Submitted:
22 April 2023
Posted:
23 April 2023
You are already at the latest version
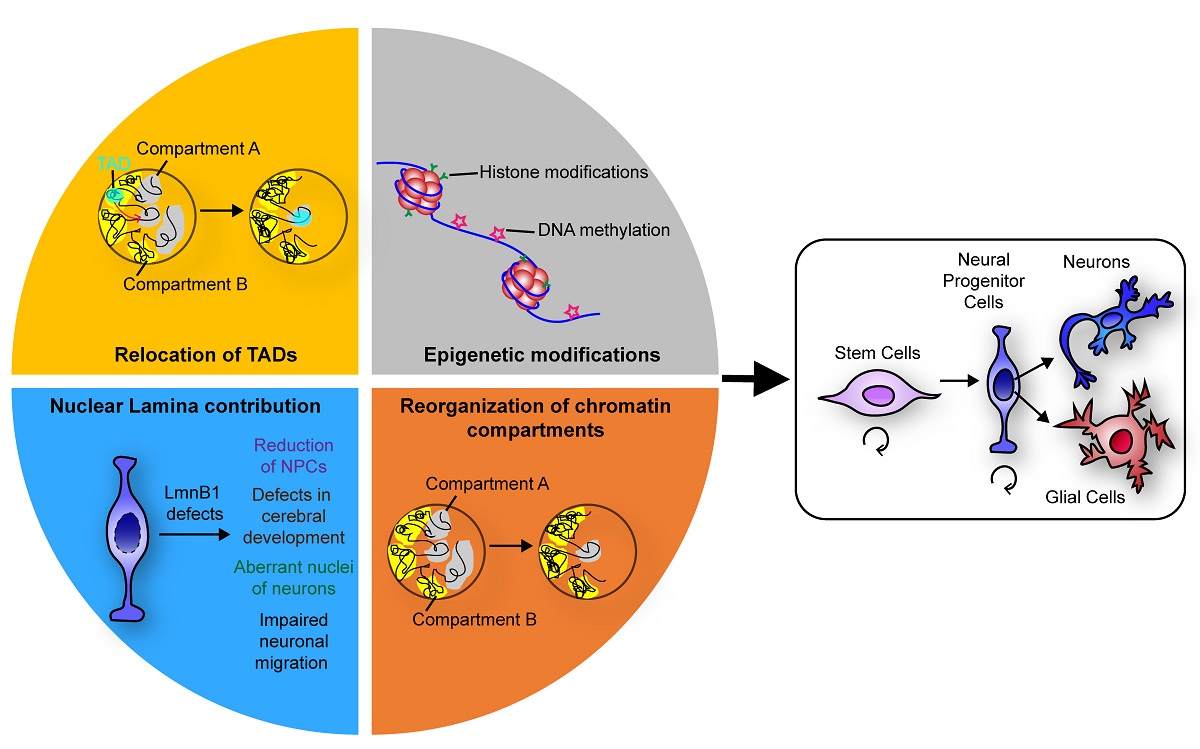
Abstract
Keywords:
1. Introduction
2. Three-dimensional organization of chromatin within the nucleus
3. Nuclear envelope: LINC complex and nuclear lamins
4. Neurogenesis: an overview
5. Chromatin structure involvement in neural development
6. Implication of nuclear lamina in neuronal development
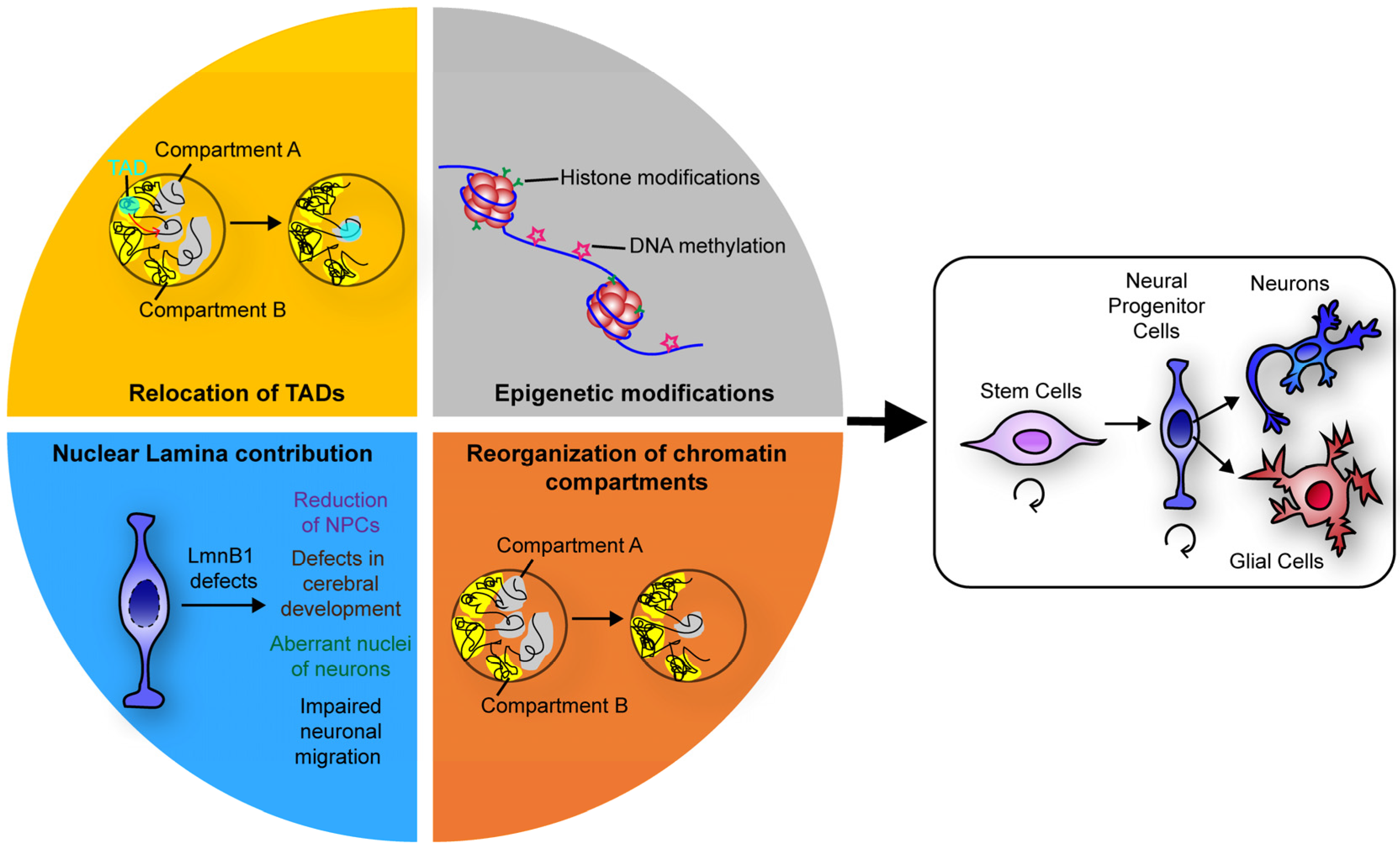
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carollo, P.S.; Barra, V. Chromatin Epigenetics and Nuclear Lamina Keep the Nucleus in Shape: Examples from Natural and Accelerated Aging. Biol. Cell 2023, 115, 2200023. [Google Scholar] [CrossRef] [PubMed]
- Maeshima, K.; Ide, S.; Babokhov, M. Dynamic Chromatin Organization without the 30-Nm Fiber. Curr. Opin. Cell Biol. 2019, 58, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Cubeñas-Potts, C.; Corces, V.G. Architectural Proteins, Transcription, and the Three-Dimensional Organization of the Genome. FEBS Lett. 2015, 589, 2923–2930. [Google Scholar] [CrossRef] [PubMed]
- Nora, E.P.; Lajoie, B.R.; Schulz, E.G.; Giorgetti, L.; Okamoto, I.; Servant, N.; Piolot, T.; van Berkum, N.L.; Meisig, J.; Sedat, J.; et al. Spatial Partitioning of the Regulatory Landscape of the X-Inactivation Centre. Nature 2012, 485, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Dixon, J.R.; Gorkin, D.U.; Ren, B. Chromatin Domains: The Unit of Chromosome Organization. Mol. Cell 2016, 62, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Bridger, J.M.; Foeger, N.; Kill, I.R.; Herrmann, H. The Nuclear Lamina. FEBS J. 2007, 274, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, M.; Dietzel, S.; Müller, S.; Solovei, I.; Fakan, S. Chromosome Territories – a Functional Nuclear Landscape. Curr. Opin. Cell Biol. 2006, 18, 307–316. [Google Scholar] [CrossRef]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef]
- Barra, V.; Chiavetta, R.F.; Titoli, S.; Provenzano, I.M.; Carollo, P.S.; Di Leonardo, A. Specific Irreversible Cell-Cycle Arrest and Depletion of Cancer Cells Obtained by Combining Curcumin and the Flavonoids Quercetin and Fisetin. Genes 2022, 13, 1125. [Google Scholar] [CrossRef]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain Organization of Human Chromosomes Revealed by Mapping of Nuclear Lamina Interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Ruthenburg, A.J.; Allis, C.D.; Wysocka, J. Methylation of Lysine 4 on Histone H3: Intricacy of Writing and Reading a Single Epigenetic Mark. Mol. Cell 2007, 25, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Pappalardo, X.G.; Barra, V. Losing DNA Methylation at Repetitive Elements and Breaking Bad. Epigenetics Chromatin 2021, 14, 25. [Google Scholar] [CrossRef] [PubMed]
- Maurer, M.; Lammerding, J. The Driving Force: Nuclear Mechanotransduction in Cellular Function, Fate, and Disease. Annu. Rev. Biomed. Eng. 2019, 21, 443–468. [Google Scholar] [CrossRef] [PubMed]
- Crisp, M.; Liu, Q.; Roux, K.; Rattner, J.B.; Shanahan, C.; Burke, B.; Stahl, P.D.; Hodzic, D. Coupling of the Nucleus and Cytoplasm: Role of the LINC Complex. J. Cell Biol. 2006, 172, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Starr, D.A.; Han, M. Role of ANC-1 in Tethering Nuclei to the Actin Cytoskeleton. Science 2002, 298, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Skepper, J.N.; Yang, F.; Davies, J.D.; Hegyi, L.; Roberts, R.G.; Weissberg, P.L.; Ellis, J.A.; Shanahan, C.M. Nesprins: A Novel Family of Spectrin-Repeat-Containing Proteins That Localize to the Nuclear Membrane in Multiple Tissues. J. Cell Sci. 2001, 114, 4485–4498. [Google Scholar] [CrossRef]
- Lygerou, Z.; Christophides, G.; Séraphin, B. A Novel Genetic Screen for SnRNP Assembly Factors in Yeast Identifies a Conserved Protein, Sad1p, Also Required for Pre-MRNA Splicing. Mol. Cell. Biol. 1999, 19, 2008–2020. [Google Scholar] [CrossRef]
- Malone, C.J.; Fixsen, W.D.; Horvitz, H.R.; Han, M. UNC-84 Localizes to the Nuclear Envelope and Is Required for Nuclear Migration and Anchoring during C. Elegans Development. Dev. Camb. Engl. 1999, 126, 3171–3181. [Google Scholar] [CrossRef]
- Guarda, A.; Bolognese, F.; Bonapace, I.M.; Badaracco, G. Interaction between the Inner Nuclear Membrane Lamin B Receptor and the Heterochromatic Methyl Binding Protein, MeCP2. Exp. Cell Res. 2009, 315, 1895–1903. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Callebaut, I.; Pezhman, A.; Courvalin, J.-C.; Worman, H.J. Domain-Specific Interactions of Human HP1-Type Chromodomain Proteins and Inner Nuclear Membrane Protein LBR *. J. Biol. Chem. 1997, 272, 14983–14989. [Google Scholar] [CrossRef] [PubMed]
- Makhija, E.; Jokhun, D.S.; Shivashankar, G.V. Nuclear Deformability and Telomere Dynamics Are Regulated by Cell Geometric Constraints. Proc. Natl. Acad. Sci. 2016, 113, E32–E40. [Google Scholar] [CrossRef] [PubMed]
- Alisafaei, F.; Jokhun, D.S.; Shivashankar, G.V.; Shenoy, V.B. Regulation of Nuclear Architecture, Mechanics, and Nucleocytoplasmic Shuttling of Epigenetic Factors by Cell Geometric Constraints. Proc. Natl. Acad. Sci. 2019, 116, 13200–13209. [Google Scholar] [CrossRef]
- Jain, N.; Iyer, K.V.; Kumar, A.; Shivashankar, G.V. Cell Geometric Constraints Induce Modular Gene-Expression Patterns via Redistribution of HDAC3 Regulated by Actomyosin Contractility. Proc. Natl. Acad. Sci. 2013, 110, 11349–11354. [Google Scholar] [CrossRef] [PubMed]
- Stiles, J.; Jernigan, T.L. The Basics of Brain Development. Neuropsychol. Rev. 2010, 20, 327–348. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Christian, K.M.; He, C.; Jin, P.; Ming, G.-L.; Song, H. Epigenetic Mechanisms in Neurogenesis. Nat. Rev. Neurosci. 2016, 17, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Nothof, S.A.; Magdinier, F.; Van-Gils, J. Chromatin Structure and Dynamics: Focus on Neuronal Differentiation and Pathological Implication. Genes 2022, 13, 639. [Google Scholar] [CrossRef]
- Golob, J.L.; Paige, S.L.; Muskheli, V.; Pabon, L.; Murry, C.E. Chromatin Remodeling during Mouse and Human Embryonic Stem Cell Differentiation. Dev. Dyn. 2008, 237, 1389–1398. [Google Scholar] [CrossRef]
- Fujita, Y.; Yamashita, T. Spatial Organization of Genome Architecture in Neuronal Development and Disease. Neurochem. Int. 2018, 119, 49–56. [Google Scholar] [CrossRef]
- Aoto, T.; Saitoh, N.; Ichimura, T.; Niwa, H.; Nakao, M. Nuclear and Chromatin Reorganization in the MHC-Oct3/4 Locus at Developmental Phases of Embryonic Stem Cell Differentiation. Dev. Biol. 2006, 298, 354–367. [Google Scholar] [CrossRef] [PubMed]
- Le Gros, M.A.; Clowney, E.J.; Magklara, A.; Yen, A.; Markenscoff-Papadimitriou, E.; Colquitt, B.; Myllys, M.; Kellis, M.; Lomvardas, S.; Larabell, C.A. Soft X-Ray Tomography Reveals Gradual Chromatin Compaction and Reorganization during Neurogenesis in Vivo. Cell Rep. 2016, 17, 2125–2136. [Google Scholar] [CrossRef] [PubMed]
- Meshorer, E.; Yellajoshula, D.; George, E.; Scambler, P.J.; Brown, D.T.; Misteli, T. Hyperdynamic Plasticity of Chromatin Proteins in Pluripotent Embryonic Stem Cells. Dev. Cell 2006, 10, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Billia, F.; Baskys, A.; Carlen, P.L.; De Boni, U. Rearrangement of Centromeric Satellite DNA in Hippocampal Neurons Exhibiting Long-Term Potentiation. Mol. Brain Res. 1992, 14, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Solovei, I.; Grandi, N.; Knoth, R.; Volk, B.; Cremer, T. Positional Changes of Pericentromeric Heterochromatin and Nucleoli in Postmitotic Purkinje Cells during Murine Cerebellum Development. Cytogenet. Genome Res. 2004, 105, 302–310. [Google Scholar] [CrossRef]
- Solovei, I.; Kreysing, M.; Lanctôt, C.; Kösem, S.; Peichl, L.; Cremer, T.; Guck, J.; Joffe, B. Nuclear Architecture of Rod Photoreceptor Cells Adapts to Vision in Mammalian Evolution. Cell 2009, 137, 356–368. [Google Scholar] [CrossRef]
- Clowney, E.J.; LeGros, M.A.; Mosley, C.P.; Clowney, F.G.; Markenskoff-Papadimitriou, E.C.; Myllys, M.; Barnea, G.; Larabell, C.A.; Lomvardas, S. Nuclear Aggregation of Olfactory Receptor Genes Governs Their Monogenic Expression. Cell 2012, 151, 724–737. [Google Scholar] [CrossRef]
- Kishi, Y.; Kondo, S.; Gotoh, Y. Transcriptional Activation of Mouse Major Satellite Regions during Neuronal Differentiation. Cell Struct. Funct. 2012, 37, 101–110. [Google Scholar] [CrossRef]
- Fraser, J.; Ferrai, C.; Chiariello, A.M.; Schueler, M.; Rito, T.; Laudanno, G.; Barbieri, M.; Moore, B.L.; Kraemer, D.C.A.; Aitken, S.; et al. Hierarchical Folding and Reorganization of Chromosomes Are Linked to Transcriptional Changes in Cellular Differentiation. Mol. Syst. Biol. 2015, 11, 852. [Google Scholar] [CrossRef]
- Decker, B.; Liput, M.; Abdellatif, H.; Yergeau, D.; Bae, Y.; Jornet, J.M.; Stachowiak, E.K.; Stachowiak, M.K. Global Genome Conformational Programming during Neuronal Development Is Associated with CTCF and Nuclear FGFR1—The Genome Archipelago Model. Int. J. Mol. Sci. 2020, 22, 347. [Google Scholar] [CrossRef]
- Stachowiak, M.K.; Stachowiak, E.K. Evidence-Based Theory for Integrated Genome Regulation of Ontogeny--An Unprecedented Role of Nuclear FGFR1 Signaling. J. Cell. Physiol. 2016, 231, 1199–1218. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shin, J.-Y.; Nakanishi, K.; Homma, S.; Kim, G.J.; Tanji, K.; Joseph, L.C.; Morrow, J.P.; Stewart, C.L.; Dauer, W.T.; et al. Postnatal Development of Mice with Combined Genetic Depletions of Lamin A/C, Emerin and Lamina-Associated Polypeptide 1. Hum. Mol. Genet. 2019, 28, 2486–2500. [Google Scholar] [CrossRef] [PubMed]
- Lochs, S.J.A.; Kefalopoulou, S.; Kind, J. Lamina Associated Domains and Gene Regulation in Development and Cancer. Cells 2019, 8, 271. [Google Scholar] [CrossRef] [PubMed]
- Peric-Hupkes, D.; Meuleman, W.; Pagie, L.; Bruggeman, S.W.M.; Solovei, I.; Brugman, W.; Gräf, S.; Flicek, P.; Kerkhoven, R.M.; van Lohuizen, M.; et al. Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions during Differentiation. Mol. Cell 2010, 38, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Shevelyov, Y.Y.; Ulianov, S.V. The Nuclear Lamina as an Organizer of Chromosome Architecture. Cells 2019, 8, 136. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, I.; Kind, J. Spatial Chromatin Organization and Gene Regulation at the Nuclear Lamina. Curr. Opin. Genet. Dev. 2019, 55, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.R.E.; Azuara, V.; Perry, P.; Sauer, S.; Dvorkina, M.; Jørgensen, H.; Roix, J.; McQueen, P.; Misteli, T.; Merkenschlager, M.; et al. Neural Induction Promotes Large-Scale Chromatin Reorganisation of the Mash1 Locus. J. Cell Sci. 2006, 119, 132–140. [Google Scholar] [CrossRef]
- Coffinier, C.; Jung, H.-J.; Nobumori, C.; Chang, S.; Tu, Y.; Barnes, R.H.; Yoshinaga, Y.; de Jong, P.J.; Vergnes, L.; Reue, K.; et al. Deficiencies in Lamin B1 and Lamin B2 Cause Neurodevelopmental Defects and Distinct Nuclear Shape Abnormalities in Neurons. Mol. Biol. Cell 2011, 22, 4683–4693. [Google Scholar] [CrossRef]
- Jung, H.-J.; Nobumori, C.; Goulbourne, C.N.; Tu, Y.; Lee, J.M.; Tatar, A.; Wu, D.; Yoshinaga, Y.; de Jong, P.J.; Coffinier, C.; et al. Farnesylation of Lamin B1 Is Important for Retention of Nuclear Chromatin during Neuronal Migration. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, E1923–1932. [Google Scholar] [CrossRef]
- Pascual-Reguant, L.; Blanco, E.; Galan, S.; Le Dily, F.; Cuartero, Y.; Serra-Bardenys, G.; Di Carlo, V.; Iturbide, A.; Cebrià-Costa, J.P.; Nonell, L.; et al. Lamin B1 Mapping Reveals the Existence of Dynamic and Functional Euchromatin Lamin B1 Domains. Nat. Commun. 2018, 9, 3420. [Google Scholar] [CrossRef]
- Lukášová, E.; Kovařík, A.; Kozubek, S. Consequences of Lamin B1 and Lamin B Receptor Downregulation in Senescence. Cells 2018, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Hu, J.; Yue, S.; Kristiani, L.; Kim, M.; Sauria, M.; Taylor, J.; Kim, Y.; Zheng, Y. Lamins Organize the Global Three-Dimensional Genome from the Nuclear Periphery. Mol. Cell 2018, 71, 802–815.e7. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Li, M.; Shao, S.; Li, C.; Ai, S.; Xue, B.; Hou, Y.; Zhang, Y.; Li, R.; Fan, X.; et al. Nuclear Peripheral Chromatin-Lamin B1 Interaction Is Required for Global Integrity of Chromatin Architecture and Dynamics in Human Cells. Protein Cell 2022, 13, 258–280. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.Y.; Yang, Y.; Weston, T.A.; Belling, J.N.; Heizer, P.; Tu, Y.; Kim, P.; Edillo, L.; Jonas, S.J.; Weiss, P.S.; et al. An Absence of Lamin B1 in Migrating Neurons Causes Nuclear Membrane Ruptures and Cell Death. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 25870–25879. [Google Scholar] [CrossRef]
- Razafsky, D.; Ward, C.; Potter, C.; Zhu, W.; Xue, Y.; Kefalov, V.J.; Fong, L.G.; Young, S.G.; Hodzic, D. Lamin B1 and Lamin B2 Are Long-Lived Proteins with Distinct Functions in Retinal Development. Mol. Biol. Cell 2016, 27, 1928–1937. [Google Scholar] [CrossRef]
- Papantonis, A.; Cook, P.R. Transcription Factories: Genome Organization and Gene Regulation. Chem. Rev. 2013, 113, 8683–8705. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




