Submitted:
19 April 2023
Posted:
20 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
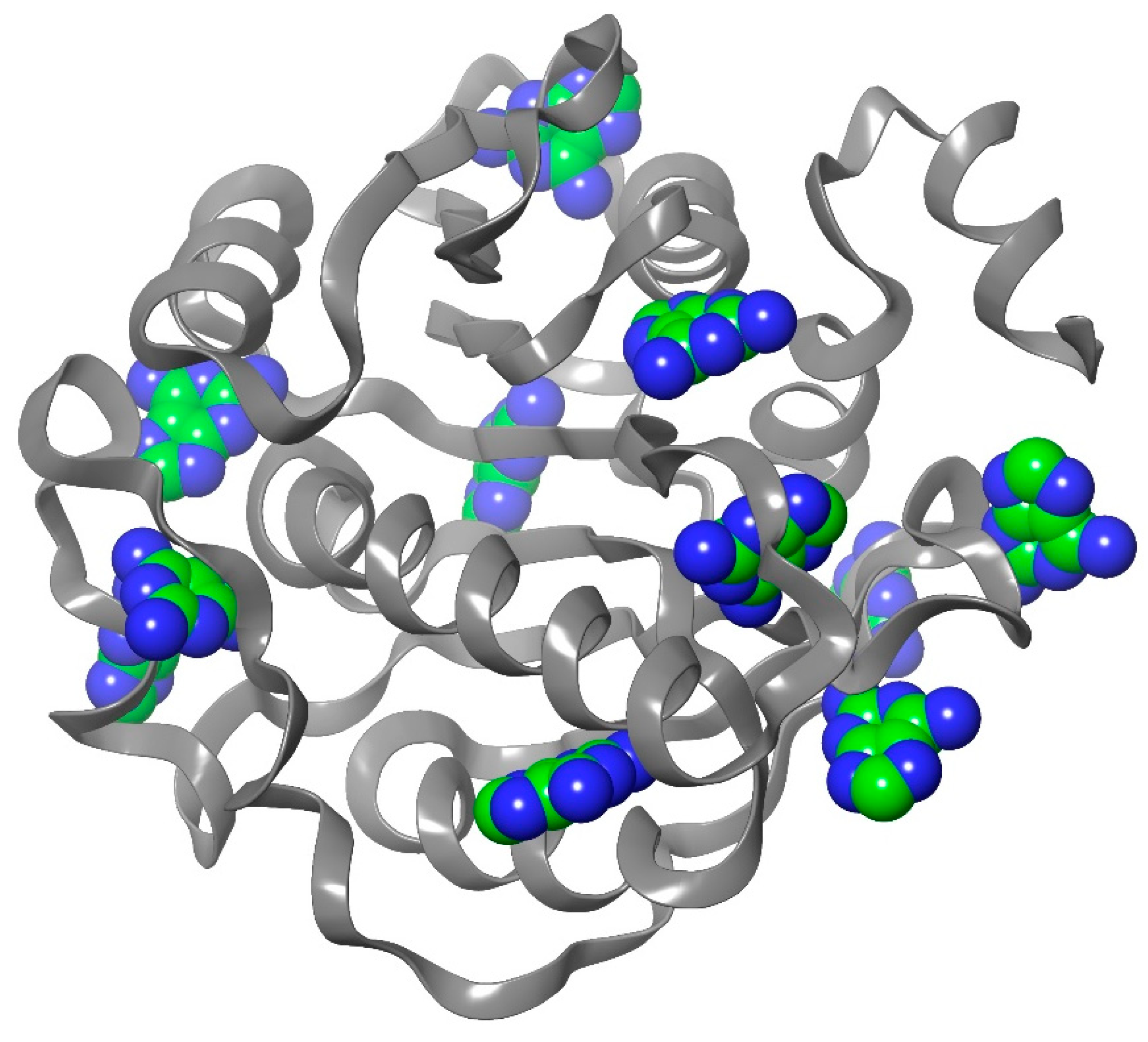
2.1. Homology modeling
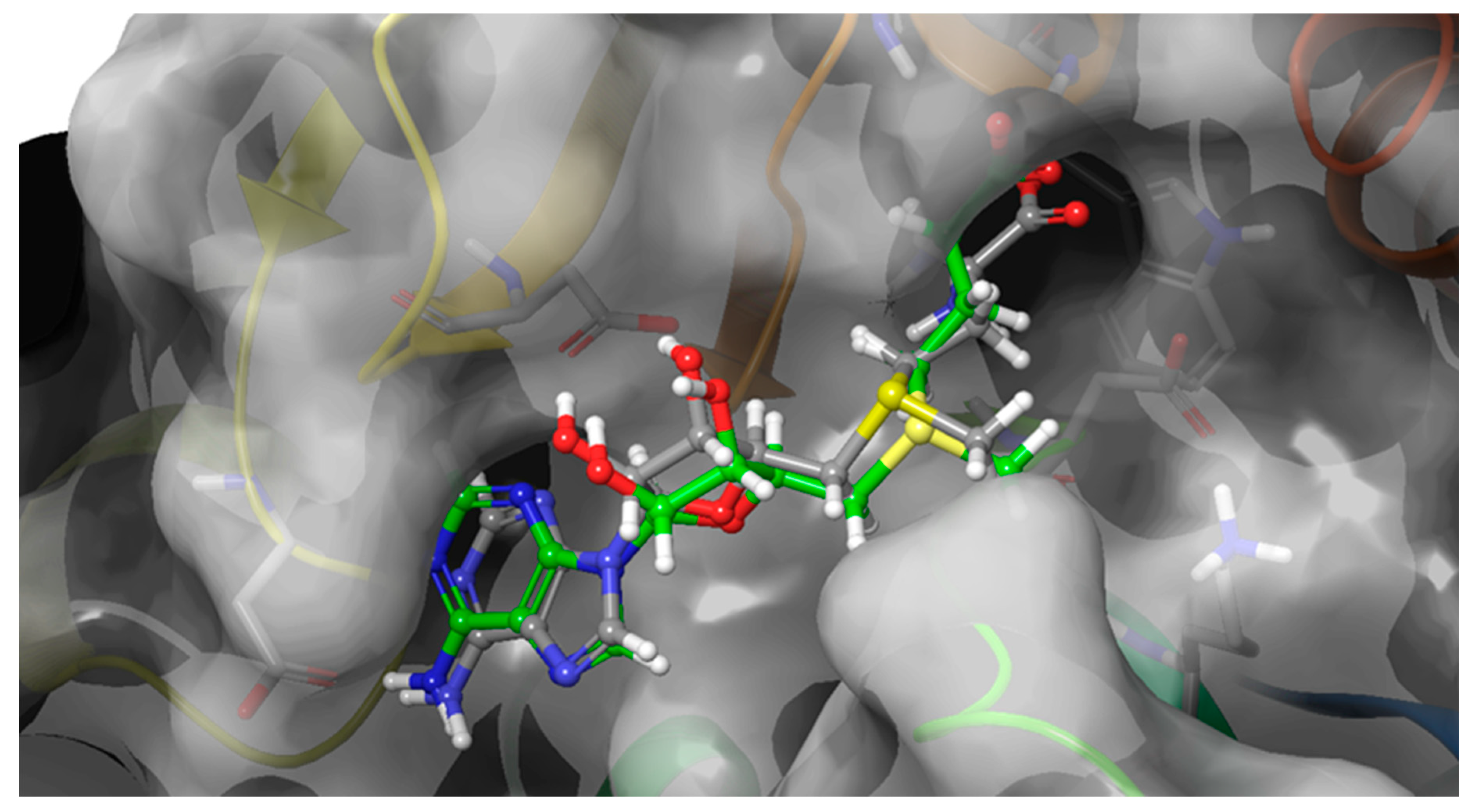
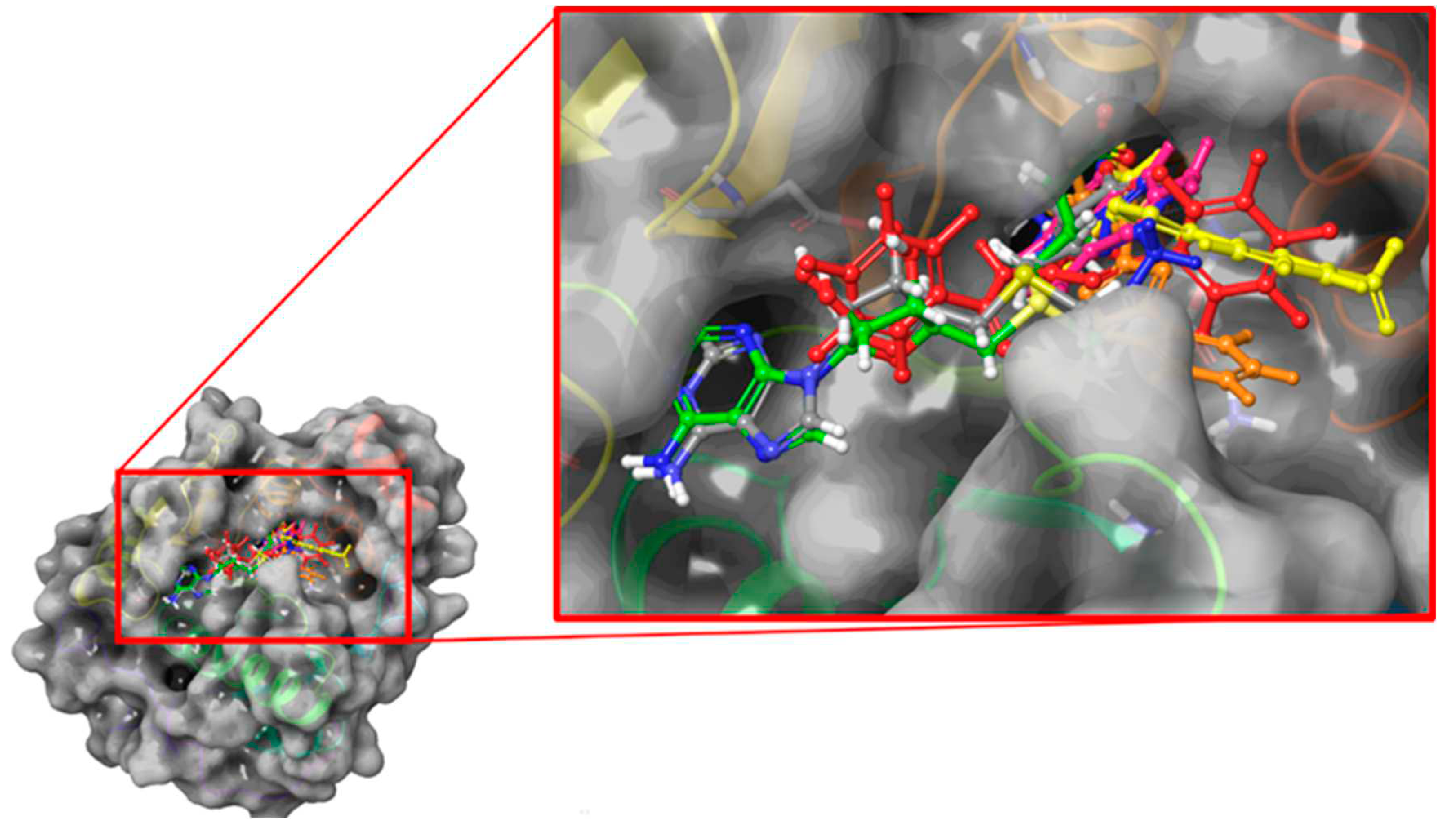
2.2. Blind Docking and semi-flexible docking analysis
2.3. Molecular Dynamics and free energy analysis
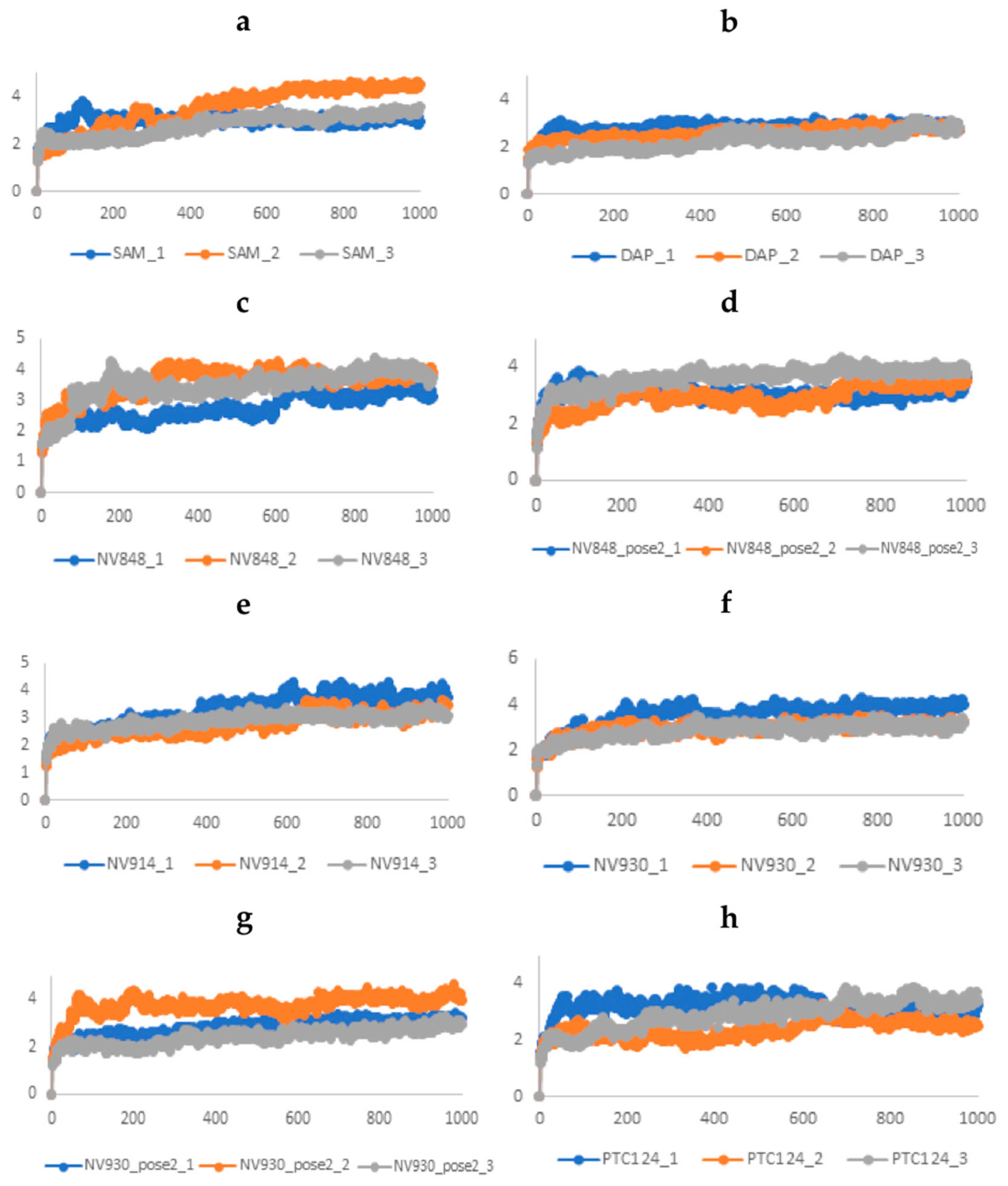
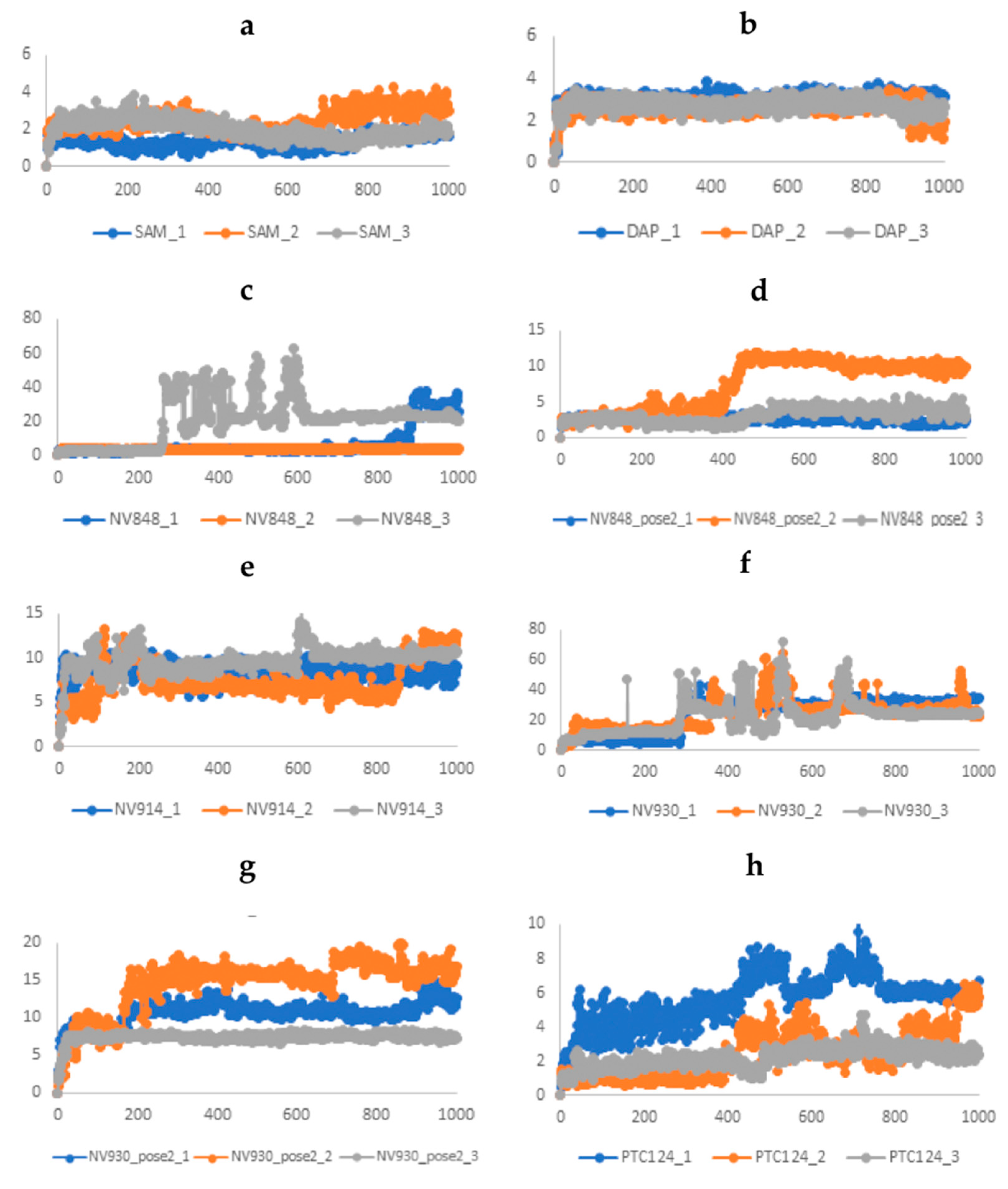
2.3.1. Stability analysis
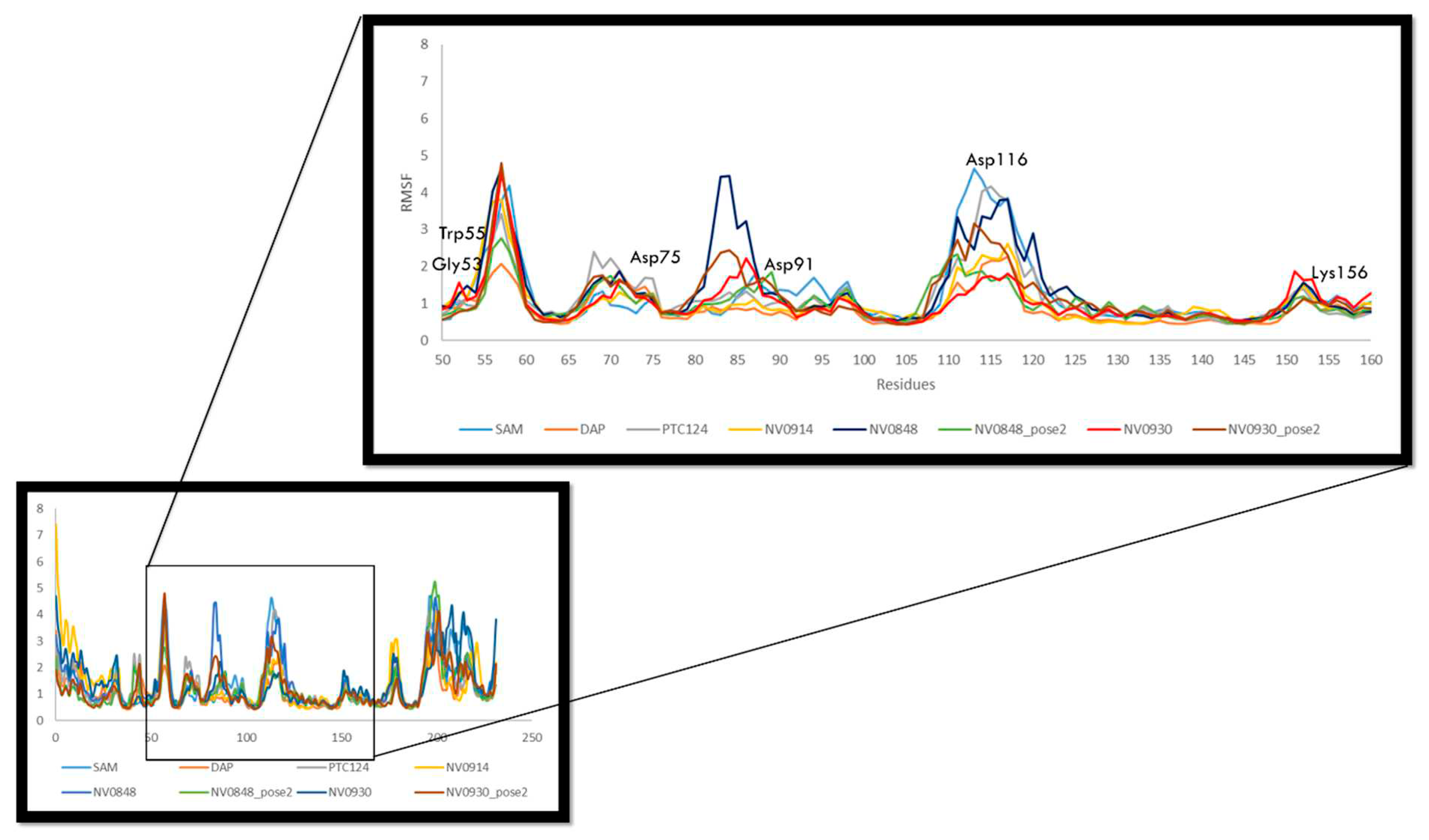
2.3.2. Residues mobility FTSJ1-ligand contact analysis
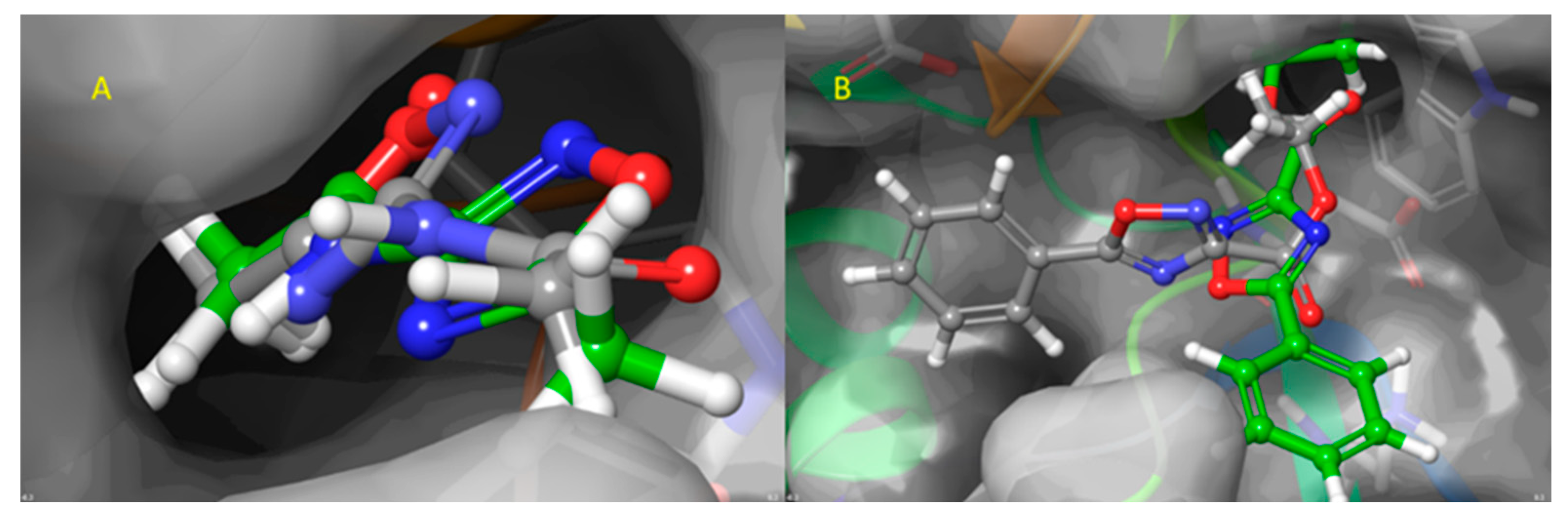
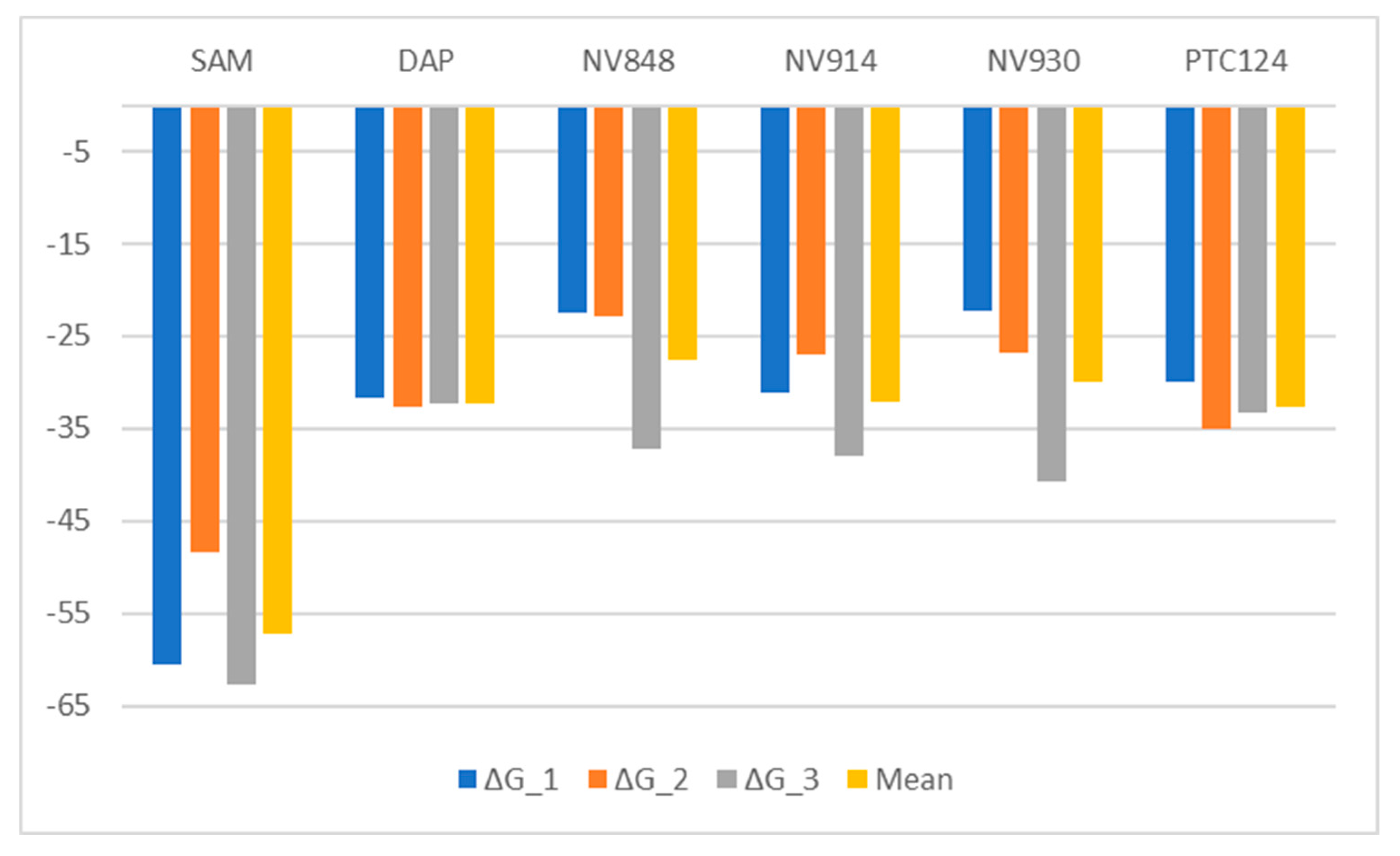
2.3.3. FTSJ1-ligand contact analysis and binding free energy analysis
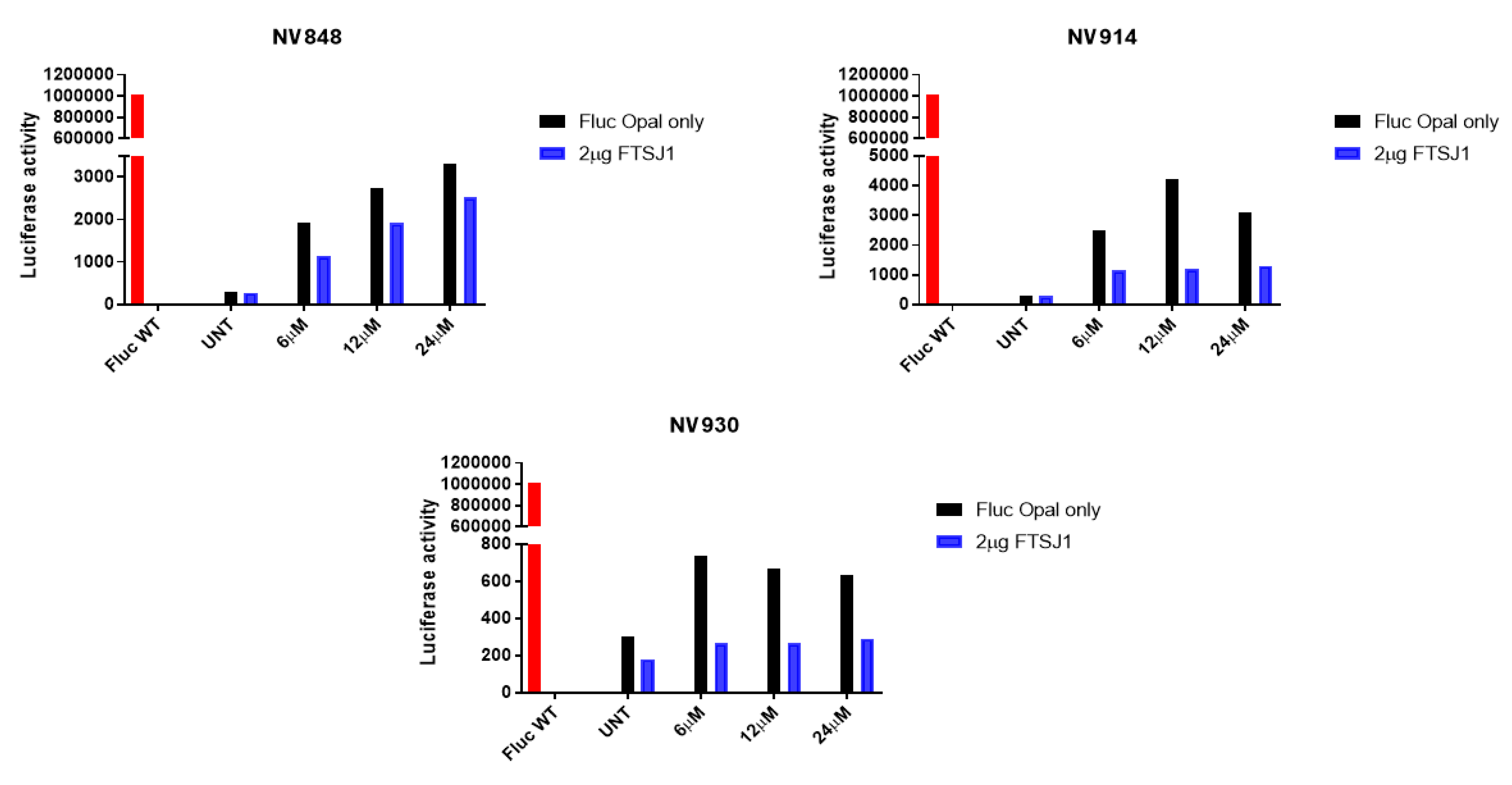
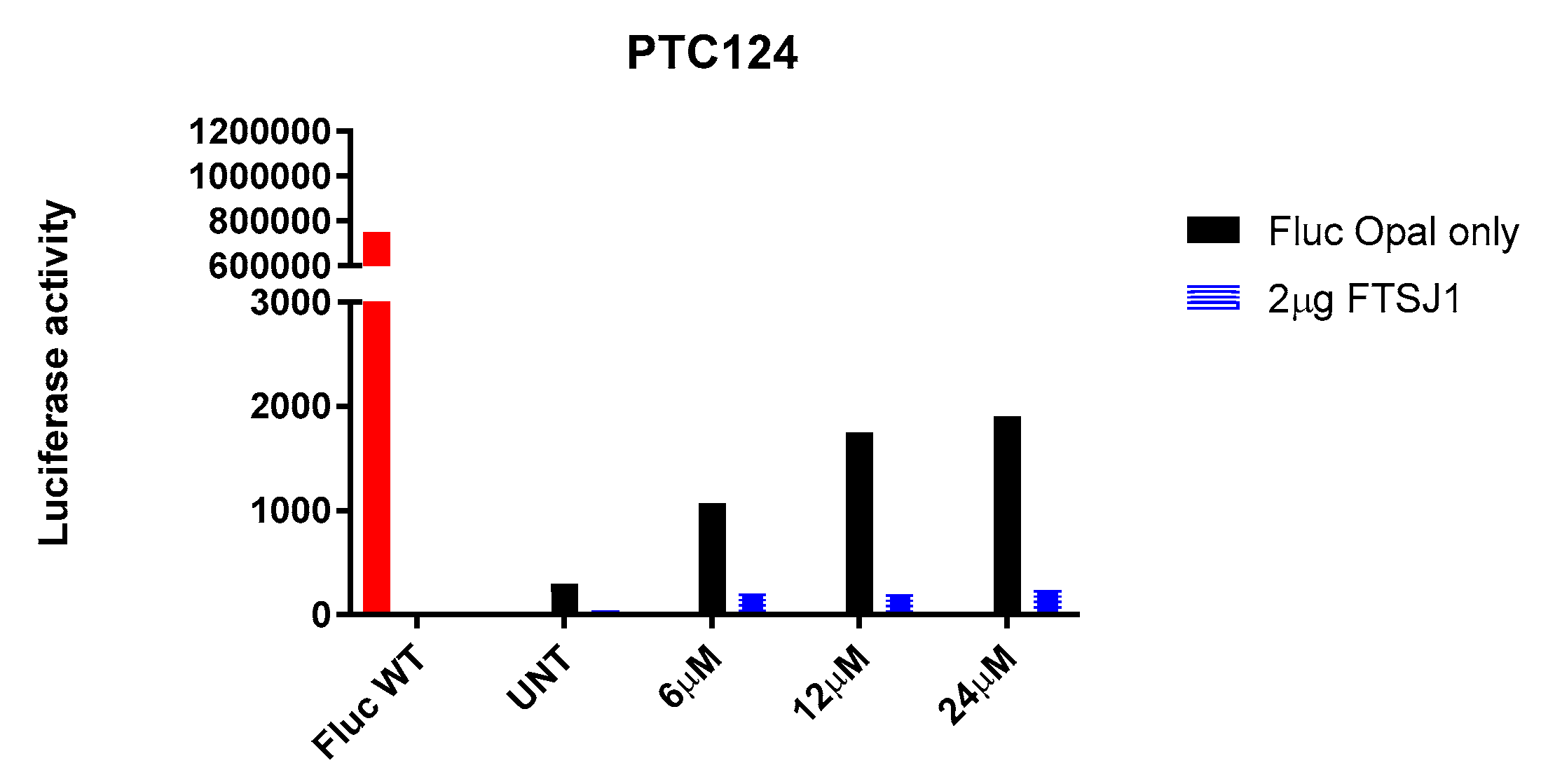
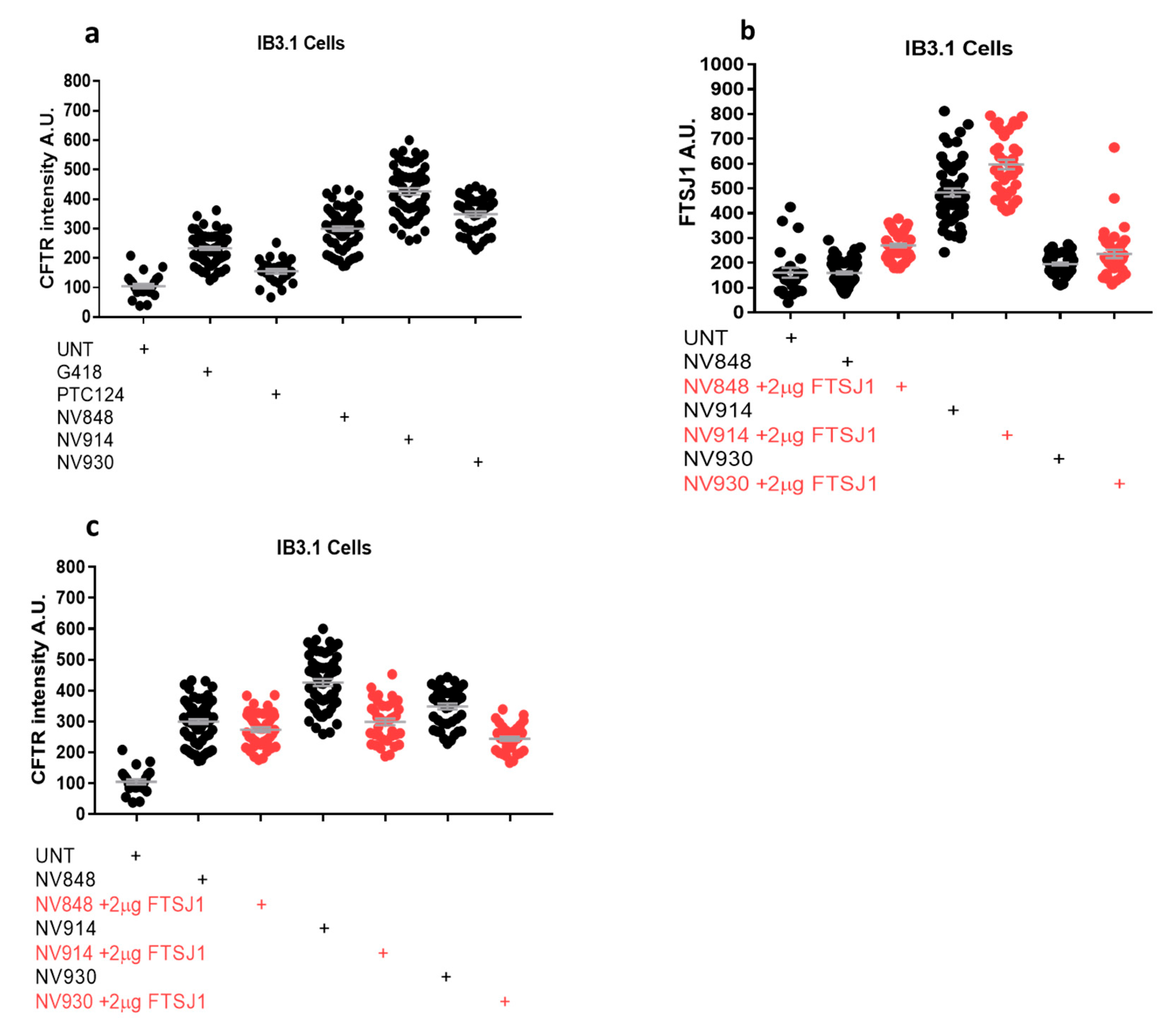
2.4. In vitro analysis
3. Materials and Methods
3.1. Homology Modeling and Protein preparation
3.2. Blind docking and semi-flexible docking
3.3. Molecular Dynamics and MM-GBSA calculations
3.4. Cell culture and NVs resuspension
3.5. Luminescence assay
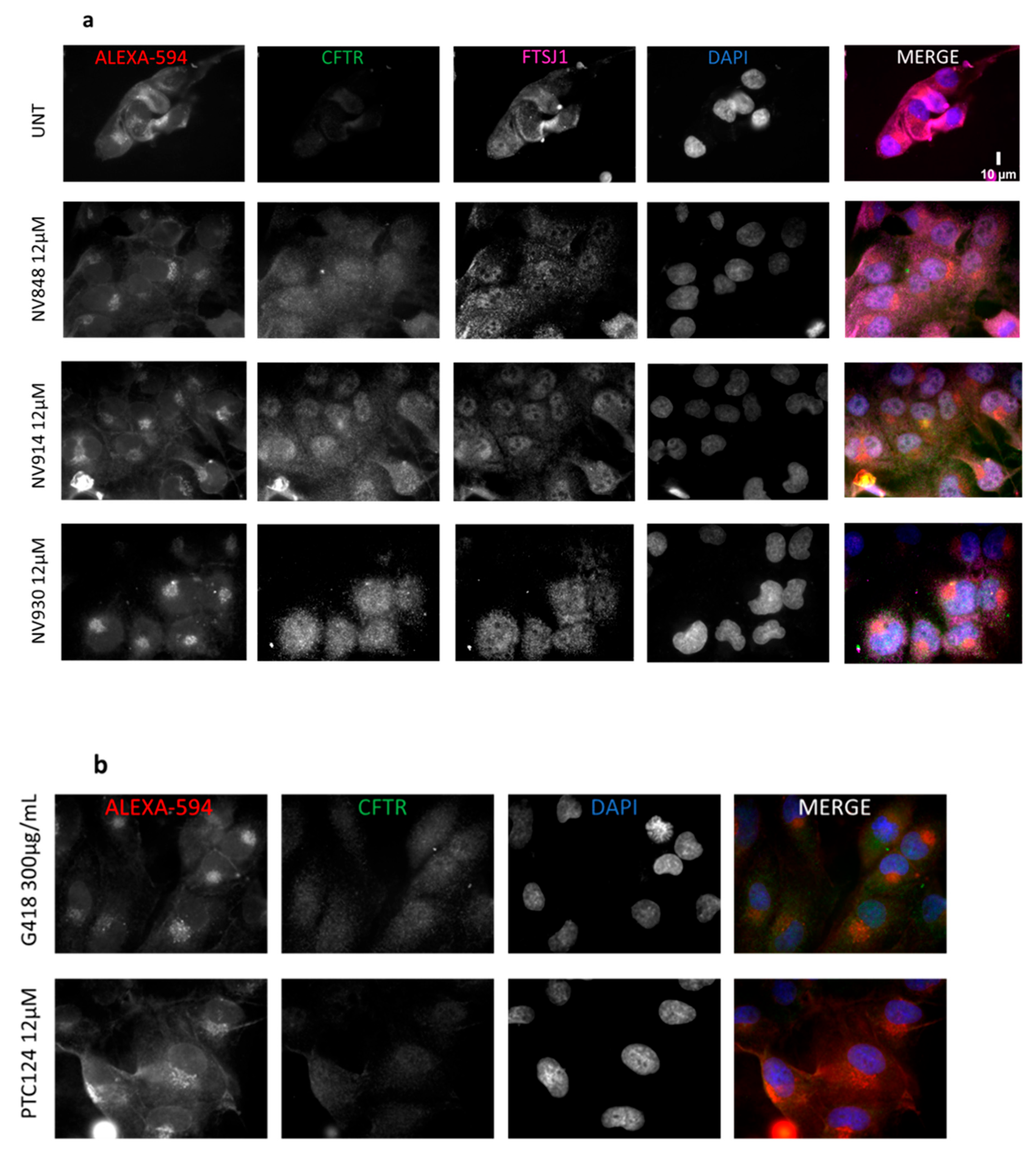
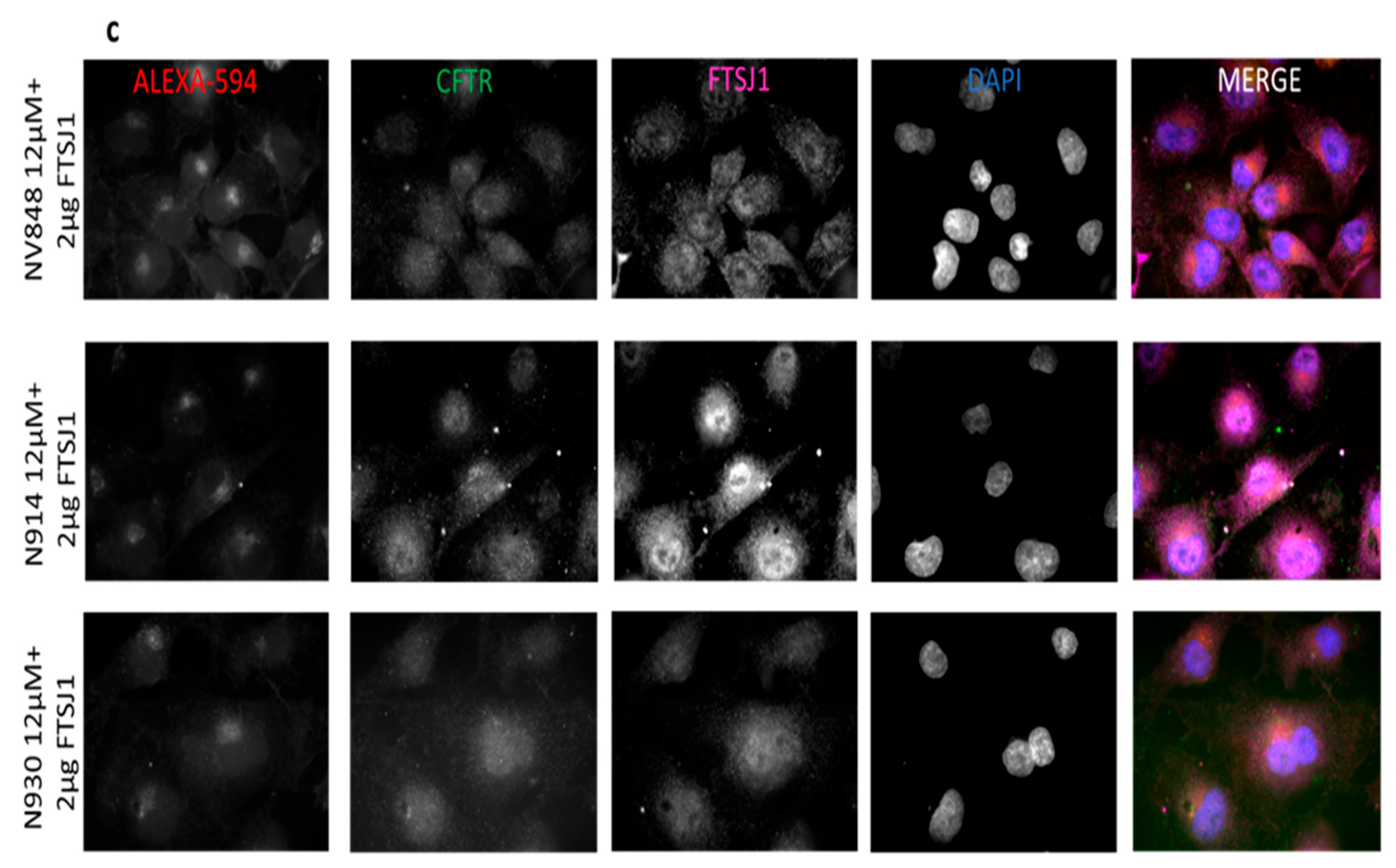
3.6. Immunofluorescence microscopy
3.7. Image Analysis of Immunofluorescence Images
4. Conclusion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crick, F. Central Dogma of Molecular Biology. Nature 1970, 227. [CrossRef]
- Palma, M.; Lejeune, F. Deciphering the Molecular Mechanism of Stop Codon Readthrough. Biological Reviews 2021, 96. [CrossRef]
- Freitag, J.; Ast, J.; Bölker, M. Cryptic Peroxisomal Targeting via Alternative Splicing and Stop Codon Read-through in Fungi. Nature 2012, 485. [CrossRef]
- Dunn, J.G.; Foo, C.K.; Belletier, N.G.; Gavis, E.R.; Weissman, J.S. Ribosome Profiling Reveals Pervasive and Regulated Stop Codon Readthrough in Drosophila Melanogaster. Elife 2013, 2013. [CrossRef]
- Loughran, G.; Chou, M.Y.; Ivanov, I.P.; Jungreis, I.; Kellis, M.; Kiran, A.M.; Baranov, P. V.; Atkins, J.F. Evidence of Efficient Stop Codon Readthrough in Four Mammalian Genes. Nucleic Acids Res 2014, 42. [CrossRef]
- Rajon, E.; Masel, J. Evolution of Molecular Error Rates and the Consequences for Evolvability. Proc Natl Acad Sci U S A 2011, 108. [CrossRef]
- Fearon, K.; McClendon, V.; Bonetti, B.; Bedwell, D.M. Premature Translation Termination Mutations Are Efficiently Suppressed in a Highly Conserved Region of Yeast Ste6p, a Member of the ATP-Binding Cassette (ABC) Transporter Family. Journal of Biological Chemistry 1994, 269. [CrossRef]
- Campofelice, A.; Lentini, L.; Di Leonardo, A.; Melfi, R.; Tutone, M.; Pace, A.; Pibiri, I. Strategies against Nonsense: Oxadiazoles as Translational Readthrough-Inducing Drugs (TRIDs). Int J Mol Sci 2019, 20, 3329. [CrossRef]
- Spelier, S.; van Doorn, E.P.M.; van der Ent, C.K.; Beekman, J.M.; Koppens, M.A.J. Readthrough Compounds for Nonsense Mutations: Bridging the Translational Gap. Trends Mol Med 2023, 29, 297–314. [CrossRef]
- Pibiri, I.; Lentini, L.; Tutone, M.; Melfi, R.; Pace, A.; Di Leonardo, A. Exploring the Readthrough of Nonsense Mutations by Non-Acidic Ataluren Analogues Selected by Ligand-Based Virtual Screening. Eur J Med Chem 2016, 122, 429–435. [CrossRef]
- Pibiri, I.; Lentini, L.; Melfi, R.; Tutone, M.; Baldassano, S.; Ricco Galluzzo, P.; Di Leonardo, A.; Pace, A. Rescuing the CFTR Protein Function: Introducing 1,3,4-Oxadiazoles as Translational Readthrough Inducing Drugs. Eur J Med Chem 2018, 159, 126–142. [CrossRef]
- Tutone, M.; Pibiri, I.; Lentini, L.; Pace, A.; Almerico, A.M. Deciphering the Nonsense Readthrough Mechanism of Action of Ataluren: An <italic Toggle="yes">in Silico</Italic> Compared Study. ACS Med Chem Lett 10, 522–527. [CrossRef]
- Tutone, M.; Pibiri, I.; Perriera, R.; Campofelice, A.; Culletta, G.; Melfi, R.; Pace, A.; Almerico, A.M.; Lentini, L. Pharmacophore-Based Design of New Chemical Scaffolds as Translational Readthrough-Inducing Drugs (TRIDs). ACS Med Chem Lett 11, 747–753. [CrossRef]
- Pibiri, I.; Melfi, R.; Tutone, M.; Di Leonardo, A.; Pace, A.; Lentini, L. Targeting Nonsense: Optimization of 1,2,4-Oxadiazole Trids to Rescue Cftr Expression and Functionality in Cystic Fibrosis Cell Model Systems. Int J Mol Sci 2020, 21. [CrossRef]
- Corrao, F.; Zizzo, M.G.; Tutone, M.; Melfi, R.; Fiduccia, I.; Carollo, P.S.; Leonardo, A. Di; Caldara, G.; Perriera, R.; Pace, A.; et al. Nonsense Codons Suppression. An Acute Toxicity Study of Three Optimized TRIDs in Murine Model, Safety and Tolerability Evaluation. Biomedicine and Pharmacotherapy 2022, 156, 113886. [CrossRef]
- Ng, M.Y.; Zhang, H.; Weil, A.; Singh, V.; Jamiolkowski, R.; Baradaran-Heravi, A.; Roberge, M.; Jacobson, A.; Friesen, W.; Welch, E.; et al. New in Vitro Assay Measuring Direct Interaction of Nonsense Suppressors with the Eukaryotic Protein Synthesis Machinery. ACS Med Chem Lett 2018, 9. [CrossRef]
- Trzaska, C.; Amand, S.; Bailly, C.; Leroy, C.; Marchand, V.; Duvernois-Berthet, E.; Saliou, J.M.; Benhabiles, H.; Werkmeister, E.; Chassat, T.; et al. 2,6-Diaminopurine as a Highly Potent Corrector of UGA Nonsense Mutations. Nat Commun 2020, 11. [CrossRef]
- Zeitlin, P.L.; Lu, L.; Rhim, J.; Cutting, G.; Stetten, G.; Kieffer, K.A.; Craig, R.; Guggino, W.B. A Cystic Fibrosis Bronchial Epithelial Cell Line: Immortalization by Adeno-12-SV40 Infection. Am J Respir Cell Mol Biol 1991, 4. [CrossRef]
- Schrödinger Maestro | Schrödinger. Schrödinger Release 2018-2 2018.
- Pérez-Sánchez, H.; Thirumal Kumar, D.; George Priya Doss, C.; Rodríguez-Schmidt, R.; Cerón-Carrasco, J.P.; Peña-García, J.; Ye, Z.W.; Yuan, S.; Günther, S. Prediction and Characterization of Influenza Virus Polymerase Inhibitors through Blind Docking and Ligand Based Virtual Screening. J Mol Liq 2021, 321. [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J Comput Chem 2009. [CrossRef]
- Stroganov, O. V.; Novikov, F.N.; Stroylov, V.S.; Kulkov, V.; Chilov, G.G. Lead Finder: An Approach to Improve Accuracy of Protein-Ligand Docking, Binding Energy Estimation, and Virtual Screening. J Chem Inf Model 2008, 48. [CrossRef]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J Med Chem 2006. [CrossRef]
- D. E. Shaw Research, New York, N. Schrödinger Release 2020-4: Desmond Molecular Dynamics System. Maestro Desmond Interoperability Tools, Schrödinger, New York 2020.
- Allegra, M.; Tutone, M.; Tesoriere, L.; Attanzio, A.; Culletta, G.; Almerico, A.M. Evaluation of the IKKβ Binding of Indicaxanthin by Induced-Fit Docking, Binding Pose Metadynamics, and Molecular Dynamics. Front Pharmacol 2021, 12. [CrossRef]
- Culletta, G.; Allegra, M.; Almerico, A.M.; Restivo, I.; Tutone, M. In Silico Design, Synthesis and Biological Evaluation of Anticancer Arylsulfonamide Endowed with Anti-Telomerase Activity. Pharmaceuticals 2022, 15. [CrossRef]
- Culletta, G.; Zappalà, M.; Ettari, R.; Almerico, A.M.; Tutone, M. Immunoproteasome and Non-Covalent Inhibition: Exploration by Advanced Molecular Dynamics and Docking Methods. Molecules 2021, 26. [CrossRef]
- Tutone, M.; Virzì, A.; Almerico, A.M. Reverse Screening on Indicaxanthin from Opuntia Ficus-Indica as Natural Chemoactive and Chemopreventive Agent. J Theor Biol 2018. [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA Methods to Estimate Ligand-Binding Affinities. Expert Opin Drug Discov 2015, 10, 449–461.
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.F.; Honig, B.; Shaw, D.E.; Friesner, R.A. A Hierarchical Approach to All-Atom Protein Loop Prediction. Proteins: Structure, Function and Genetics 2004, 55, 351–367. [CrossRef]
- Auld, D.S.; Thorne, N.; Maguire, W.F.; Inglese, J. Mechanism of PTC124 Activity in Cell-Based Luciferase Assays of Nonsense Codon Suppression. Proc Natl Acad Sci U S A 2009, 106. [CrossRef]
- Lentini, L.; Melfi, R.; Di Leonardo, A.; Spinello, A.; Barone, G.; Pace, A.; Palumbo Piccionello, A.; Pibiri, I. Toward a Rationale for the PTC124 (Ataluren) Promoted Readthrough of Premature Stop Codons: A Computational Approach and GFP-Reporter Cell-Based Assay. Mol Pharm 2014. [CrossRef]
- Pibiri, I.; Lentini, L.; Melfi, R.; Gallucci, G.; Pace, A.; Spinello, A.; Barone, G.; Di Leonardo, A. Enhancement of Premature Stop Codon Readthrough in the CFTR Gene by Ataluren (PTC124) Derivatives. Eur J Med Chem 2015. [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat Methods 2012, 9.
- Welch, E.M.; Barton, E.R.; Zhuo, J.; Tomizawa, Y.; Friesen, W.J.; Trifillis, P.; Paushkin, S.; Patel, M.; Trotta, C.R.; Hwang, S.; et al. PTC124 Targets Genetic Disorders Caused by Nonsense Mutations. Nature 2007. [CrossRef]
- Lentini, L.; Melfi, R.; Cancemi, P.; Pibiri, I.; Di Leonardo, A. Caffeine Boosts Ataluren’s Readthrough Activity. Heliyon 2019, 5. [CrossRef]
| Docking | MD | |||||||
| Replica 1 | Replica 2 | Replica 3 | ||||||
| Cpd* | Residue | Int.** | Residue | Int.** | Residue | Int.** | Residue | Int.** |
| DAP | Trp55 | Pi-pi | Asp116 | HB | Asp116 | HB | Asp116 | HB |
| Lys156 | Pi-cation | Trp55 | HPhob | Trp55 | HPhob | Trp55 | HPhob | |
| Asp116 | vdW | Asp47 | HB | Asp47 | HB | Asp47 | HB | |
| Asp47 | vdW | Leu48 | HPhob | Leu48 | HPhob | Leu48 | HPhob | |
| Gly53 | HB | Gly53 | HB | |||||
| Ser54 | HB | |||||||
| NV848 | Trp55 | HB, pi-cation | Trp55 | HB, HPhob | Asp47 | WB, HPhob | Lys28 | HB, ionic |
| Gly53 | vdW | Gly53 | HB | Leu48 | HB, HPhob, Ionic | Gly53 | HB | |
| Ser54 | vdW | Ser54 | HB | Gly53 | HB | Ser54 | HB | |
| Asp116 | vdW | Asp116 | WB | Ser54 | HB, Hphobic | Trp55 | HB, Hphob | |
| Cys115 | WB, HB | Asp116 | HB, WB | |||||
| Asp116 | WB, HB | Lys156 | Hphob | |||||
| NV914 | Asp75 | vdW | Cys49 | HB, Hphob | Cys49 | HB, Hphob | Cys49 | WB, HB |
| Asp116 | vdW | Leu76 | HB, HPhob | Leu76 | HPhob | Asp91 | WB | |
| Lys156 | vdW | Asp91 | WB | Ile92 | HPhob, WB | Ala118 | HB, WB | |
| Ala118 | vdW, HB | Ala118 | HB, WB | Ala118 | HB, WB | Asp120 | HB, WB | |
| Cys49 | vdW | |||||||
| NV930 | Asp75 | vdW | Leu48 | HPhob | Asp91 | HB, Hphobic. WB | Ile92 | Hphobic, WB |
| Ala118 | HB, vdW | Cys49 | HB, HPhob | Ile92 | WB | Ala118 | HB, Hphobic, WB | |
| Lys156 | vdW | Val74 | HPhob | Tyr130 | Hphobic | Leu135 | Hphobic | |
| Asp116 | vdW | Asp75 | WB | Gln134 | HB, WB | Ala139 | ||
| Cys49 | vdW | Ile92 | HB, HPhob | Leu135 | Hphobic | |||
| Leu135 | vdW | Leu135 | HPhob | Tyr218 | Hphobic, WB | |||
| Ile142 | HPhob | |||||||
| PTC124 | Ser25 | HB, vdW | Arg24 | HB, WB | Arg24 | HB, WB | Ser25 | HB, WB |
| Arg24 | Salt bridge | Ser25 | HB, WB | Ser25 | HB, WB | Arg24 | HB, WB | |
| Lys28 | Salt bridge | Pro52 | Hphobic | Lys28 | HB, WB, Ionic, Hphobic | Lys28 | HB, WB, Hphobic | |
| Lys156 | Pi-cation | Trp55 | Hphobic | Trp55 | Hphobic | Trp55 | Hphobic | |
| Cys49 | vdW | Ala118 | Hphobic | Lys156 | HB, Hphobic, ionic, WB | Lys156 | Hphobic, ionic, WB | |
| Pro52 | vdW | Arg186 | HB, ionic, WB | Arg186 | HB, ionic, WB | |||
| Gly53 | vdW | |||||||
| Trp55 | vdW | |||||||
| Arg186 | Salt bridge | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





