colitis. BMC Microbiol 2012, 12, 105. https://doi.org/10.1186/1471-2180-12-105. [CrossRef]
- Wei, Z.; Cao, S.; Liu, S.; Yao, Z.; Sun, T.; Li, Y.; Li, J.; Zhang, D.; Zhou, Y. Could gut microbiota serve as prognostic biomarker associated with colorectal cancer patients' survival? A pilot study on relevant mechanism. Oncotarget 2016, 7, 46158-46172. https://doi.org/10.18632/oncotarget.10064. [CrossRef]
- Bell, H.N.; Rebernick, R.J.; Goyert, J.; Singhal, R.; Kuljanin, M.; Kerk, S.A.; Huang, W.; Das, N.K.; Andren, A.; Solanki, S.; et al. Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance. Cancer Cell 2022, 40, 185-200 e186. https://doi.org/10.1016/j.ccell.2021.12.001. [CrossRef]
- Azad, M.A.K.; Sarker, M.; Li, T.; Yin, J. Probiotic Species in the Modulation of Gut Microbiota: An Overview. Biomed Res Int 2018, 2018, 9478630. https://doi.org/10.1155/2018/9478630. [CrossRef]
- Legesse Bedada, T.; Feto, T.K.; Awoke, K.S.; Garedew, A.D.; Yifat, F.T.; Birri, D.J. Probiotics for cancer alternative prevention and treatment. Biomed Pharmacother 2020, 129, 110409. https://doi.org/10.1016/j.biopha.2020.110409. [CrossRef]
- Ciecierska, A.; Drywien, M.E.; Hamulka, J.; Sadkowski, T. Nutraceutical functions of beta-glucans in human nutrition. Rocz Panstw Zakl Hig 2019, 70, 315-324. https://doi.org/10.32394/rpzh.2019.0082. [CrossRef]
- Wang, H.; Chen, K.; Ning, M.; Wang, X.; Wang, Z.; Yue, Y.; Yuan, Y.; Yue, T. Intake of Pro- and/or Prebiotics as a Promising Approach for Prevention and Treatment of Colorectal Cancer. Mol Nutr Food Res 2023, 67, e2200474. https://doi.org/10.1002/mnfr.202200474. [CrossRef]
- Zhang, J.; Wu, K.; Shi, C.; Li, G. Cancer Immunotherapy: Fecal Microbiota Transplantation Brings Light. Curr Treat Options Oncol 2022, 23, 1777-1792. https://doi.org/10.1007/s11864-022-01027-2. [CrossRef]
- Xu, H.; Cao, C.; Ren, Y.; Weng, S.; Liu, L.; Guo, C.; Wang, L.; Han, X.; Ren, J.; Liu, Z. Antitumor effects of fecal microbiota transplantation: Implications for microbiome modulation in cancer treatment. Front Immunol 2022, 13, 949490. https://doi.org/10.3389/fimmu.2022.949490. [CrossRef]
- Ting, N.L.; Lau, H.C.; Yu, J. Cancer pharmacomicrobiomics: targeting microbiota to optimise cancer therapy outcomes. Gut 2022, 71, 1412-1425. https://doi.org/10.1136/gutjnl-2021-326264. [CrossRef]
- Zhao, Y.; Zhan, J.; Wang, Y.; Wang, D. The Relationship Between Plant-Based Diet and Risk of Digestive System Cancers: A Meta-Analysis Based on 3,059,009 Subjects. Front Public Health 2022, 10, 892153. https://doi.org/10.3389/fpubh.2022.892153. [CrossRef]
- Kato, I.; Sun, J. Microbiome and Diet in Colon Cancer Development and Treatment. Cancer J 2023, 29, 89-97. https://doi.org/10.1097/PPO.0000000000000649. [CrossRef]
- Gomes, S.; Teixeira-Guedes, C.; Silva, E.; Baltazar, F.; Preto, A. Colon microbiota modulation by dairy-derived diet: new strategy for prevention and treatment of colorectal cancer. Food Funct 2022, 13, 9183-9194. https://doi.org/10.1039/d2fo01720b. [CrossRef]
- Jia, F.; Yu, Q.; Wang, R.; Zhao, L.; Yuan, F.; Guo, H.; Shen, Y.; He, F. Optimized Antimicrobial Peptide Jelleine-I Derivative Br-J-I Inhibits Fusobacterium Nucleatum to Suppress Colorectal Cancer Progression. Int J Mol Sci 2023, 24. https://doi.org/10.3390/ijms24021469. [CrossRef]
- Jang, H.I.; Rhee, K.J.; Eom, Y.B. Antibacterial and antibiofilm effects of alpha-humulene against Bacteroides fragilis. Can J Microbiol 2020, 66, 389-399. https://doi.org/10.1139/cjm-2020-0004. [CrossRef]
- Li, S.; Liu, J.; Zheng, X.; Ren, L.; Yang, Y.; Li, W.; Fu, W.; Wang, J.; Du, G. Tumorigenic bacteria in colorectal cancer: mechanisms and treatments. Cancer Biol Med 2021, 19, 147-162. https://doi.org/10.20892/j.issn.2095-3941.2020.0651. [CrossRef]
- Alam, Z.; Shang, X.; Effat, K.; Kanwal, F.; He, X.; Li, Y.; Xu, C.; Niu, W.; War, A.R.; Zhang, Y. The potential role of prebiotics, probiotics, and synbiotics in adjuvant cancer therapy especially colorectal cancer. J Food Biochem 2022, 46, e14302. https://doi.org/10.1111/jfbc.14302. [CrossRef]
- Yaseen, A.; Ladenheim, A.; Olson, K.A.; Libertini, S.J.; McPherson, J.D.; Matsukuma, K. Whole exome sequencing of a gut-associated lymphoid tissue neoplasm points to precursor or early form of sporadic colon carcinoma. Pathol Res Pract 2021, 220, 153406. https://doi.org/10.1016/j.prp.2021.153406. [CrossRef]
- Muzes, G.; Bohusne Barta, B.; Sipos, F. Colitis and Colorectal Carcinogenesis: The Focus on Isolated Lymphoid Follicles. Biomedicines 2022, 10. https://doi.org/10.3390/biomedicines10020226. [CrossRef]
- Morbe, U.M.; Jorgensen, P.B.; Fenton, T.M.; von Burg, N.; Riis, L.B.; Spencer, J.; Agace, W.W. Human gut-associated lymphoid tissues (GALT); diversity, structure, and function. Mucosal Immunol 2021, 14, 793-802. https://doi.org/10.1038/s41385-021-00389-4. [CrossRef]
- Barone, F.; Patel, P.; Sanderson, J.D.; Spencer, J. Gut-associated lymphoid tissue contains the molecular machinery to support T-cell-dependent and T-cell-independent class switch recombination. Mucosal Immunol 2009, 2, 495-503. https://doi.org/10.1038/mi.2009.106. [CrossRef]
- Cosola, C.; Rocchetti, M.T.; Gesualdo, L. Gut Microbiota, the Immune System, and Cytotoxic T Lymphocytes. Methods Mol Biol 2021, 2325, 229-241. https://doi.org/10.1007/978-1-0716-1507-2_16. [CrossRef]
- Ohue, Y.; Nishikawa, H. Regulatory T (Treg) cells in cancer: Can Treg cells be a new therapeutic target? Cancer Sci 2019, 110, 2080-2089. https://doi.org/10.1111/cas.14069. [CrossRef]
- Law, A.M.K.; Valdes-Mora, F.; Gallego-Ortega, D. Myeloid-Derived Suppressor Cells as a Therapeutic Target for Cancer. Cells 2020, 9. https://doi.org/10.3390/cells9030561. [CrossRef]
- Pandey, P.; Khan, F.; Qari, H.A.; Upadhyay, T.K.; Alkhateeb, A.F.; Oves, M. Revolutionization in Cancer Therapeutics via Targeting Major Immune Checkpoints PD-1, PD-L1 and CTLA-4. Pharmaceuticals (Basel) 2022, 15. https://doi.org/10.3390/ph15030335. [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduct Target Ther 2021, 6, 263. https://doi.org/10.1038/s41392-021-00658-5. [CrossRef]
- Gardner, A.; de Mingo Pulido, A.; Ruffell, B. Dendritic Cells and Their Role in Immunotherapy. Front Immunol 2020, 11, 924. https://doi.org/10.3389/fimmu.2020.00924. [CrossRef]
- Korhonen, E.A.; Murtomaki, A.; Jha, S.K.; Anisimov, A.; Pink, A.; Zhang, Y.; Stritt, S.; Liaqat, I.; Stanczuk, L.; Alderfer, L.; et al. Lymphangiogenesis requires Ang2/Tie/PI3K signaling for VEGFR3 cell-surface expression. J Clin Invest 2022, 132. https://doi.org/10.1172/JCI155478. [CrossRef]
- Roy, S.; Banerjee, P.; Ekser, B.; Bayless, K.; Zawieja, D.; Alpini, G.; Glaser, S.S.; Chakraborty, S. Targeting Lymphangiogenesis and Lymph Node Metastasis in Liver Cancer. Am J Pathol 2021, 191, 2052-2063. https://doi.org/10.1016/j.ajpath.2021.08.011. [CrossRef]
- Zhang, Y.; Wang, X. Targeting the Wnt/beta-catenin signaling pathway in cancer. J Hematol Oncol 2020, 13, 165. https://doi.org/10.1186/s13045-020-00990-3. [CrossRef]
- Malki, A.; ElRuz, R.A.; Gupta, I.; Allouch, A.; Vranic, S.; Al Moustafa, A.E. Molecular Mechanisms of Colon Cancer Progression and Metastasis: Recent Insights and Advancements. Int J Mol Sci 2020, 22. https://doi.org/10.3390/ijms22010130. [CrossRef]
- Boonekamp, K.E.; Heo, I.; Artegiani, B.; Asra, P.; van Son, G.; de Ligt, J.; Clevers, H. Identification of novel human Wnt target genes using adult endodermal tissue-derived organoids. Dev Biol 2021, 474, 37-47. https://doi.org/10.1016/j.ydbio.2021.01.009. [CrossRef]
- Aghabozorgi, A.S.; Bahreyni, A.; Soleimani, A.; Bahrami, A.; Khazaei, M.; Ferns, G.A.; Avan, A.; Hassanian, S.M. Role of adenomatous polyposis coli (APC) gene mutations in the pathogenesis of colorectal cancer; current status and perspectives. Biochimie 2019, 157, 64-71. https://doi.org/10.1016/j.biochi.2018.11.003. [CrossRef]
- Steinhart, Z.; Angers, S. Wnt signaling in development and tissue homeostasis. Development 2018, 145. https://doi.org/10.1242/dev.146589. [CrossRef]
- Lecarpentier, Y.; Schussler, O.; Hebert, J.L.; Vallee, A. Multiple Targets of the Canonical WNT/beta-Catenin Signaling in Cancers. Front Oncol 2019, 9, 1248. https://doi.org/10.3389/fonc.2019.01248. [CrossRef]
- Yoshida, G.J. Regulation of heterogeneous cancer-associated fibroblasts: the molecular pathology of activated signaling pathways. J Exp Clin Cancer Res 2020, 39, 112. https://doi.org/10.1186/s13046-020-01611-0. [CrossRef]
- Wang, J.; Cai, H.; Liu, Q.; Xia, Y.; Xing, L.; Zuo, Q.; Zhang, Y.; Chen, C.; Xu, K.; Yin, P.; et al. Cinobufacini Inhibits Colon Cancer Invasion and Metastasis via Suppressing Wnt/beta-Catenin Signaling Pathway and EMT. Am J Chin Med 2020, 48, 703-718. https://doi.org/10.1142/S0192415X20500354. [CrossRef]
- Kasprzak, A. Angiogenesis-Related Functions of Wnt Signaling in Colorectal Carcinogenesis. Cancers (Basel) 2020, 12. https://doi.org/10.3390/cancers12123601. [CrossRef]
- Yang, P.; Yu, D.; Zhou, J.; Zhuang, S.; Jiang, T. TGM2 interference regulates the angiogenesis and apoptosis of colorectal cancer via Wnt/beta-catenin pathway. Cell Cycle 2019, 18, 1122-1134. https://doi.org/10.1080/15384101.2019.1609831. [CrossRef]
- Zhang, J.; Fan, J.; Zeng, X.; Nie, M.; Luan, J.; Wang, Y.; Ju, D.; Yin, K. Hedgehog signaling in gastrointestinal carcinogenesis and the gastrointestinal tumor microenvironment. Acta Pharm Sin B 2021, 11, 609-620. https://doi.org/10.1016/j.apsb.2020.10.022. [CrossRef]
- Li, X.; Xiang, Y.; Li, F.; Yin, C.; Li, B.; Ke, X. WNT/beta-Catenin Signaling Pathway Regulating T Cell-Inflammation in the Tumor Microenvironment. Front Immunol 2019, 10, 2293. https://doi.org/10.3389/fimmu.2019.02293. [CrossRef]
- Flanagan, D.J.; Woodcock, S.A.; Phillips, C.; Eagle, C.; Sansom, O.J. Targeting ligand-dependent wnt pathway dysregulation in gastrointestinal cancers through porcupine inhibition. Pharmacol Ther 2022, 238, 108179. https://doi.org/10.1016/j.pharmthera.2022.108179. [CrossRef]
- Liu, Y.; Qi, X.; Donnelly, L.; Elghobashi-Meinhardt, N.; Long, T.; Zhou, R.W.; Sun, Y.; Wang, B.; Li, X. Mechanisms and inhibition of Porcupine-mediated Wnt acylation. Nature 2022, 607, 816-822. https://doi.org/10.1038/s41586-022-04952-2. [CrossRef]
- Chua, K.; Sim, A.Y.L.; Yeo, E.Y.M.; Bin Masroni, M.S.; Naw, W.W.; Leong, S.M.; Lee, K.W.; Lim, H.J.; Virshup, D.M.; Lee, V.K.M. ETC-159, an Upstream Wnt inhibitor, Induces Tumour Necrosis via Modulation of Angiogenesis in Osteosarcoma. Int J Mol Sci 2023, 24. https://doi.org/10.3390/ijms24054759. [CrossRef]
- Diamond, J.R.; Becerra, C.; Richards, D.; Mita, A.; Osborne, C.; O'Shaughnessy, J.; Zhang, C.; Henner, R.; Kapoun, A.M.; Xu, L.; et al. Phase Ib clinical trial of the anti-frizzled antibody vantictumab (OMP-18R5) plus paclitaxel in patients with locally advanced or metastatic HER2-negative breast cancer. Breast Cancer Res Treat 2020, 184, 53-62. https://doi.org/10.1007/s10549-020-05817-w. [CrossRef]
- Sun, Y.; Wang, W.; Zhao, C. Frizzled Receptors in Tumors, Focusing on Signaling, Roles, Modulation Mechanisms, and Targeted Therapies. Oncol Res 2021, 28, 661-674. https://doi.org/10.3727/096504020X16014648664459. [CrossRef]
- Shirai, F.; Mizutani, A.; Yashiroda, Y.; Tsumura, T.; Kano, Y.; Muramatsu, Y.; Chikada, T.; Yuki, H.; Niwa, H.; Sato, S.; et al. Design and Discovery of an Orally Efficacious Spiroindolinone-Based Tankyrase Inhibitor for the Treatment of Colon Cancer. J Med Chem 2020, 63, 4183-4204. https://doi.org/10.1021/acs.jmedchem.0c00045. [CrossRef]
- Katoh, M. Multi-layered prevention and treatment of chronic inflammation, organ fibrosis and cancer associated with canonical WNT/beta-catenin signaling activation (Review). Int J Mol Med 2018, 42, 713-725. https://doi.org/10.3892/ijmm.2018.3689. [CrossRef]
- Moon, J.H.; Hong, S.W.; Kim, J.E.; Shin, J.S.; Kim, J.S.; Jung, S.A.; Ha, S.H.; Lee, S.; Kim, J.; Lee, D.H.; et al. Targeting beta-catenin overcomes MEK inhibition resistance in colon cancer with KRAS and PIK3CA mutations. Br J Cancer 2019, 120, 941-951. https://doi.org/10.1038/s41416-019-0434-5. [CrossRef]
- He, G.; Nie, J.J.; Liu, X.; Ding, Z.; Luo, P.; Liu, Y.; Zhang, B.W.; Wang, R.; Liu, X.; Hai, Y.; et al. Zinc oxide nanoparticles inhibit osteosarcoma metastasis by downregulating beta-catenin via HIF-1alpha/BNIP3/LC3B-mediated mitophagy pathway. Bioact Mater 2023, 19, 690-702. https://doi.org/10.1016/j.bioactmat.2022.05.006. [CrossRef]
- Braggio, D.A.; Costas, C.d.F.F.; Koller, D.; Jin, F.; Zewdu, A.; Lopez, G.; Batte, K.; Casadei, L.; Welliver, M.; Horrigan, S.K.; et al. Preclinical efficacy of the Wnt/beta-catenin pathway inhibitor BC2059 for the treatment of desmoid tumors. PLoS One 2022, 17, e0276047. https://doi.org/10.1371/journal.pone.0276047. [CrossRef]
- Shah, A.H.; Gami, A.J.; Desai, N.H.; Gandhi, J.S.; Trivedi, P.P. Tumor budding as a prognostic indicator in colorectal carcinoma: a retrospective study of primary colorectal carcinoma cases in a tertiary care center. Indian J Surg Oncol 2022, 13, 459-467. https://doi.org/10.1007/s13193-022-01498-7. [CrossRef]
- Okuyama, K.; Suzuki, K.; Yanamoto, S. Relationship between Tumor Budding and Partial Epithelial-Mesenchymal Transition in Head and Neck Cancer. Cancers (Basel) 2023, 15. https://doi.org/10.3390/cancers15041111. [CrossRef]
- Gao, L.F.; Zhong, Y.; Long, T.; Wang, X.; Zhu, J.X.; Wang, X.Y.; Hu, Z.Y.; Li, Z.G. Tumor bud-derived CCL5 recruits fibroblasts and promotes colorectal cancer progression via CCR5-SLC25A24 signaling. J Exp Clin Cancer Res 2022, 41, 81. https://doi.org/10.1186/s13046-022-02300-w. [CrossRef]
- Moller, T.; James, J.P.; Holmstrom, K.; Sorensen, F.B.; Lindebjerg, J.; Nielsen, B.S. Co-Detection of miR-21 and TNF-alpha mRNA in Budding Cancer Cells in Colorectal Cancer. Int J Mol Sci 2019, 20. https://doi.org/10.3390/ijms20081907. [CrossRef]
- Bradley, C.A.; Dunne, P.D.; Bingham, V.; McQuaid, S.; Khawaja, H.; Craig, S.; James, J.; Moore, W.L.; McArt, D.G.; Lawler, M.; et al. Transcriptional upregulation of c-MET is associated with invasion and tumor budding in colorectal cancer. Oncotarget 2016, 7, 78932-78945. https://doi.org/10.18632/oncotarget.12933. [CrossRef]
- Mielcarska, S.; Kula, A.; Dawidowicz, M.; Kiczmer, P.; Chrabanska, M.; Rynkiewicz, M.; Wziatek-Kuczmik, D.; Swietochowska, E.; Waniczek, D. Assessment of the RANTES Level Correlation and Selected Inflammatory and Pro-Angiogenic Molecules Evaluation of Their Influence on CRC Clinical Features: A Preliminary Observational Study. Medicina (Kaunas) 2022, 58. https://doi.org/10.3390/medicina58020203. [CrossRef]
- Gonzalez, L.O.; Eiro, N.; Fraile, M.; Sanchez, R.; Andicoechea, A.; Fernandez-Francos, S.; Schneider, J.; Vizoso, F.J. Joint Tumor Bud-MMP/TIMP Count at the Invasive Front Improves the Prognostic Evaluation of Invasive Breast Carcinoma. Biomedicines 2021, 9. https://doi.org/10.3390/biomedicines9020196. [CrossRef]
- Zhao, H.; Ming, T.; Tang, S.; Ren, S.; Yang, H.; Liu, M.; Tao, Q.; Xu, H. Wnt signaling in colorectal cancer: pathogenic role and therapeutic target. Mol Cancer 2022, 21, 144. https://doi.org/10.1186/s12943-022-01616-7. [CrossRef]
- Gulley, J.L.; Schlom, J.; Barcellos-Hoff, M.H.; Wang, X.J.; Seoane, J.; Audhuy, F.; Lan, Y.; Dussault, I.; Moustakas, A. Dual inhibition of TGF-beta and PD-L1: a novel approach to cancer treatment. Mol Oncol 2022, 16, 2117-2134. https://doi.org/10.1002/1878-0261.13146. [CrossRef]
- Fendler, A.; Bauer, D.; Busch, J.; Jung, K.; Wulf-Goldenberg, A.; Kunz, S.; Song, K.; Myszczyszyn, A.; Elezkurtaj, S.; Erguen, B.; et al. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat Commun 2020, 11, 929. https://doi.org/10.1038/s41467-020-14700-7. [CrossRef]
- Li, M.; Younis, M.H.; Zhang, Y.; Cai, W.; Lan, X. Clinical summary of fibroblast activation protein inhibitor-based radiopharmaceuticals: cancer and beyond. Eur J Nucl Med Mol Imaging 2022, 49, 2844-2868. https://doi.org/10.1007/s00259-022-05706-y. [CrossRef]
- Huang, C.Y.; Chung, C.L.; Hu, T.H.; Chen, J.J.; Liu, P.F.; Chen, C.L. Recent progress in TGF-beta inhibitors for cancer therapy. Biomed Pharmacother 2021, 134, 111046. https://doi.org/10.1016/j.biopha.2020.111046. [CrossRef]
- Mondal, S.; Adhikari, N.; Banerjee, S.; Amin, S.A.; Jha, T. Matrix metalloproteinase-9 (MMP-9) and its inhibitors in cancer: A minireview. Eur J Med Chem 2020, 194, 112260. https://doi.org/10.1016/j.ejmech.2020.112260. [CrossRef]
- Fuerst, R.; Choi, J.Y.; Knapinska, A.M.; Cameron, M.D.; Ruiz, C.; Delmas, A.; Sundrud, M.S.; Fields, G.B.; Roush, W.R. Development of a putative Zn(2+)-chelating but highly selective MMP-13 inhibitor. Bioorg Med Chem Lett 2022, 76, 129014. https://doi.org/10.1016/j.bmcl.2022.129014. [CrossRef]
- Sanyal, S.; Amin, S.A.; Banerjee, P.; Gayen, S.; Jha, T. A review of MMP-2 structures and binding mode analysis of its inhibitors to strategize structure-based drug design. Bioorg Med Chem 2022, 74, 117044. https://doi.org/10.1016/j.bmc.2022.117044. [CrossRef]
- Secinti, I.E.; Ozgur, T.; Dede, I. PD-L1 Expression in Colorectal Adenocarcinoma Is Associated With the Tumor Immune Microenvironment and Epithelial-Mesenchymal Transition. Am J Clin Pathol 2022, 158, 506-515. https://doi.org/10.1093/ajcp/aqac077. [CrossRef]
- Zhang, L.; Li, Z.; Skrzypczynska, K.M.; Fang, Q.; Zhang, W.; O'Brien, S.A.; He, Y.; Wang, L.; Zhang, Q.; Kim, A.; et al. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell 2020, 181, 442-459 e429. https://doi.org/10.1016/j.cell.2020.03.048. [CrossRef]
- Zhou, W.; Guo, S.; Liu, M.; Burow, M.E.; Wang, G. Targeting CXCL12/CXCR4 Axis in Tumor Immunotherapy. Curr Med Chem 2019, 26, 3026-3041. https://doi.org/10.2174/0929867324666170830111531. [CrossRef]
- Alghamri, M.S.; Banerjee, K.; Mujeeb, A.A.; Mauser, A.; Taher, A.; Thalla, R.; McClellan, B.L.; Varela, M.L.; Stamatovic, S.M.; Martinez-Revollar, G.; et al. Systemic Delivery of an Adjuvant CXCR4-CXCL12 Signaling Inhibitor Encapsulated in Synthetic Protein Nanoparticles for Glioma Immunotherapy. ACS Nano 2022, 16, 8729-8750. https://doi.org/10.1021/acsnano.1c07492. [CrossRef]
- Kobayashi, H.; Choyke, P.L. Near-Infrared Photoimmunotherapy of Cancer. Acc Chem Res 2019, 52, 2332-2339. https://doi.org/10.1021/acs.accounts.9b00273. [CrossRef]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol 2018, 11, 64. https://doi.org/10.1186/s13045-018-0605-5. [CrossRef]
- Ma, Z.; Guo, D.; Wang, Q.; Liu, P.; Xiao, Y.; Wu, P.; Wang, Y.; Chen, B.; Liu, Z.; Liu, Q. Lgr5-mediated p53 Repression through PDCD5 leads to doxorubicin resistance in Hepatocellular Carcinoma. Theranostics 2019, 9, 2967-2983. https://doi.org/10.7150/thno.30562. [CrossRef]
- Xia, L.; Oyang, L.; Lin, J.; Tan, S.; Han, Y.; Wu, N.; Yi, P.; Tang, L.; Pan, Q.; Rao, S.; et al. The cancer metabolic reprogramming and immune response. Mol Cancer 2021, 20, 28. https://doi.org/10.1186/s12943-021-01316-8. [CrossRef]
- Li, Z.; Sun, C.; Qin, Z. Metabolic reprogramming of cancer-associated fibroblasts and its effect on cancer cell reprogramming. Theranostics 2021, 11, 8322-8336. https://doi.org/10.7150/thno.62378. [CrossRef]
- Chifman, J.; Pullikuth, A.; Chou, J.W.; Bedognetti, D.; Miller, L.D. Conservation of immune gene signatures in solid tumors and prognostic implications. BMC Cancer 2016, 16, 911. https://doi.org/10.1186/s12885-016-2948-z. [CrossRef]
- Uddin, M.N.; Li, M.; Wang, X. Identification of Transcriptional Signatures of Colon Tumor Stroma by a Meta-Analysis. J Oncol 2019, 2019, 8752862. https://doi.org/10.1155/2019/8752862. [CrossRef]
- Jasso, G.J.; Jaiswal, A.; Varma, M.; Laszewski, T.; Grauel, A.; Omar, A.; Silva, N.; Dranoff, G.; Porter, J.A.; Mansfield, K.; et al. Colon stroma mediates an inflammation-driven fibroblastic response controlling matrix remodeling and healing. PLoS Biol 2022, 20, e3001532. https://doi.org/10.1371/journal.pbio.3001532. [CrossRef]
- Kim, M.S.; Ha, S.E.; Wu, M.; Zogg, H.; Ronkon, C.F.; Lee, M.Y.; Ro, S. Extracellular Matrix Biomarkers in Colorectal Cancer. Int J Mol Sci 2021, 22. https://doi.org/10.3390/ijms22179185. [CrossRef]
- Sandberg, T.P.; Stuart, M.; Oosting, J.; Tollenaar, R.; Sier, C.F.M.; Mesker, W.E. Increased expression of cancer-associated fibroblast markers at the invasive front and its association with tumor-stroma ratio in colorectal cancer. BMC Cancer 2019, 19, 284. https://doi.org/10.1186/s12885-019-5462-2. [CrossRef]
- Stefani, C.; Miricescu, D.; Stanescu, S., II; Nica, R.I.; Greabu, M.; Totan, A.R.; Jinga, M. Growth Factors, PI3K/AKT/mTOR and MAPK Signaling Pathways in Colorectal Cancer Pathogenesis: Where Are We Now? Int J Mol Sci 2021, 22. https://doi.org/10.3390/ijms221910260. [CrossRef]
- Kobayashi, H.; Gieniec, K.A.; Lannagan, T.R.M.; Wang, T.; Asai, N.; Mizutani, Y.; Iida, T.; Ando, R.; Thomas, E.M.; Sakai, A.; et al. The Origin and Contribution of Cancer-Associated Fibroblasts in Colorectal Carcinogenesis. Gastroenterology 2022, 162, 890-906. https://doi.org/10.1053/j.gastro.2021.11.037. [CrossRef]
- Liang, Y.; Lv, Z.; Huang, G.; Qin, J.; Li, H.; Nong, F.; Wen, B. Prognostic significance of abnormal matrix collagen remodeling in colorectal cancer based on histologic and bioinformatics analysis. Oncol Rep 2020, 44, 1671-1685. https://doi.org/10.3892/or.2020.7729. [CrossRef]
- Lindgren, M.; Rask, G.; Jonsson, J.; Berglund, A.; Lundin, C.; Jonsson, P.; Ljuslinder, I.; Nystrom, H. Type IV Collagen in Human Colorectal Liver Metastases-Cellular Origin and a Circulating Biomarker. Cancers (Basel) 2022, 14. https://doi.org/10.3390/cancers14143396. [CrossRef]
- Buttacavoli, M.; Di Cara, G.; Roz, E.; Pucci-Minafra, I.; Feo, S.; Cancemi, P. Integrated Multi-Omics Investigations of Metalloproteinases in Colon Cancer: Focus on MMP2 and MMP9. Int J Mol Sci 2021, 22. https://doi.org/10.3390/ijms222212389. [CrossRef]
- Cui, G.; Cai, F.; Ding, Z.; Gao, L. MMP14 predicts a poor prognosis in patients with colorectal cancer. Hum Pathol 2019, 83, 36-42. https://doi.org/10.1016/j.humpath.2018.03.030. [CrossRef]
- Tian, L.; Zhao, Z.F.; Xie, L.; Zhu, J.P. Taurine up-regulated 1 accelerates tumorigenesis of colon cancer by regulating miR-26a-5p/MMP14/p38 MAPK/Hsp27 axis in vitro and in vivo. Life Sci 2019, 239, 117035. https://doi.org/10.1016/j.lfs.2019.117035. [CrossRef]
- Xuefeng, X.; Hou, M.X.; Yang, Z.W.; Agudamu, A.; Wang, F.; Su, X.L.; Li, X.; Shi, L.; Terigele, T.; Bao, L.L.; et al. Epithelial-mesenchymal transition and metastasis of colon cancer cells induced by the FAK pathway in cancer-associated fibroblasts. J Int Med Res 2020, 48, 300060520931242. https://doi.org/10.1177/0300060520931242. [CrossRef]
- Hu, L.; Wang, J.; Wang, Y.; Wu, L.; Wu, C.; Mao, B.; Maruthi Prasad, E.; Wang, Y.; Chin, Y.E. LOXL1 modulates the malignant progression of colorectal cancer by inhibiting the transcriptional activity of YAP. Cell Commun Signal 2020, 18, 148. https://doi.org/10.1186/s12964-020-00639-1. [CrossRef]
- Wang, J.; Li, R.; Li, M.; Wang, C. Fibronectin and colorectal cancer: signaling pathways and clinical implications. J Recept Signal Transduct Res 2021, 41, 313-320. https://doi.org/10.1080/10799893.2020.1817074. [CrossRef]
- Zhang, Z.; Fang, C.; Wang, Y.; Zhang, J.; Yu, J.; Zhang, Y.; Wang, X.; Zhong, J. COL1A1: A potential therapeutic target for colorectal cancer expressing wild-type or mutant KRAS. Int J Oncol 2018, 53, 1869-1880. https://doi.org/10.3892/ijo.2018.4536. [CrossRef]
- Kamali Zonouzi, S.; Pezeshki, P.S.; Razi, S.; Rezaei, N. Cancer-associated fibroblasts in colorectal cancer. Clin Transl Oncol 2022, 24, 757-769. https://doi.org/10.1007/s12094-021-02734-2. [CrossRef]
- Mortezapour, M.; Tapak, L.; Bahreini, F.; Najafi, R.; Afshar, S. Identification of key genes in colorectal cancer diagnosis by co-expression analysis weighted gene co-expression network analysis. Comput Biol Med 2023, 157, 106779. https://doi.org/10.1016/j.compbiomed.2023.106779. [CrossRef]
- Thibaudin, M.; Limagne, E.; Hampe, L.; Ballot, E.; Truntzer, C.; Ghiringhelli, F. Targeting PD-L1 and TIGIT could restore intratumoral CD8 T cell function in human colorectal cancer. Cancer Immunol Immunother 2022, 71, 2549-2563. https://doi.org/10.1007/s00262-022-03182-9. [CrossRef]
- Yang, Z.; Wu, G.; Zhang, X.; Gao, J.; Meng, C.; Liu, Y.; Wei, Q.; Sun, L.; Wei, P.; Bai, Z.; et al. Current progress and future perspectives of neoadjuvant anti-PD-1/PD-L1 therapy for colorectal cancer. Front Immunol 2022, 13, 1001444. https://doi.org/10.3389/fimmu.2022.1001444. [CrossRef]
- Zeynep, O.; Funda, C.; Evrim, Y.; Deniz, A.; Bulent, Y.; Fatih, Y.N. PD-L1 and PD-L2 expression in colorectal cancer. Indian J Pathol Microbiol 2023, 66, 31-37. https://doi.org/10.4103/ijpm.ijpm_814_21. [CrossRef]
- Cheruku, S.; Rao, V.; Pandey, R.; Rao Chamallamudi, M.; Velayutham, R.; Kumar, N. Tumor-associated macrophages employ immunoediting mechanisms in colorectal tumor progression: Current research in Macrophage repolarization immunotherapy. Int Immunopharmacol 2023, 116, 109569. https://doi.org/10.1016/j.intimp.2022.109569. [CrossRef]
- Liu, J.N.; Kong, X.S.; Huang, T.; Wang, R.; Li, W.; Chen, Q.F. Clinical Implications of Aberrant PD-1 and CTLA4 Expression for Cancer Immunity and Prognosis: A Pan-Cancer Study. Front Immunol 2020, 11, 2048. https://doi.org/10.3389/fimmu.2020.02048. [CrossRef]
- Chen, J.; Ye, X.; Pitmon, E.; Lu, M.; Wan, J.; Jellison, E.R.; Adler, A.J.; Vella, A.T.; Wang, K. IL-17 inhibits CXCL9/10-mediated recruitment of CD8(+) cytotoxic T cells and regulatory T cells to colorectal tumors. J Immunother Cancer 2019, 7, 324. https://doi.org/10.1186/s40425-019-0757-z. [CrossRef]
- Shang, S.; Yang, Y.W.; Chen, F.; Yu, L.; Shen, S.H.; Li, K.; Cui, B.; Lv, X.X.; Zhang, C.; Yang, C.; et al. TRIB3 reduces CD8(+) T cell infiltration and induces immune evasion by repressing the STAT1-CXCL10 axis in colorectal cancer. Sci Transl Med 2022, 14, eabf0992. https://doi.org/10.1126/scitranslmed.abf0992. [CrossRef]
- Bess, S.N.; Greening, G.J.; Rajaram, N.; Muldoon, T.J. Macrophage-targeted anti-CCL2 immunotherapy enhances tumor sensitivity to 5-fluorouracil in a Balb/c-CT26 murine colon carcinoma model measured using diffuse reflectance spectroscopy. BMC Immunol 2022, 23, 20. https://doi.org/10.1186/s12865-022-00493-5. [CrossRef]
- Li, J.; Yang, J.; Xing, R.; Wang, Y. A novel inflammation-related signature for predicting prognosis and characterizing the tumor microenvironment in colorectal cancer. Aging (Albany NY) 2023, 15, 2554-2581. https://doi.org/10.18632/aging.204630. [CrossRef]
- Soylemez, Z.; Arikan, E.S.; Solak, M.; Arikan, Y.; Tokyol, C.; Seker, H. Investigation of the expression levels of CPEB4, APC, TRIP13, EIF2S3, EIF4A1, IFNg, PIK3CA and CTNNB1 genes in different stage colorectal tumors. Turk J Med Sci 2021, 51, 661-674. https://doi.org/10.3906/sag-2010-18. [CrossRef]
- Wang, R.; Ma, Y.; Zhan, S.; Zhang, G.; Cao, L.; Zhang, X.; Shi, T.; Chen, W. B7-H3 promotes colorectal cancer angiogenesis through activating the NF-kappaB pathway to induce VEGFA expression. Cell Death Dis 2020, 11, 55. https://doi.org/10.1038/s41419-020-2252-3. [CrossRef]
- Chen, C.; Huang, Z.; Mo, X.; Song, Y.; Li, X.; Li, X.; Zhang, M. The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J Exp Clin Cancer Res 2020, 39, 91. https://doi.org/10.1186/s13046-020-01594-y. [CrossRef]
- Chen, C.; Huang, Z.; Mo, X.; Song, Y.; Li, X.; Li, X.; Zhang, M. Retraction Note: The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J Exp Clin Cancer Res 2022, 41, 114. https://doi.org/10.1186/s13046-022-02342-0. [CrossRef]
- Zhong, M.; Li, N.; Qiu, X.; Ye, Y.; Chen, H.; Hua, J.; Yin, P.; Zhuang, G. TIPE regulates VEGFR2 expression and promotes angiogenesis in colorectal cancer. Int J Biol Sci 2020, 16, 272-283. https://doi.org/10.7150/ijbs.37906. [CrossRef]
- Cai, X.; Wei, B.; Li, L.; Chen, X.; Yang, J.; Li, X.; Jiang, X.; Lv, M.; Li, M.; Lin, Y.; et al. Therapeutic Potential of Apatinib Against Colorectal Cancer by Inhibiting VEGFR2-Mediated Angiogenesis and beta-Catenin Signaling. Onco Targets Ther 2020, 13, 11031-11044. https://doi.org/10.2147/OTT.S266549. [CrossRef]
- Gaibar, M.; Galan, M.; Romero-Lorca, A.; Anton, B.; Malon, D.; Moreno, A.; Fernandez-Santander, A.; Novillo, A. Genetic Variants of ANGPT1, CD39, FGF2 and MMP9 Linked to Clinical Outcome of Bevacizumab Plus Chemotherapy for Metastatic Colorectal Cancer. Int J Mol Sci 2021, 22. https://doi.org/10.3390/ijms22031381. [CrossRef]
- Munakata, S.; Ueyama, T.; Ishihara, H.; Komiyama, H.; Tsukamoto, R.; Kawai, M.; Takahashi, M.; Kojima, Y.; Tomiki, Y.; Sakamoto, K. Angiopoietin-2 as a Prognostic Factor in Patients with Incurable Stage IV Colorectal Cancer. J Gastrointest Cancer 2021, 52, 237-242. https://doi.org/10.1007/s12029-020-00392-1. [CrossRef]
- Jary, M.; Hasanova, R.; Vienot, A.; Asgarov, K.; Loyon, R.; Tirole, C.; Bouard, A.; Orillard, E.; Klajer, E.; Kim, S.; et al. Molecular description of ANGPT2 associated colorectal carcinoma. Int J Cancer 2020, 147, 2007-2018. https://doi.org/10.1002/ijc.32993. [CrossRef]
- Wang, J.; Yu, S.; Chen, G.; Kang, M.; Jin, X.; Huang, Y.; Lin, L.; Wu, D.; Wang, L.; Chen, J. A novel prognostic signature of immune-related genes for patients with colorectal cancer. J Cell Mol Med 2020, 24, 8491-8504. https://doi.org/10.1111/jcmm.15443. [CrossRef]
- Caiado, H.; Conceicao, N.; Tiago, D.; Marreiros, A.; Vicente, S.; Enriquez, J.L.; Vaz, A.M.; Antunes, A.; Guerreiro, H.; Caldeira, P.; et al. Data on the evaluation of FGF2 gene expression in Colorectal Cancer. Data Brief 2020, 31, 105765. https://doi.org/10.1016/j.dib.2020.105765. [CrossRef]
- Cheng, X.; Jin, Z.; Ji, X.; Shen, X.; Feng, H.; Morgenlander, W.; Ou, B.; Wu, H.; Gao, H.; Ye, F.; et al. ETS variant 5 promotes colorectal cancer angiogenesis by targeting platelet-derived growth factor BB. Int J Cancer 2019, 145, 179-191. https://doi.org/10.1002/ijc.32071. [CrossRef]
- Sae-Lim, S.; Soontornworajit, B.; Rotkrua, P. Inhibition of Colorectal Cancer Cell Proliferation by Regulating Platelet-Derived Growth Factor B Signaling with a DNA Aptamer. Asian Pac J Cancer Prev 2019, 20, 487-494. https://doi.org/10.31557/APJCP.2019.20.2.487. [CrossRef]
- Zheng, Y.; Fu, Y.; Wang, P.P.; Ding, Z.Y. Neoantigen: A Promising Target for the Immunotherapy of Colorectal Cancer. Dis Markers 2022, 2022, 8270305. https://doi.org/10.1155/2022/8270305. [CrossRef]
- Gilazieva, Z.; Ponomarev, A.; Rizvanov, A.; Solovyeva, V. The Dual Role of Mesenchymal Stromal Cells and Their Extracellular Vesicles in Carcinogenesis. Biology (Basel) 2022, 11. https://doi.org/10.3390/biology11060813. [CrossRef]
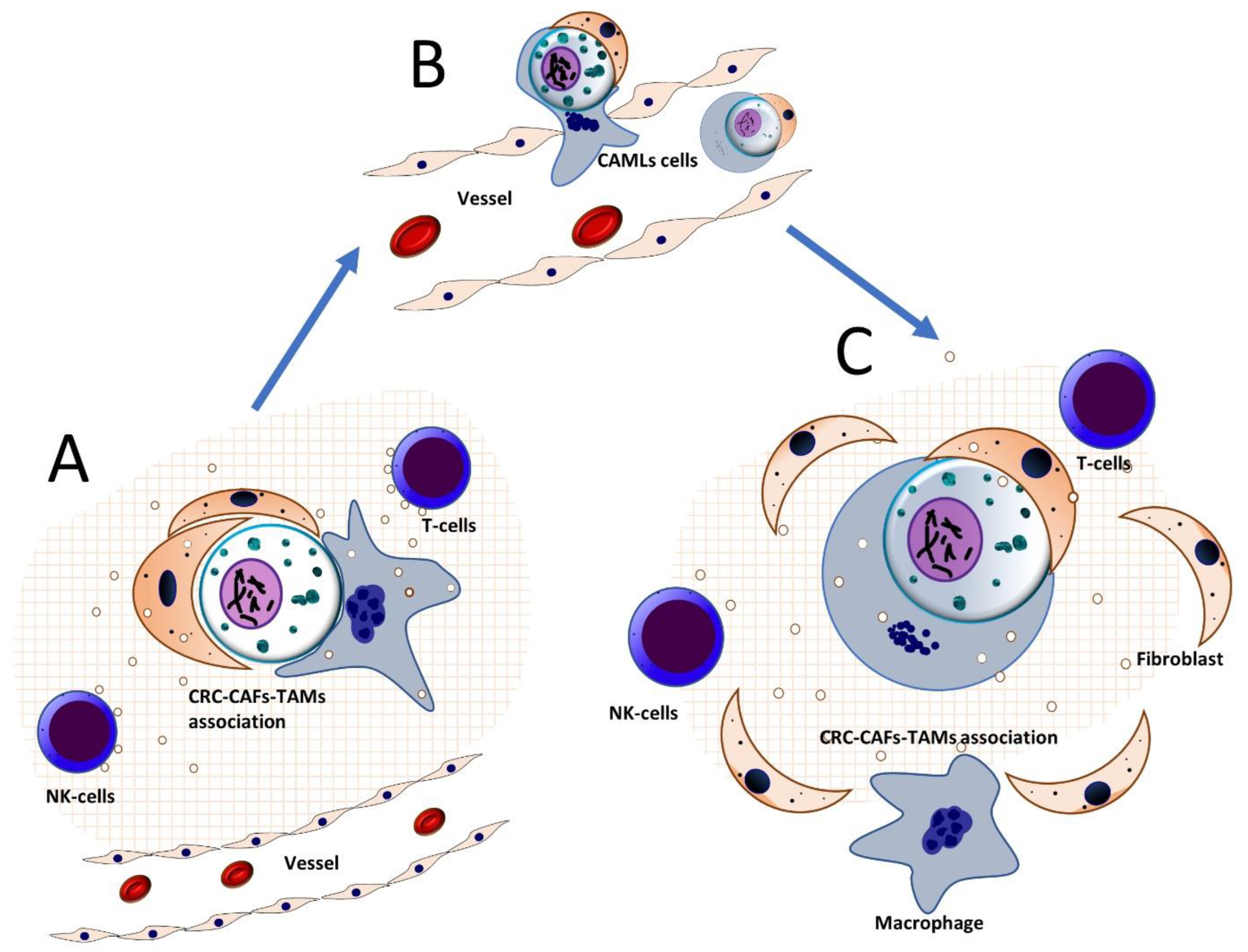
- Currais, P.; Rosa, I.; Claro, I. Colorectal cancer carcinogenesis: From bench to bedside. World J Gastrointest Oncol 2022, 14, 654-663. https://doi.org/10.4251/wjgo.v14.i3.654. [CrossRef]
- Tang, C.M.; Adams, D.L. Clinical Applications of Cancer-Associated Cells Present in the Blood of Cancer Patients. Biomedicines 2022, 10. https://doi.org/10.3390/biomedicines10030587. [CrossRef]
- Kasabwala, D.M.; Bergan, R.C.; Gardner, K.P.; Lapidus, R.; Tsai, S.; Aldakkak, M.; Adams, D.L. Micronuclei in Circulating Tumor Associated Macrophages Predicts Progression in Advanced Colorectal Cancer. Biomedicines 2022, 10. https://doi.org/10.3390/biomedicines10112898. [CrossRef]
- Sutton, T.L.; Patel, R.K.; Anderson, A.N.; Bowden, S.G.; Whalen, R.; Giske, N.R.; Wong, M.H. Circulating Cells with Macrophage-like Characteristics in Cancer: The Importance of Circulating Neoplastic-Immune Hybrid Cells in Cancer. Cancers (Basel) 2022, 14. https://doi.org/10.3390/cancers14163871. [CrossRef]
| Pro-Cancer Bacterias | Anti-Cancer Bacterial Species |
|---|---|
| Fusobacterium nucleatum [5,6] | Bifidobacterium longum [13] |
| Bacteroides fragilis [14] | Lactobacillus acidophilus [15,16] |
| Enterococcus faecalis [17] | Lactobacillus rhamnosus [16,18] |
| Escherichia coli [19] | Faecalibacterium prausnitzii [20] |
| Peptostreptococcus anaerobius [21] | Bifidobacterium breve [13] |
| Streptococcus gallolyticus [22] | Lactobacillus reuteri [16] |
| Clostridium septicum [23] | Bifidobacterium adolescentis [13] |
| Ruminococcus gnavus [13] | Lactobacillus plantarum [16] |
| Gene | Full Name | Role in Colorectal Cancer Stroma | References |
|---|---|---|---|
| MMP2 | Matrix Metalloproteinase 2 | ECM remodeling, degrades various ECM components, facilitates tumor cell invasion and metastasis. | [118,119] |
| MMP9 | Matrix Metalloproteinase 9 | ECM remodeling, degrades collagen and other ECM components, promotes tumor cell invasion, supports angiogenesis. | [118,119] |
| MMP14 | Matrix Metalloproteinase 14 | ECM remodeling, involved in the cleavage of cell surface proteins and the breakdown of ECM components, promotes tumor invasion and angiogenesis. | [120,121] |
| LOX | Lysyl Oxidase | ECM remodeling, catalyzes the cross-linking of collagens and elastin, contributes to the stiffening of the tumor microenvironment and promotes tumor progression. | [122,123] |
| FN1 | Fibronectin | ECM remodeling, involved in cell adhesion, migration, and proliferation; its increased expression is associated with tumor progression and poor prognosis in colorectal cancer. | [124] |
| COL1A1 | Collagen Type I Alpha 1 Chain | ECM remodeling, major structural component of the ECM, its increased expression is associated with tumor progression and poor prognosis in colorectal cancer. | [125] |
| COL3A1 | Collagen Type III Alpha 1 Chain | ECM remodeling, another structural component of the ECM, its increased expression is associated with tumor progression and poor prognosis in colorectal cancer. | [126] |
| COL5A1 | Collagen Type V Alpha 1 Chain | ECM remodeling, another structural component of the ECM, its increased expression is associated with tumor progression and poor prognosis in colorectal cancer. | [127] |
| CD274 | Programmed Death-Ligand 1 (PD-L1) | Immune checkpoint molecule, inhibits T cell activation, promotes immune evasion by tumor cells. | [128-130] |
| PDCD1 | Programmed Cell Death Protein 1 (PD-1) | Immune checkpoint receptor, dampens immune response, allows tumor cells to escape immune surveillance. | [128-130] |
| CTLA4 | Cytotoxic T-Lymphocyte-Associated Protein 4 | Immune checkpoint receptor, inhibits T cell activation, contributes to immune evasion by tumor cells. | [131,132] |
| CXCL9 | Chemokine (C-X-C motif) Ligand 9 | Recruits immune cells, such as T cells and natural killer cells, to the tumor microenvironment; enhanced anti-tumor immunity. | [133] |
| CXCL10 | Chemokine (C-X-C motif) Ligand 10 | Recruits immune cells, such as T cells and natural killer cells, to the tumor microenvironment; enhanced anti-tumor immunity. | [133,134] |
| CCL2 | Chemokine (C-C motif) Ligand 2 | Recruitment of monocytes, macrophages, and other immune cells to the tumor site; altered expression associated with immune cell infiltration and tumor progression. | [135,136] |
| IFNG | Interferon Gamma | Activates and modulates immune response against tumor cells, affects expression of immune checkpoint molecules and other immune-related genes. | [137] |
| VEGFA | Vascular Endothelial Growth Factor A | Promotes growth of new blood vessels from existing vasculature, stimulates endothelial cell proliferation, migration, and survival. | [138-140] |
| VEGFR2 | Vascular Endothelial Growth Factor Receptor 2 (KDR) | Primary receptor for VEGFA on endothelial cells, activation by VEGFA leads to a signaling cascade promoting angiogenesis and vascular permeability. | [141,142] |
| ANGPT1 | Angiopoietin-1 | Regulates angiogenesis by binding to the endothelial cell receptor tyrosine kinase, Tie2, promotes vessel maturation and stability. | [143,144] |
| ANGPT2 | Angiopoietin-2 | Acts as an antagonist of ANGPT1, binds to Tie2, promotes vessel destabilization and sprouting angiogenesis. | [144,145] |
| FGF2 | Fibroblast Growth Factor 2 (bFGF) | Regulates angiogenesis, stimulates endothelial cell proliferation, migration, and differentiation, acts synergistically with VEGFA to promote blood vessel formation. | [146,147] |
| PDGFB | Platelet-Derived Growth Factor B | Promotes recruitment of pericytes to newly formed blood vessels, essential for blood vessel maturation and stabilization. | [148,149] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





