Submitted:
15 April 2023
Posted:
17 April 2023
You are already at the latest version
Abstract
Keywords:
Introduction
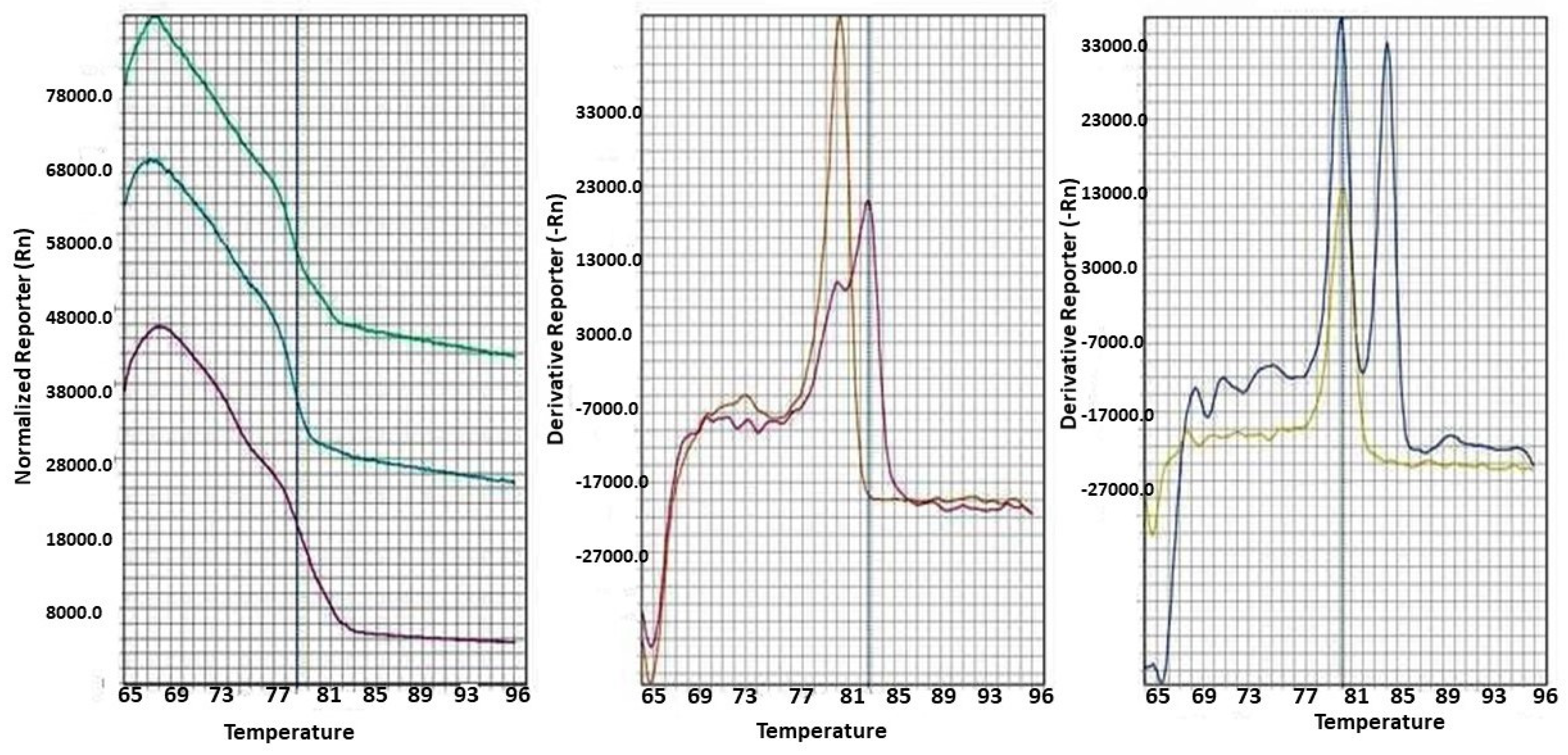
Materials and Methods
Tissue and ctDNA extraction:
Bisulfite modification
Statistical analysis
Results
Discussion
Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Guerrero MA, Schreinemakers JM, Vriens MR, Suh I, Hwang J, Shen WT, et al. Clinical spectrum of pheochromocytoma. Journal of the American College of Surgeons. 2009;209(6):727-732. [CrossRef]
- Neumann HP, Young Jr WF, Eng C. Pheochromocytoma and paraganglioma. New England Journal of Medicine. 2019;381(6):552-565.
- Beard C, Sheps S, Kurland L, Carney J, Lie J, editors. Occurrence of pheochromocytoma in Rochester, Minnesota, 1950 through 1979. Mayo Clinic Proceedings; 1983.
- Sutton M, Sheps S, Lie J, editors. Prevalence of clinically unsuspected pheochromocytoma. Review of a 50-year autopsy series. Mayo Clinic Proceedings; 1981. [CrossRef]
- Aghamir SMK, Heshmat R, Ebrahimi M, Khatami F. Liquid Biopsy: The Unique Test for Chasing the Genetics of Solid Tumors. Epigenetics Insights. 2020;13:2516865720904052. [CrossRef]
- Wang L, Li Y, Guan X, Zhao J, Shen L, Liu J. Exosomal double-stranded DNA as a biomarker for the diagnosis and preoperative assessment of pheochromocytoma and paraganglioma. Molecular cancer. 2018;17(1):1-6. [CrossRef]
- Khatami F, Tavangar SM. Current diagnostic status of pheochromocytomaand future perspective: A mini review. Iranian journal of pathology. 2017;12(3):313. [CrossRef]
- Kulis M, Esteller M. DNA methylation and cancer. Advances in genetics. 70: Elsevier; 2010. p. 27-56.
- Khatami F, Mohammadamoli M, Tavangar SM. Genetic and epigenetic differences of benign and malignant pheochromocytomas and paragangliomas (PPGLs). Endocrine regulations. 2018;52(1):41-54. [CrossRef]
- Nazar E, Khatami F, Saffar H, Tavangar SM. The Emerging Role of Succinate Dehyrogenase Genes (SDHx) in Tumorigenesis. International Journal of Hematology-Oncology and Stem Cell Research. 2019;13(2):72. [CrossRef]
- Backman S, Maharjan R, Falk-Delgado A, Crona J, Cupisti K, Stålberg P, et al. Global DNA Methylation Analysis Identifies Two Discrete clusters of Pheochromocytoma with Distinct Genomic and Genetic Alterations. Sci Rep. 2017;7:44943-44943. [CrossRef]
- Fatemeh Khatami BL, Ramin Heshmat, Shirzad Nasiri, Hiva Saffar, Gita Shafiee, Azam Mossafa, Seyed Mohammad Tavangar. Promoter Methylation of Four Tumor Suppressor Genes in Human Papillary Thyroid Carcinoma. Iranian Journal of Pathology. 2018;In Press. [CrossRef]
- Brigliadori G, Foca F, Dall’Agata M, Rengucci C, Melegari E, Cerasoli S, et al. Defining the cutoff value of MGMT gene promoter methylation and its predictive capacity in glioblastoma. Journal of Neuro-Oncology. 2016;128(2):333-339. [CrossRef]
- Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harbor perspectives in biology. 2016;8(9):a019505. [CrossRef]
- Chatterjee A, Rodger EJ, Eccles MR, editors. Epigenetic drivers of tumourigenesis and cancer metastasis. Seminars in cancer biology; 2018: Elsevier. [CrossRef]
- Ma M, Zhu H, Zhang C, Sun X, Gao X, Chen G. “Liquid biopsy”—ctDNA detection with great potential and challenges. Annals of translational medicine. 2015;3(16). [CrossRef]
- Khatami F, Larijani B, Tavangar SM. The presence of tumor extrachomosomal circular DNA (ecDNA) as a component of liquid biopsy in blood. Medical hypotheses. 2018;114:5-7. [CrossRef]
- Khatami F, Aghaii M, Aghamir SMK. Prime editing: The state-of-the-art of genome editing. Meta Gene. 2020;24:100661. [CrossRef]
- Tamehri Zadeh SS, Taheri D, Shivarani S, Khatami F, Kazemi R. Liquid Biopsy in Prostate Cancer Diagnosis and Prognosis: A Narrative Review. Translational Research In Urology. 2020;2(4):139-146. [CrossRef]
- Letouzé E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer cell. 2013;23(6):739-752. [CrossRef]
- Loriot C, Burnichon N, Gadessaud N, Vescovo L, Amar L, Libé R, et al. Epithelial to mesenchymal transition is activated in metastatic pheochromocytomas and paragangliomas caused by SDHB gene mutations. The Journal of Clinical Endocrinology & Metabolism. 2012;97(6):E954-E962. [CrossRef]
- Astuti D, Morris M, Krona C, Abel F, Gentle D, Martinsson T, et al. Investigation of the role of SDHB inactivation in sporadic phaeochromocytoma and neuroblastoma. British journal of cancer. 2004;91(10):1835-1841. [CrossRef]
- Grau E, Oltra S, Orellana C, Hernández-Martí M, Castel V. There is no evidence that the SDHB gene is involved in neuroblastoma development. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics. 2005;15(7-8):393-398. [CrossRef]
- Loriot C, Domingues M, Berger A, Menara M, Ruel M, Morin A, et al. Deciphering the molecular basis of invasiveness in Sdhb-deficient cells. Oncotarget. 2015;6(32):32955-32965. [CrossRef]
- Ricci R, Martini M, Ravegnini G, Cenci T, Milione M, Lanza P, et al. Preferential MGMT methylation could predispose a subset of KIT/PDGFRA-WT GISTs, including SDH-deficient ones, to respond to alkylating agents. Clinical Epigenetics. 2019;11(1):2. [CrossRef]
- Liu Y, Pang Y, Caisova V, Ding J, Yu D, Zhou Y, et al. Targeting NRF2-governed glutathione synthesis for SDHB-mutated pheochromocytoma and paraganglioma. Cancers. 2020;12(2):280. [CrossRef]
- Lamy C, Hadoux J, Durand S, Alghuzlan A, Riviere J, Lefevre D, et al. Preclinical evaluation of new therapeutic strategies on SDHB invalidated clones from human pheochromocytoma cells. AACR; 2019. [CrossRef]
- Rashedi S. Landscape of Circular Ribonucleic Acids in Urological Cancers. Translational Research Urology. 2021;3(2):45-47. [CrossRef]
- Gill AJ, Benn DE, Chou A, Clarkson A, Muljono A, Meyer-Rochow GY, et al. Immunohistochemistry for SDHB triages genetic testing of SDHB, SDHC, and SDHD in paraganglioma-pheochromocytoma syndromes. Human pathology. 2010;41(6):805-814. [CrossRef]
- Remacha L, Comino-Méndez I, Richter S, Contreras L, Currás-Freixes M, Pita G, et al. Targeted exome sequencing of Krebs cycle genes reveals candidate cancer–predisposing mutations in pheochromocytomas and paragangliomas. Clinical Cancer Research. 2017;23(20):6315-6324. [CrossRef]
- Shi C, Zeng Z, Zhao D, Hanzhong L, Miao Q, Zhu W, et al. Application of SDHB and SDHC immunohistochemistry in the differentiation of malignant and benign pheochromocytoma and paraganglioma. Chinese Journal of Endocrinology and Metabolism. 2018;34(6):472-478.
- Karimaei S, Oliveira Reis L. Cytotoxicity and Apoptotic Effect of Nisin as an Effective Bacteriocin on the Cancer Cells. Translational Research Urology. 2020;2(2):45-47. [CrossRef]
- Richter S, Klink B, Nacke B, de Cubas AA, Mangelis A, Rapizzi E, et al. Epigenetic mutation of the succinate dehydrogenase C promoter in a patient with two paragangliomas. The Journal of Clinical Endocrinology & Metabolism. 2016;101(2):359-363. [CrossRef]
- Smestad J, Hamidi O, Wang L, Holte MN, Al Khazal F, Erber L, et al. Characterization and metabolic synthetic lethal testing in a new model of SDH-loss familial pheochromocytoma and paraganglioma. Oncotarget. 2018;9(5):6109. [CrossRef]
- Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Molecular and cellular biology. 2003;23(6):1863-1873. [CrossRef]
- De Cubas AA, Korpershoek E, Inglada-Pérez L, Letouzé E, Currás-Freixes M, Fernández AF, et al. DNA methylation profiling in pheochromocytoma and paraganglioma reveals diagnostic and prognostic markers. Clinical Cancer Research. 2015;21(13):3020-3030. [CrossRef]
- Goncalves J, Lussey-Lepoutre C, Favier J, Gimenez-Roqueplo A-P, Castro-Vega LJ, editors. Emerging molecular markers of metastatic pheochromocytomas and paragangliomas. Annales d'endocrinologie; 2019: Elsevier. [CrossRef]
- Björklund P, Backman S. Epigenetics of pheochromocytoma and paraganglioma. Molecular and Cellular Endocrinology. 2018;469:92-97. [CrossRef]
- Oishi T, Iino K, Okawa Y, Kakizawa K, Matsunari S, Yamashita M, et al. DNA methylation analysis in malignant pheochromocytoma and paraganglioma. Journal of clinical & translational endocrinology. 2017;7:12-20. [CrossRef]
- Job S, Georges A, Burnichon N, Buffet A, Amar L, Bertherat J, et al. Transcriptome analysis of lncRNAs in pheochromocytomas and paragangliomas. The Journal of Clinical Endocrinology & Metabolism. 2020;105(3):898-907. [CrossRef]
- Nicolas M, Dahia P. Predictors of outcome in phaeochromocytomas and paragangliomas. F1000Research. 2017;6. [CrossRef]

| Gene | Part | Forward Primer | Reverse Primer | Tann | Number of CpG sites in the amplicon |
|---|---|---|---|---|---|
|
RDBP |
a | 5' GGTAAGTTTTTTGTTTTTTAT 3' | 5' TTTAAATACATATAATTCA 3' | 56°C | 15 |
| b | 5' GGATATAGTTTGGTTTAAG 3' | 5’ ACATCTTTCTCCACTATTAC 3’ | 52°C | 9 | |
| SDHB | a |
5' GTTAGTGTTTTAGTGGATGT 3' |
5' AAACTCACCTACAAACAAAC 3' | 57°C | 17 |
| b | 5' GGGAAGTTAAATGGGT 3' | 5' TCCACTAAAACCCACT 3' | 55°C | 14 | |
| SDHC | a |
5' GTAATTAGTTAGGTAGAG 3' |
5' ACTAAATCACCTCAACA 3' | 50°C | 14 |
| b |
5'TAGATGTAGATTTTGAGTTA 3' |
5'ACTCTACTAACTAATTTAC 3' | 49°C | 6 |
| Variables | PCCs/PGLs cases (n= 12) | Controls (n=12) | P-value |
|---|---|---|---|
| Age (years) | 41.25 (±10.532) | 42.42 (±11.828) | - |
|
Gender: Female Male |
8 (66.7%) 4 (33.3%) |
8 (66.7%) 4 (33.3%) |
- |
| Weight (kg) | 61.58 (±5.299) | 74.00 (±11.201) | 0. 002 |
| Height (cm) | 165.50 (±9.200) | 165.32 (±9.55) | 0.370 |
| BMI | 23.68 (±3.884) | 27.05 (±3.833) | 0.043 |
| SBP (mm/Hg) | 13.72(±1.190) | 11.50 (±1.167) | <.001 |
| DBP (mm/Hg) | 10.36(±1.68) | 7.91 (±1.37) | 0.001 |
| Educated (After High School) | 7 (58.3%) | 10 (83.3%) | 0.146 |
| Malignant | 5(41.6%) | - | - |
| Tumor size | 0.8-12 cm | - | - |
| Norepinephrine level | 4.2 ± 1.5% | - | - |
| Epinephrine | 58.02(±5.12) | - | - |
| Norepinephrine | 61.3 (±3.459) | - | - |
| Dopamine | 220 (±2.023) | - | - |
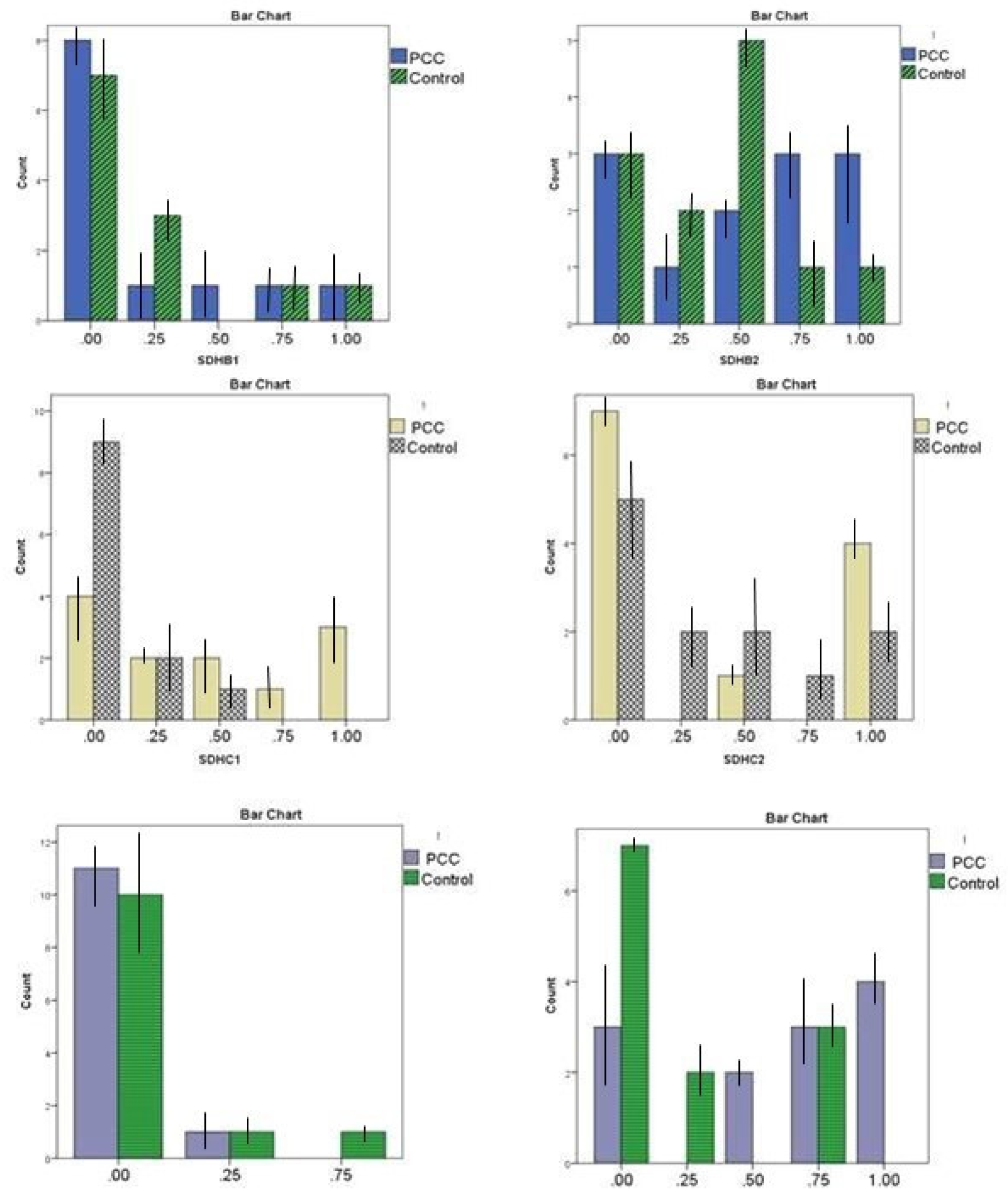
| Promoter Region | Methylation | PCCs/PGLs patients Number (percentage) | Control (percentage) | P-value |
|
SDHBa |
0-12.5% methylated (non-methylated) | 8 (66.6%) | 7 (58.3%) | 0.886 |
| 12.5 ≤, <25% methylated | 1 (8.33%) | 3 (25.0%) | ||
| 25≤, <50% methylated | 1 (8.33%) | 0 (0.0%) | ||
| 50≤, <75 % methylated | 1 (8.33%) | 1 (8.33%) | ||
| 75- 100% methylated | 1 (8.33%) | 1 (8.33%) | ||
|
SDHBb |
0-12.5% methylated (non-methylated) | 3 (25.0%) | 3 (25.0%) | 0.507 |
| 12.5 <, ≥25% methylated | 1 (8.33%) | 2 (16.6%) | ||
| 25≤, <50% methylated | 2 (16.6%) | 5 (41.6%) | ||
| 50≤, <75 % methylated | 3 (25.0%) | 1 (8.33%) | ||
| 75- 100% methylated | 3 (25.0%) | 1 (8.33%) | ||
|
SDHCa |
0-12.5% methylated (non-methylated) | 4 (33.3%) | 9 (75.0%) | 0.026* |
| 12.5 ≤, <25% methylated | 2 (16.6%) | 2 (16.6%) | ||
| 25≤, <50% methylated | 2 (16.6%) | 1 (8.33%) | ||
| 50≤, <75 % methylated | 1 (8.33%) | 0 (0.0%) | ||
| 75- 100% methylated | 3 (25.0%) | 0 (0.0%) | ||
|
SDHCb |
0-12.5% methylated (non-methylated) | 7 (58.3%) | 5 (41.6%) | 0.750 |
| 12.5 ≤, <25% methylated | 0 (0.0%) | 2 (16.6%) | ||
| 25≤, <50% methylated | 1 (8.33%) | 2 (16.6%) | ||
| 50≤, <75 % methylated | 0 (0.0%) | 1 (8.33%) | ||
| 75- 100% methylated | 4 (33.3%) | 2 (16.6%) | ||
| RDBPa | 0-12.5% methylated (non-methylated) | 11 (891.6%) | 10 (83.3%) | 0.987 |
| 12.5 ≤, <25% methylated | 1 (8.33%) | 1 (8.33%) | ||
| 25≤, <50% methylated | 0 (0.0%) | 1 (8.33%) | ||
| 50≤, <75 % methylated | 0 (0.0%) | 0 (0.0%) | ||
| 75- 100% methylated | 0 (0.0%) | 0 (0.0%) | ||
| RDBPb | 0-12.5% methylated (non-methylated) | 3 (25.0%) | 7 (58.3%) | 0.032* |
| 12.5 ≤, <25% methylated | 0 (0.0%) | 2 (16.6%) | ||
| 25≤, <50% methylated | 2 (16.6%) | 0 (0.0%) | ||
| 50≤, <75 % methylated | 3 (25.0%) | 3 (25.0%) | ||
| 75- 100% methylated | 4 (33.3%) | 0 (0.0%) |
| RDBP1 | RDBP2 | SDHB1 | SDHB2 | |
|---|---|---|---|---|
| SDHCa | .567 | .668 | .715 | .919 * |
| SDHCb | .624 | .824 * | .805 | .649 |
| The ctDNA promoter Locus | AUC (95 % CI) | Sensitivity | Specificity |
|---|---|---|---|
| SDHBa | 0.476 (0.240-0.712) | 25% | 83.3% |
| SDHBb | 0.622 (0.388 - 0.855) | 66.6% | 41.6% |
| SDHCa | 0.757 (0.558 - 0.956) | 50% | 91.6% |
| SDHCb | 0.483 (0.242–0.723) | 41.6% | 58.3% |
| RDBPa | 0.455 (0.220 – 0.690) | 21% | 91.6% |
| RDBPb | 0.750 (0.549 - 0.951) | 75% | 75% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





