Submitted:
12 April 2023
Posted:
13 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
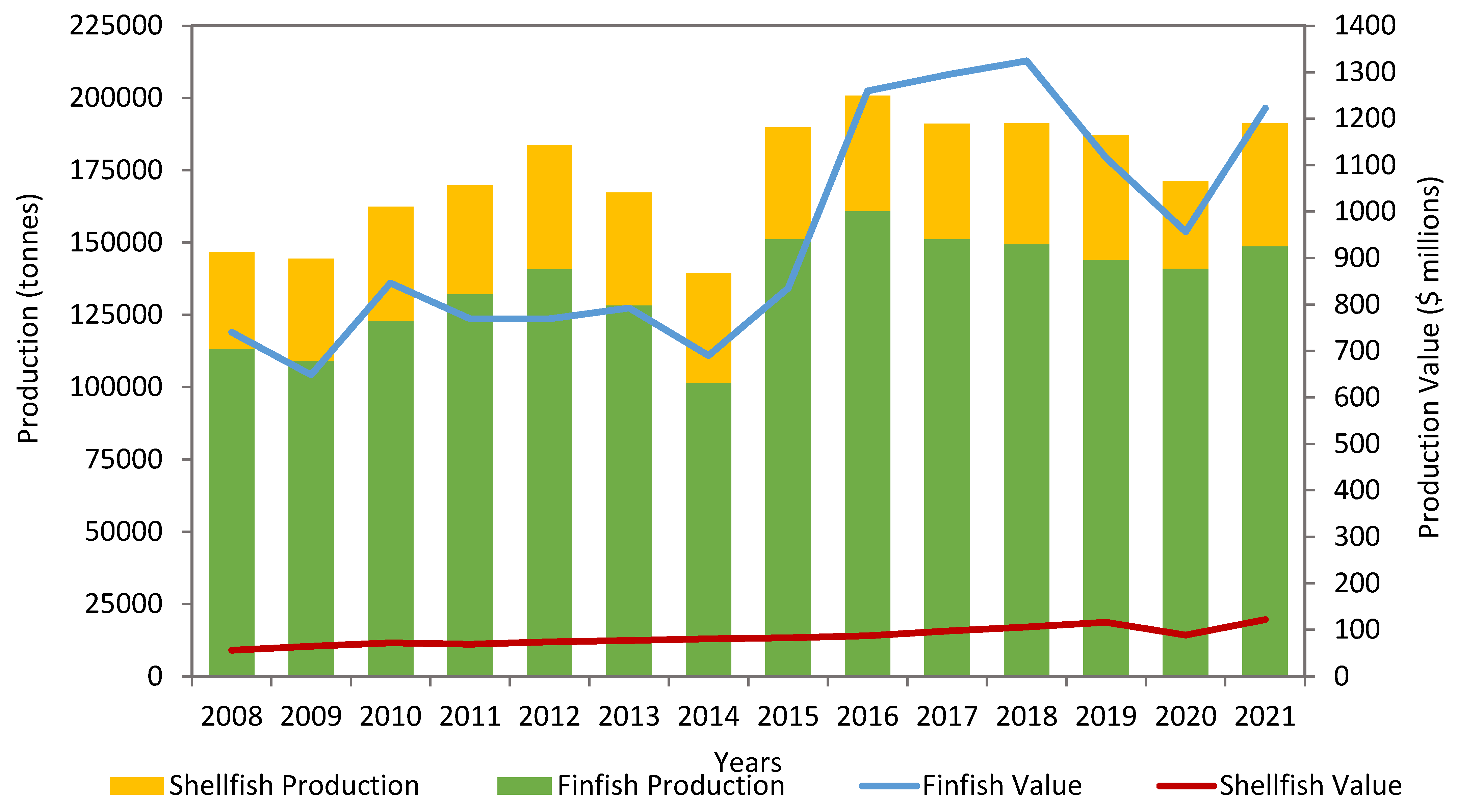
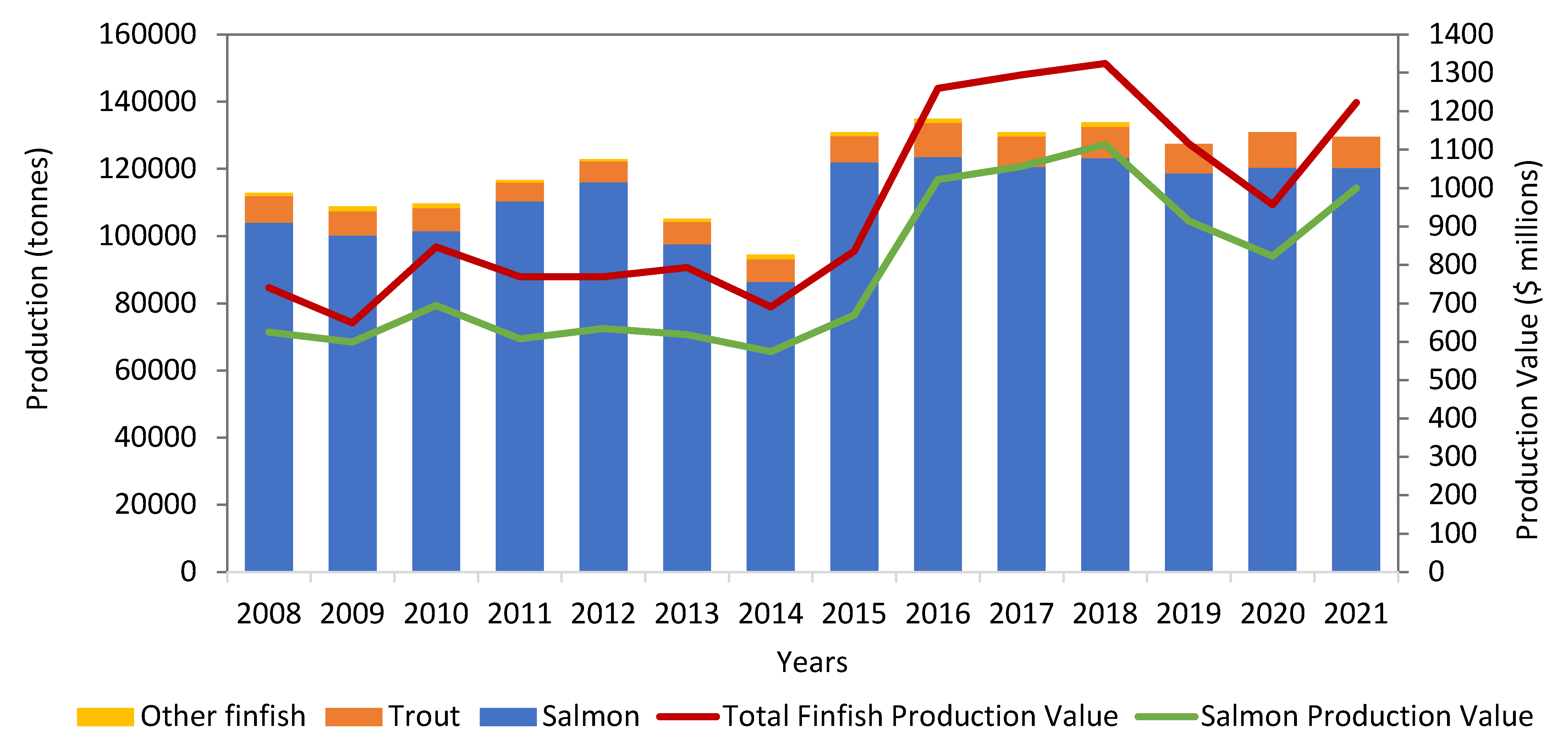
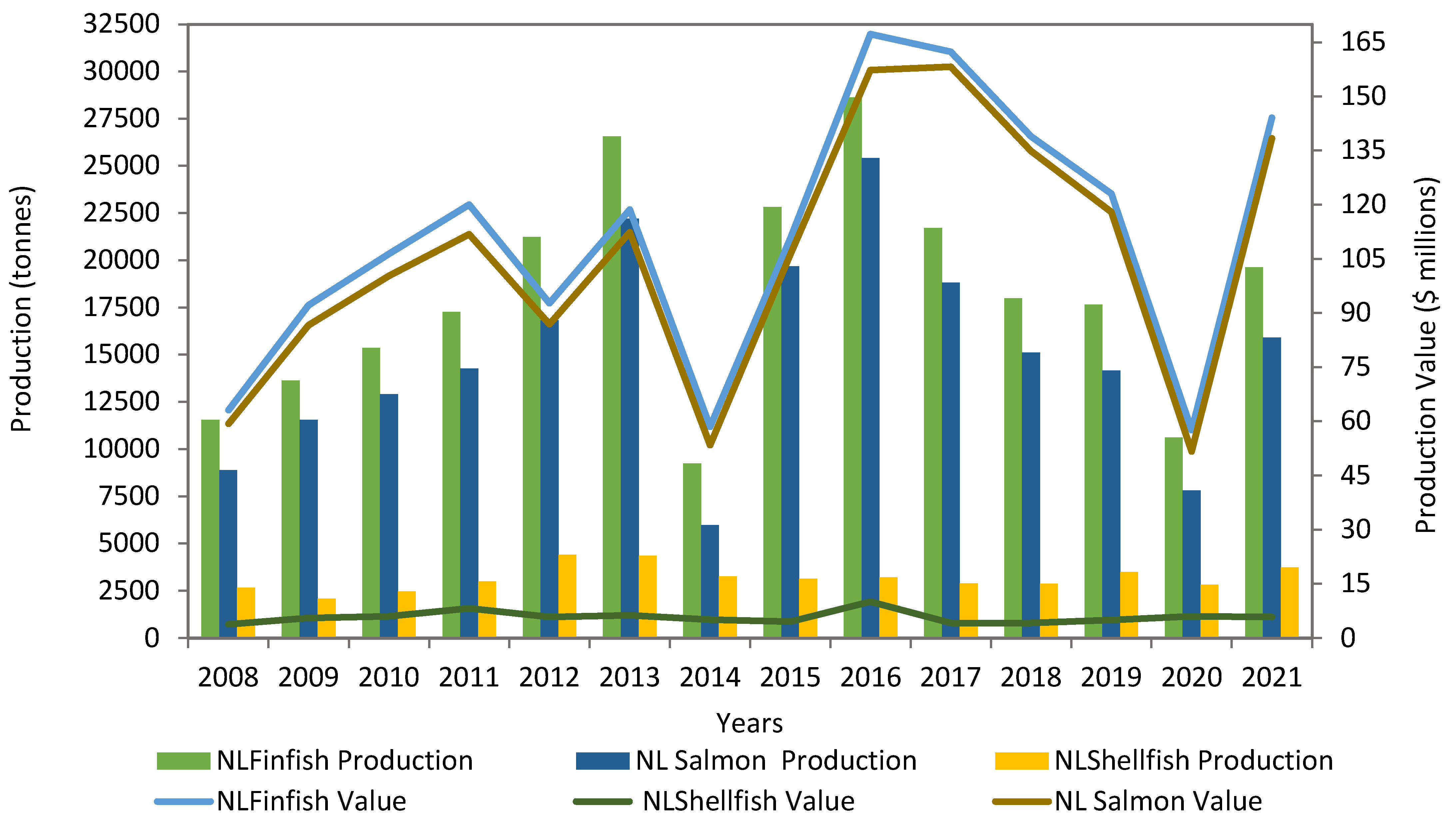
2. Salmon Production
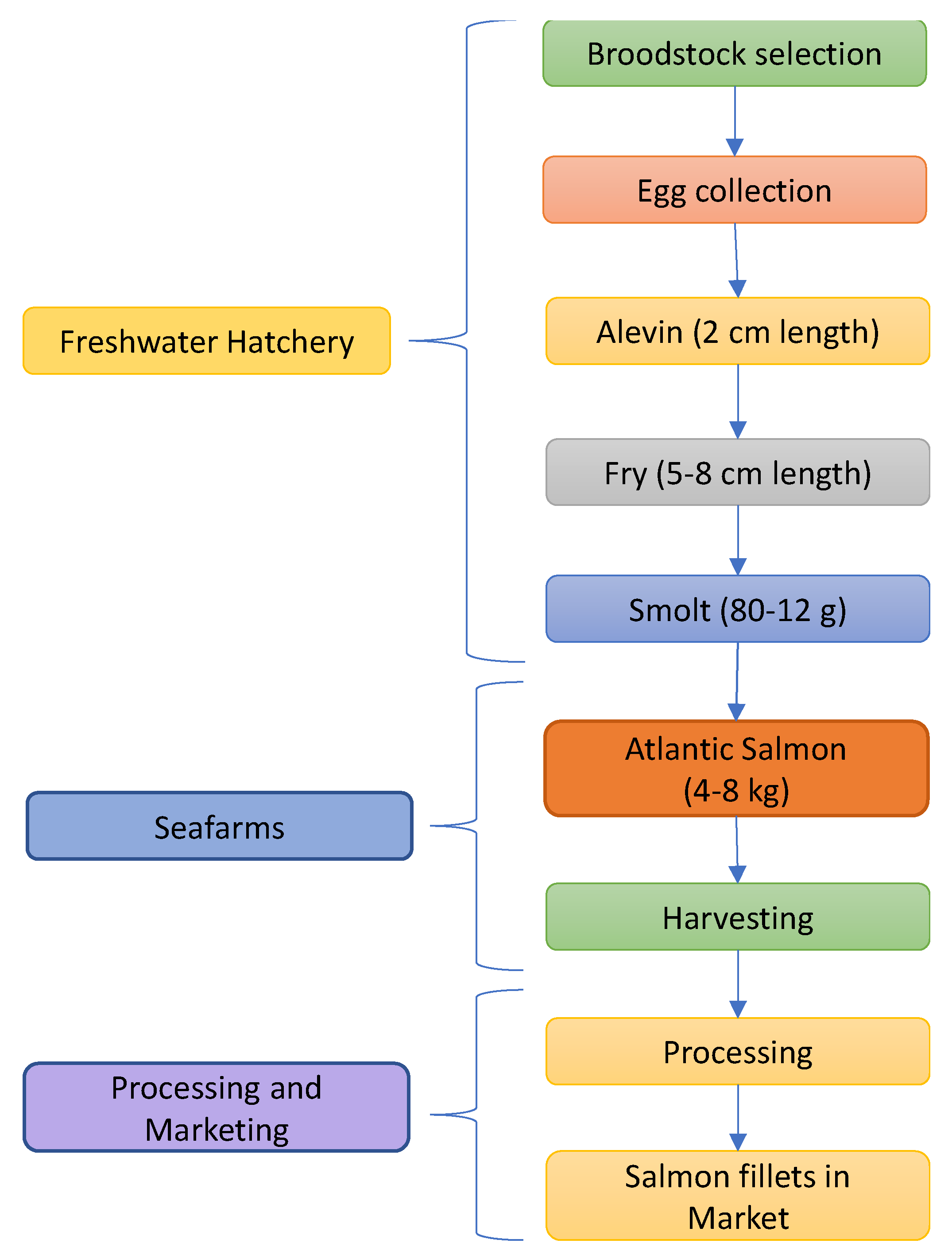
3. Atlantic Salmon Production Lifecycle
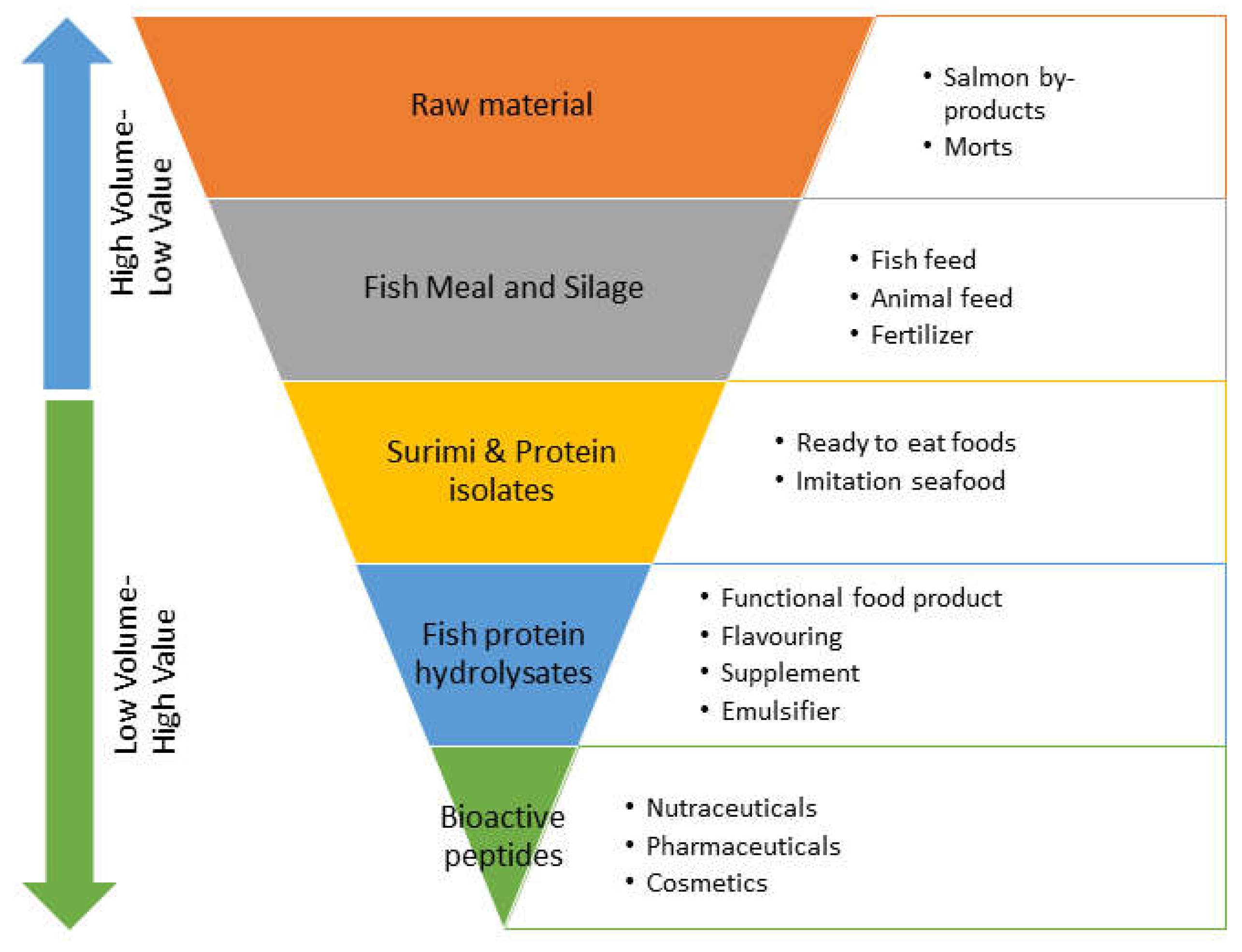
4. Atlantic Salmon Processing and Utilization of By-Products
5. Chemical Composition of Atlantic Salmon
5.1. Lipid and fatty acid composition
5.2. Protein and amino acid composition
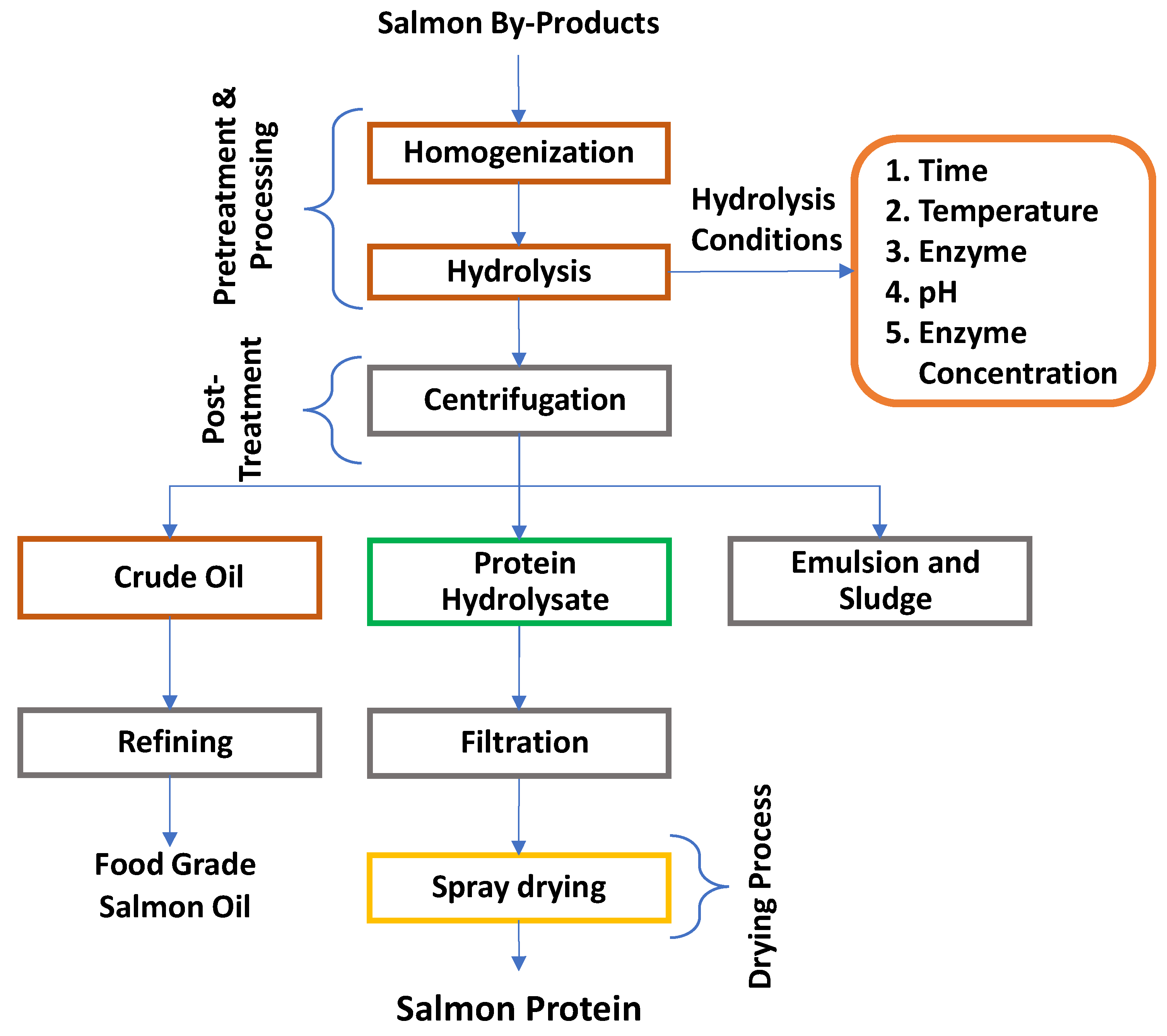
6. Production of Protein Hydrolysates
6.1. Chemical hydrolysis
6.2. Enzymatic hydrolysis
6.3. Ultrasound-assisted enzymatic extraction (UAEE)
6.4. High-pressure processing-enzymatic hydrolysis
7. Composition of Salmon Protein Hydrolysate
7.1. Functional properties of salmon protein hydrolysate
7.1.1. Solubility of proteins
7.1.2. Water holding capacity (WHC)
7.1.3. Emulsifying properties
7.1.4. Fat absorption capacity
7.2. Sensory properties of salmon protein hydrolysate
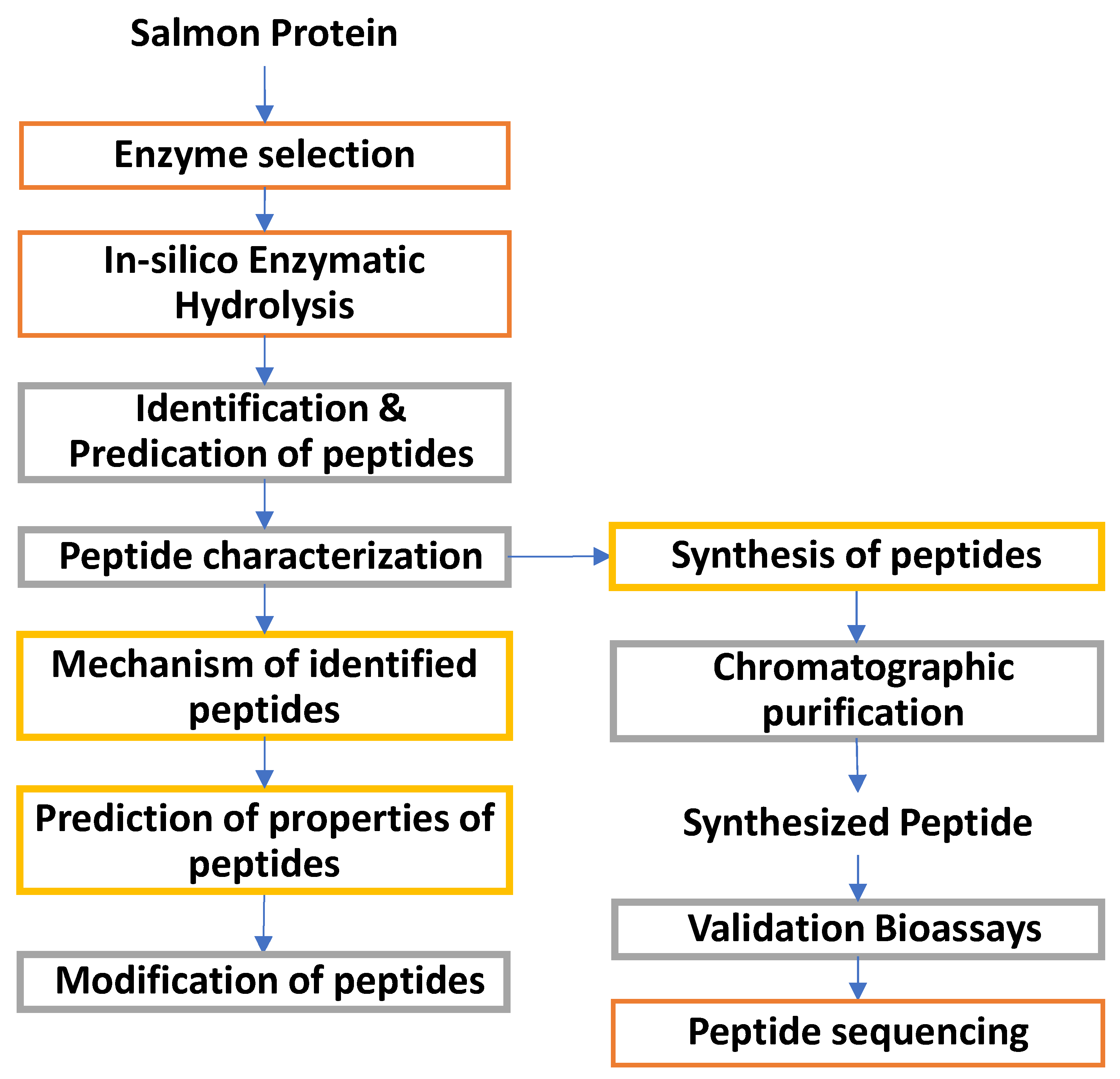
8. Bioactive Peptides
8.1. Empirical production of bioactive peptides and their functions
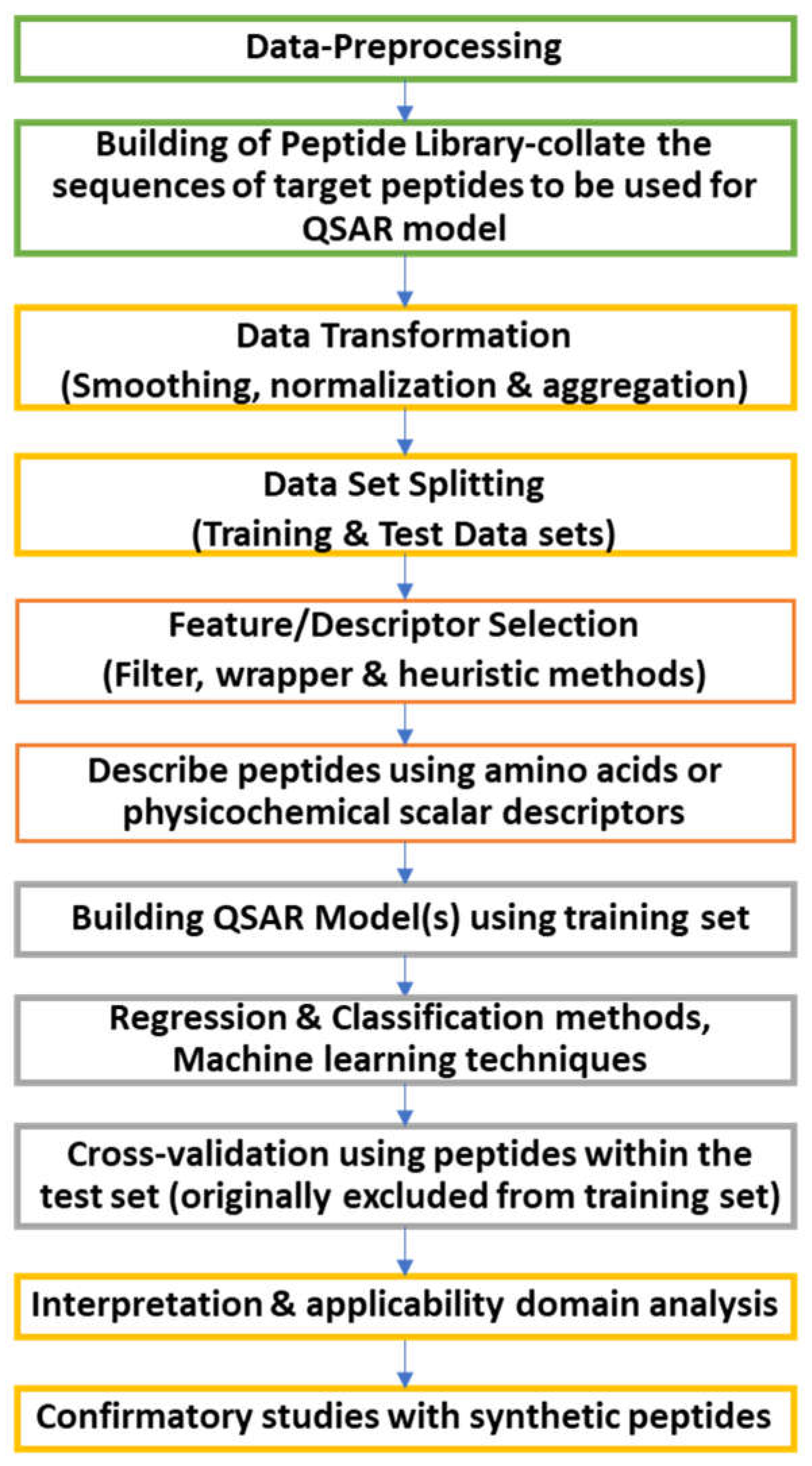
8.2. In-silico production of bioactive peptides
8.3. Computational characterization of peptides
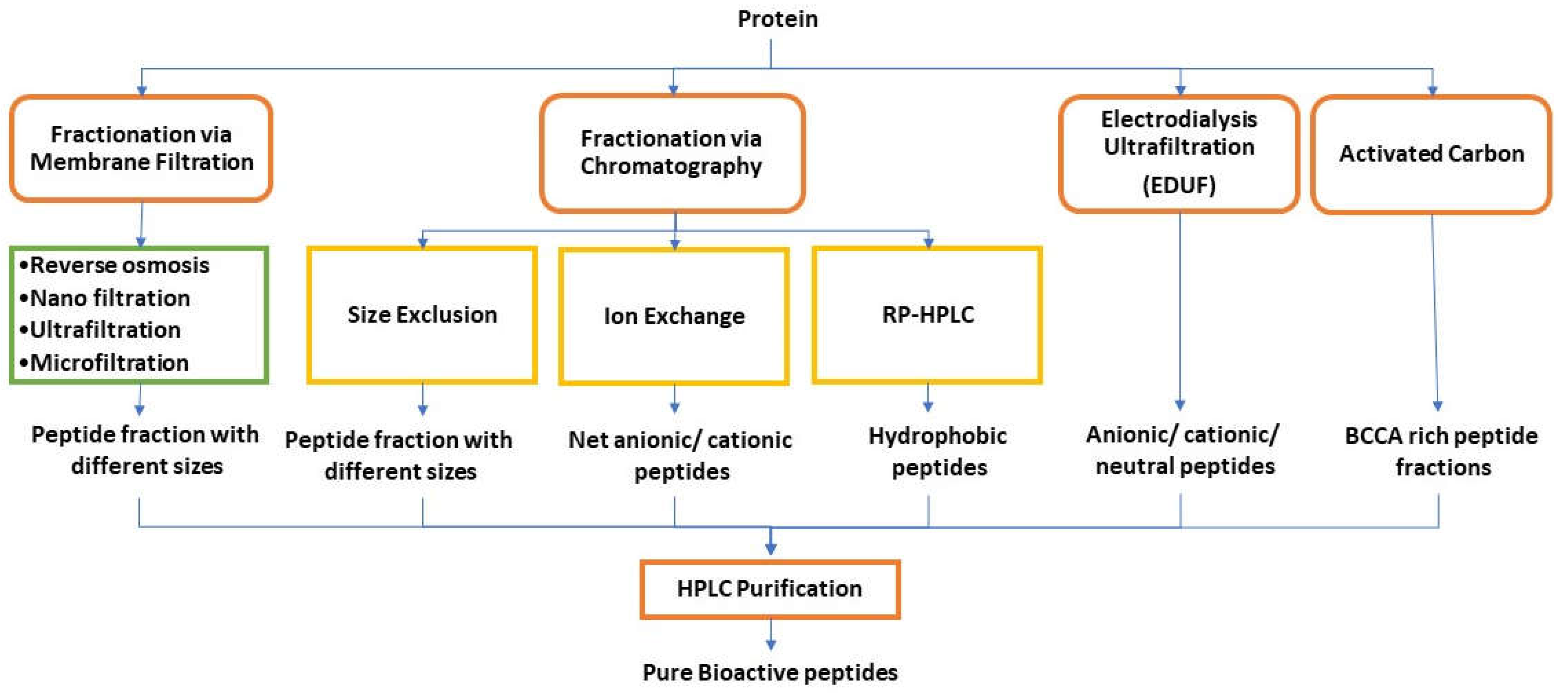
9. Purification and Separation of Peptides
9.1. Selective precipitation
9.2. Membrane filtration
9.3. Chromatographic techniques
9.3.1. Ion-exchange chromatography (IEC)
9.3.2. Size Exclusion chromatography (SEC)
9.3.3. Reversed-phase high-performance liquid chromatography (RP-HPLC)
10. Identification and Characterization of Peptides
11. Health Effects and Commercial Approval
12. Conclusion
Author Contributions
Funding
Conflicts of Interest
References
- Dekkers, E.; Raghavan, S.; Kristinsson, H.G.; Marshall, M.R. Oxidative stability of mahi mahi red muscle dipped in tilapia protein hydrolysates. Food Chem. 2011, 124, 640–645. [Google Scholar] [CrossRef]
- Chalamaiah, M.; Dinesh, K.B.; Hemalatha, R.; Jyothirmayi, T. Fish protein hydrolysates: Proximate composition, amino acid composition, antioxidant activities and applications: A review. Food Chem. 2012, 135, 3020–3038. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.R.; Newton, R.W.; Tlusty, M.; Little, D.C. The rise of aquaculture by-products: Increasing food production, value, and sustainability through strategic utilisation. Mar. Policy 2018, 90, 115–124. [Google Scholar] [CrossRef]
- Coppola, D.; Lauritano, C.; Esposito, F.P.; Riccio, G.; Rizzo, C.; de Pascale, D. Fish waste: From problem to valuable resource. Mar. Drugs 2021, 19, 116. [Google Scholar] [CrossRef]
- Ghaly, A.E.; Ramakrishnan, V.V.; Brooks, M.S.; Budge, S.M.; Dave, D. Fish processing wastes as a potential source of proteins, amino acids and oils: A critical review. J. Microb. Biochem. Technol. 2013, 5, 107–129. [Google Scholar]
- Routray, W.; Dave, D.; Ramakrishnan, V.V.; Murphy, W. Production of high quality fish oil by enzymatic protein hydrolysis from cultured atlantic salmon by-products: Investigation on effect of various extraction parameters using central composite rotatable design. Waste Biomass Valorization 2018, 9, 2003–2014. [Google Scholar] [CrossRef]
- Amarowicz, R.; Shahidi, F. Antioxidant activity of peptide fractions of capelin protein hydrolysates. Food Chem. 1997, 58, 355–359. [Google Scholar] [CrossRef]
- Kristinsson, H.G.; and Rasco, B.A. Kinetics of the hydrolysis of Atlantic salmon (Salmo salar) muscle proteins by alkaline proteases and a visceral serine protease mixture. Process Biochem. 2000, 36, 131–139. [Google Scholar] [CrossRef]
- Kristinsson, H.G.; Rasco, B.A. Hydrolysis of salmon muscle proteins by an enzyme mixture extracted from Atlantic salmon (Salmo salar) pyloric caeca. J. Food Biochem. 2000, 24, 177–187. [Google Scholar] [CrossRef]
- Thorkelsson, G.; Kristinsson, H. Bioactive peptides from marine sources. State of art. Report to the NORA fund. Peptides 2009, 1–19. [Google Scholar]
- Harnedy, P.A.; FitzGerald, R.J. Bioactive peptides from marine processing waste and shellfish: A review. J. Funct. Foods 2012, 4, 6–24. [Google Scholar] [CrossRef]
- Chalamaiah, M.; Yu, W.; Wu, J. Immunomodulatory and anticancer protein hydrolysates (peptides) from food proteins: A review. Food Chem. 2018, 245, 205–222. [Google Scholar] [CrossRef] [PubMed]
- Chalamaiah, M.; Keskin, U.S.; Hong, H.; Wu, J. Regulatory requirements of bioactive peptides (protein hydrolysates) from food proteins. J. Funct. Foods 2019, 58, 123–129. [Google Scholar] [CrossRef]
- Yarnpakdee, S.; Benjakul, S.; Kristinsson, H.G. Lipid oxidation and fishy odour in protein hydrolysate derived from Nile tilapia (Oreochromis niloticus) protein isolate as influenced by haemoglobin. J. Sci. Food Agric. 2014, 94, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Yarnpakdee, S.; Benjakul, S.; Kristinsson, H.G.; Kishimura, H. Antioxidant and sensory properties of protein hydrolysate derived from Nile tilapia (Oreochromis niloticus) by one- and two-step hydrolysis. J. Food Sci. Technol. 2015, 52, 3336–3349. [Google Scholar] [CrossRef] [PubMed]
- Opheim, M.; Šližyte, R.; Sterten, H.; Provan, F.; Larssen, E.; Kjos, N.P. Hydrolysis of Atlantic salmon (Salmo salar) rest raw materials - Effect of raw material and processing on composition, nutritional value, and potential bioactive peptides in the hydrolysates. Process Biochem. 2015, 50, 1247–1257. [Google Scholar] [CrossRef]
- Neves, A.C.; Harnedy, P.A.; O’Keeffe, M.B.; Alashi, M.A.; Aluko, R.E.; FitzGerald, R.J. Peptide identification in a salmon gelatin hydrolysate with antihypertensive, dipeptidyl peptidase IV inhibitory and antioxidant activities. Food Res. Int. 2017, 100, 112–120. [Google Scholar] [CrossRef]
- Neves, A.C.; Harnedy, P.A.; O’Keeffe, M.B.; FitzGerald, R.J. Bioactive peptides from Atlantic salmon (Salmo salar) with angiotensin converting enzyme and dipeptidyl peptidase IV inhibitory, and antioxidant activities. Food Chem. 2017, 218, 396–405. [Google Scholar] [CrossRef]
- Kristinsson, H.G.; Rasco, B.A. Biochemical and functional properties of Atlantic salmon (Salmo salar) muscle proteins hydrolyzed with various alkaline proteases. J. Agric. Food Chem. 2000, 48, 657–666. [Google Scholar] [CrossRef]
- Ishak, N.H.; Sarbon, N.M. A review of protein hydrolysates and bioactive peptides deriving from wastes generated by fish processing. Food Bioprocess Technol. 2018, 11, 2–16. [Google Scholar] [CrossRef]
- DFO. Aquaculture production quantities and value. In: Fish Ocean Canada, 2021. https://www.dfo-mpo.gc.ca/stats/aqua/aqua-prod-eng.htm. Accessed 2 Jul 2021.
- GovNL. Aquaculture - Newfoundland & Labrador Canada. In: Invest Aquac. 2021. https://www.findnewfoundlandlabrador.com/invest/aquaculture/. Accessed 2 Jul 2021.
- Seafish. Fishmeal and fish oil figures. 2011. Seafish 30.
- DFO. State of salmon aquaculture technologies. 2019. 2019.
- MOWI. Salmon Farming Industry Handbook 2019. 2019.
- ACCFA. Growing our fish. In: Atl. Fish Farmers. 2021. https://www.atlanticfishfarmers.com/growing-our-fish%0Ahttps://www.atlanticfishfarmers.com/growing-our-fish#:~:text=Stocking,between 15%2C000 and 30%2C000 smolts. Accessed 2 Jul 2021.
- Borderías, A.J; a Sánchez-alonso, I. First processing steps and the quality of wild and farmed fish. J. Food Sci. 2011, 76, R1–R5. [Google Scholar] [CrossRef] [PubMed]
- Frida, N.-U.; Nesse, A.G. Smoother harvest of farmed salmon, value-adding or costly? Norwegian University of Science and Technology 2014. [Google Scholar]
- Dave, D.; Routray, W. Current scenario of Canadian fishery and corresponding underutilized species and fishery byproducts: A potential source of omega-3 fatty acids. J. Clean. Prod. 2018, 180, 617–641. [Google Scholar] [CrossRef]
- Kim, Y.S.; Park, J.W. Negative roles of salt in gelation properties of fish protein isolate. J Food Sci. 2008, 73, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Nolsøe, H.; Undeland, I. The acid and alkaline solubilization process for the isolation of muscle proteins: State of the art. Food Bioproc. Tech. 2009, 2, 1–27. [Google Scholar] [CrossRef]
- Ghaly, A.E.; Dave, D.; Brooks, M.S.; Budge, S. Production of biodiesel by enzymatic transesterification: Review. Am. J. Biochem. Biotechnol. 2010, 6, 54–76. [Google Scholar] [CrossRef]
- Aas, T.S.; Åsgård, T.; Ytrestøyl, T. Chemical composition of whole body and fillet of slaughter sized Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss) farmed in Norway in 2020. Aquac. Rep. 2022, 25, 101252. [Google Scholar] [CrossRef]
- Dave, D.; Ramakrishnan, V.V.; Trenholm, S.; Manuel, H.; Pohling, J.; Murphy, W. Marine oils as potential feedstock for biodiesel production: Physicochemical characterization. J Bioprocess Biotech. 2014, 4, 1–12. [Google Scholar] [CrossRef]
- Malcorps, W.; Newton, W.R.; Sprague, M.; Glencross, B.D.; Little, C.D. Nutritional characterisation of European aquaculture processing by-products to facilitate strategic utilisation. Front Sustain Food Syst. 2021, 5, 720595. [Google Scholar] [CrossRef]
- Kralovec, J.A.; Zhang, S.; Zhang, W.; Barrow, C.J. A review of the progress in enzymatic concentration and microencapsulation of omega-3 rich oil from fish and microbial sources. Food Chem. 2012, 131, 639–644. [Google Scholar] [CrossRef]
- Liu, Y.; Ramakrishnan, V.V.; Dave, D. Lipid class and fatty acid composition of oil extracted from Atlantic salmon by-products under different optimization parameters of enzymatic hydrolysis. Biocatal Agric Biotechnol. 2020, 30, 101866. [Google Scholar] [CrossRef]
- Polvi, S.M.; Ackman, R.G. Atlantic salmon (Salmo salar) muscle lipids and their response to alternative dietary fatty acid sources. J. Agric. Food Chem. 1992, 40, 1001–1007. [Google Scholar] [CrossRef]
- Głowacz-Różyńska, A.; Tynek, M.; Malinowska-Pańczyk, E.; Martysiak-Żurowska, D.; Pawłowicz, R.; Kołodziejska, I. Comparison of oil yield and quality obtained by different extraction procedures from salmon (Salmo salar) processing byproducts. Eur. J. Lipid Sci. Technol. 2016, 118, 1759–1767. [Google Scholar] [CrossRef]
- Liaset, B.; Julshamn, K.; Espe, M. Chemical composition and theoretical nutritional evaluation of the produced fractions from enzymic hydrolysis of salmon frames with ProtamexTM. Process Biochem. 2003, 38, 1747–1759. [Google Scholar] [CrossRef]
- Haq, M.; Ahmed, R.; Cho, Y.J.; Chun, B.S. Quality Properties and bio-potentiality of edible oils from Atlantic salmon by-products extracted by supercritial carbon dioxide and conventional methods. Waste Biomass Valorization 2017, 8, 1953–1967. [Google Scholar] [CrossRef]
- Colombo, S.M.; Mazal, X. Investigation of the nutritional composition of different types of salmon available to Canadian consumers. J. Agric. Food Res. 2020, 2, 100056. [Google Scholar] [CrossRef]
- Fomena-Temgoua, N.S.; Sun, Z.; Okoye, C.O.; Pan, H. Fatty acid profile, physicochemical composition, and sensory properties of Atlantic salmon fish (Salmo salar) during Different culinary treatments. J. Food Qual. 2022, 7425142. [Google Scholar] [CrossRef]
- Cheung, R.C.F.; Ng, T.B.; Wong, J.H. Marine peptides: Bioactivities and applications. Mar. Drugs 2015, 13, 4006–4043. [Google Scholar] [CrossRef]
- Senadheera, T.R.L.; Hossain, A.; Dave, D.; Shahidi, F. Functional and physiochemical properties of protein isolates from different body parts of North Atlantic sea cucumber (Cucumaria frondosa). Food Biosci 2023, 52, 102511. [Google Scholar] [CrossRef]
- Hossain, A.; Dave, D.; Shahidi, F. Antioxidant potential of sea cucumbers and their beneficial effects on human health. Mar. Drugs 2002, 20, 521. [Google Scholar] [CrossRef]
- Kristinsson, H.G.; Rasco, B.A. Fish protein hydrolysates: Production, biochemical, and functional properties. Crit. Rev. Food Sci. Nutr. 2000, 40, 43–81. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Bond, J.S. Proteases: Multifunctional enzymes in life and disease. J. Biol. Chem. 2008, 283, 30433–30437. [Google Scholar] [CrossRef] [PubMed]
- Senadheera, T.R.L.; Dave, D.; Shahidi, F. Antioxidant potential and physicochemical properties of protein hydrolysates from body parts of North Atlantic sea cucumber (Cucumaria frondosa). Food Prod. Process. Nutr. 2021, 3, 1–22. [Google Scholar] [CrossRef]
- Shen, H.B.; Chou, K.C. Identification of proteases and their types. Anal. Biochem. 2009, 385, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, M.; Undeland, I. Physicochemical and gel-forming properties of protein isolated from salmon, cod and herring by-products using the pH-shift method. LWT - Food Sci. Technol 2019, 101, 678–684. [Google Scholar] [CrossRef]
- Xiong, Y.L. Muscle proteins. In Muscle proteins. In: Proteins in Food Processing; Elsevier Ltd., 2004; pp. 100–122. [Google Scholar]
- Ramakrishnan, V.V.; Ghaly, A.E.; Brooks, M.S.; Budge, S.M. Extraction of proteins from mackerel fish processing waste using Alcalase enzyme. J. Bioprocess. Biotech. 2013, 3, 1–9. [Google Scholar]
- Dave, D.; Vegneshwaran, V.R.; Julia, P.; Sukhinder, K.C.; Sheila, T.; Heather, M.; Wade, M. Investigation on oil extraction methods and its influence on omega-3 content from cultured salmon. J. Food Process. Technol. 2014, 5, 1000401. [Google Scholar]
- Hou, Y.; Wu, Z.; Dai, Z.; Wang, G.; Wu, G. Protein hydrolysates in animal nutrition: Industrial production, bioactive peptides, and functional significance. J. Anim. Sci. Biotechnol. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Petrova, I.; Tolstorebrov, I.; Eikevik, T.M. Production of fish protein hydrolysates step by step: Technological aspects, equipment used, major energy costs and methods of their minimizing. Int. Aquat. Res. 2018, 10, 223–241. [Google Scholar] [CrossRef]
- Kadam, S.U.; Tiwari, B.K.; Álvarez, C.; O’Donnell, C.P. Ultrasound applications for the extraction, identification and delivery of food proteins and bioactive peptides. Trends Food Sci. Technol. 2015, 46, 60–67. [Google Scholar] [CrossRef]
- Grosso, C.; Valentão, P.; Ferreres, F.; Andrade, P.B. Alternative and efficient extraction methods for marine-derived compounds. Mar. Drugs 2015, 13, 3182–3230. [Google Scholar] [CrossRef] [PubMed]
- Gbogouri, G.A.; Linder, M.; Fanni, J.; Parmentier, M. Influence of hydrolysis degree on the functional properties of salmon byproducts hydrolysates. J. Food Sci. 2004, 69, C615–C622. [Google Scholar] [CrossRef]
- See, S.F.; Hoo, L.L.; Babji, A.S. Optimization of enzymatic hydrolysis of salmon (Salmo salar) skin by Alcalase. Int. Food Res. J. 2011, 18, 1359–1365. [Google Scholar]
- Zhang, X.; Dai, Z.; Zhang, Y.; Dong, Y.; Hu, X. Structural characteristics and stability of salmon skin protein hydrolysates obtained with different proteases. LWT - Food Sci. Technol 2022, 153, 112460. [Google Scholar] [CrossRef]
- Slizyte, R.; Rommi, K.; Mozuraityte, R.; Eck, P.; Five, K.; Rustad, T. Bioactivities of fish protein hydrolysates from defatted salmon backbones. Biotechnol. Rep. 2016, 11, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Aspevik, T.; Egede-Nissen, H.; Oterhals, Å. A systematic approach to the comparison of cost efficiency of endopeptidases for the hydrolysis of Atlantic salmon (Salmo salar) by-products. Food Technol. Biotechnol 2016, 54, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Liaset, B.; Nortvedt, R.; Lied, E.; Espe, M. Studies on the nitrogen recovery in enzymic hydrolysis of Atlantic salmon (Salmo salar, L.) frames by Protamex™ protease. Process Biochem 2002, 37, 1263–1269. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Sotelo, C.G.; Sanz, N.; Pérez-Martín, R.I.; Amado, I.R.; Valcarcel, J. Valorization of aquaculture by-products of salmonids to produce enzymatic hydrolysates: Process optimization, chemical characterization and evaluation of bioactives. Mar. Drugs 2019, 17, 676. [Google Scholar] [CrossRef]
- Idowu, A.T.; Benjakul, S.; Sinthusamran, S.; Sookchoo, P.; Kishimura, H. Protein hydrolysate from salmon frames: Production, characteristics and antioxidative activity. J. Food Biochem. 2019, 43, e12734. [Google Scholar] [CrossRef]
- Aspevik, T.; Steinsholm, S.; Vang, B.; Carlehög, M.; Arnesen, J.A.; Kousoulaki, K. Nutritional and sensory properties of protein hydrolysates based on salmon (Salmo salar), mackerel (Scomber scombrus), and herring (Clupea harengus) heads and backbones. Front. Nutr. 2021, 8, 695151. [Google Scholar] [CrossRef]
- Kangsanant, S.; Murkovic, M.; Thongraung, C. Antioxidant and nitric oxide inhibitory activities of tilapia (Oreochromis niloticus) protein hydrolysate: Effect of ultrasonic pretreatment and ultrasonic-assisted enzymatic hydrolysis. Int. J. Food Sci. Technol. 2014, 49, 1932–1938. [Google Scholar] [CrossRef]
- Álvarez, C.; Lélu, P.; Lynch, S.A.; Tiwari, B.K. Optimised protein recovery from mackerel whole fish by using sequential acid/alkaline isoelectric solubilization precipitation (ISP) extraction assisted by ultrasound. LWT - Food Sci. Technol 2018, 88, 210–216. [Google Scholar] [CrossRef]
- Misir, G.B.; Koral, S. Effects of ultrasound treatment on structural, chemical and functional properties of protein hydrolysate of rainbow trout (Oncorhynchus mykiss) by-products. Ital J Food Sci. 2019, 31, 205–223. [Google Scholar]
- Tian, J.; Wang, Y.; Zhu, Z.; Zeng, Q.; Xin, M. Recovery of tilapia (Oreochromis niloticus) protein isolate by high-intensity ultrasound-aided alkaline isoelectric solubilization/precipitation process. Food Bioproc. Tech. 2015, 8, 758–769. [Google Scholar] [CrossRef]
- Elamin, W.M.; Endan, J.B.; Yosuf, Y.A.; Shamsudin, R.; Ahmedov, A. High pressure processing technology and equipment evolution: A review. J. Eng. Sci. Technol. Rev. 2015, 8, 75–83. [Google Scholar] [CrossRef]
- Hossain, A.; Dave, D.; Shahidi, F. Effect of high-pressure processing (hpp) on phenolics of North Atlantic sea cucumber (Cucumaria frondosa). J. Agric. Food Chem. 2022, 70, 3489–3501. [Google Scholar] [CrossRef] [PubMed]
- Marciniak, A.; Suwal, S.; Naderi, N.; Pouliot, Y.; Doyen, A. Enhancing enzymatic hydrolysis of food proteins and production of bioactive peptides using high hydrostatic pressure technology. Trends Food Sci. Technol. 2018, 80, 187–198. [Google Scholar] [CrossRef]
- Rivalain, N.; Roquain, J.; Demazeau, G. Development of high hydrostatic pressure in biosciences: Pressure effect on biological structures and potential applications in Biotechnologies. Biotechnol. Adv 2010, 28, 659–672. [Google Scholar] [CrossRef]
- Boonyaratanakornkit, B.B.; Park, C.B.; Clark, D.S. Pressure effects on intra- and intermolecular interactions within proteins. Biochim. Biophys. Acta - Protein Struc. Mol. Enzymol 2002, 1595, 235–249. [Google Scholar] [CrossRef]
- Royer, C.A. Insights into the role of hydration in protein structure and stability obtained through hydrostatic pressure studies. Braz. J. Med. Biol. Res. 2005, 38, 1167–1173. [Google Scholar] [CrossRef]
- Silva, J.L.; Foguel, D. Hydration, cavities and volume in protein folding, aggregation and amyloid assembly. Phys. Biol. 2009, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Grigera, J.R.; McCarthy, A.N. The behavior of the hydrophobic effect under pressure and protein denaturation. Biophysical J. 2010, 98, 1626–1631. [Google Scholar] [CrossRef] [PubMed]
- Knorr, D.; Heinz, V.; Buckow, R. High pressure application for food biopolymers. Biochim. Biophys. Acta - Proteins Proteom 2006, 1764, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, G.; Schwalbe, H. Protein NMR spectroscopy: Hydrogen bonds under pressure. Nat. Chem. 2012, 4, 693–695. [Google Scholar] [CrossRef] [PubMed]
- Alemán, A.; Giménez, B.; Gómez-Guillén, M.C. ’ Montero, P. Enzymatic hydrolysis of fish gelatin under high pressure treatment. Int. J. Food Sci. Technol. 2011, 46, 1129–1136. [Google Scholar] [CrossRef]
- Hemker, A.K.; Nguyen, L.T.; Karwe, M.; Salvi, D. Effects of pressure-assisted enzymatic hydrolysis on functional and bioactive properties of tilapia (Oreochromis niloticus) by-product protein hydrolysates. LWT - Food Sci. Technol. 2020, 122, 109003. [Google Scholar] [CrossRef]
- Nakajima, K.; Yoshie-Stark, Y.; Ogushi, M. Comparison of ACE inhibitory and DPPH radical scavenging activities of fish muscle hydrolysates. Food Chem. 2009, 114, 844–851. [Google Scholar] [CrossRef]
- Harnedy, P.A.; Parthsarathy, V.; McLaughlin, C.M.; O’Keeffe, M.B.; Allsopp, P.J.; McSorley, E.M.; O’Harte, F.P.M.; FitzGerald, R.J. Atlantic salmon (Salmo salar) co-product-derived protein hydrolysates: A source of antidiabetic peptides. Food Res. Int. 2018, 106, 598–606. [Google Scholar] [CrossRef]
- Abdollahi, M.; Undeland, I. Structural, functional, and sensorial properties of protein isolate produced from salmon, cod, and herring by-products. Food Bioproc. Tech. 2018, 11, 1733–1749. [Google Scholar] [CrossRef]
- Ucak, I.; Afreen, M.; Montesano, D.; Carrillo, C.; Tomasevic, I.; Simal-Gandara, J.; Barba, F.J. Functional and bioactive properties of peptides derived from marine side streams. Mar. Drugs 2021, 19, 71. [Google Scholar] [CrossRef]
- Zayas, J.F. Functionality of proteins in food; Springer: Berlin/Heidelberg, Germany, 1997. [Google Scholar]
- Jones, S.M. Quality and Grading of Carcasses of Meat Animals. CRC Press Jungbauer, A. and Hahn, R. 2009, Chapter 22 Ion-Exchange Chromatography. In: Methods in Enzymology. Academic Press Inc., pp 349–371.
- Munasinghe, D.M.S.; Sakai, T. Sodium chloride (0.8 M) as a better protein extractant for fish meat quality assessments. J. Food Sci 2003, 68, 1059–1062. [Google Scholar] [CrossRef]
- Chan, S.K.W.; Roth, B.; Jessen, F.; Jakobsen, A.N.; Lerfall, J. Water holding properties of Atlantic salmon. Compr. Rev. Food Sci. Food Saf. 2021, 21, 477–498. [Google Scholar] [CrossRef] [PubMed]
- Lam, R.S.H.; Nickerson, M.T. Food proteins: A review on their emulsifying properties using a structure-function approach. Food Chem. 2013, 141, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.A.; Timilsena, Y.P.; Adhikari, B. Food proteins, structure, and function. Ref. Modul. Food Sci. 2016. [Google Scholar]
- Damodaran, S.; Parkin, K.L.; Fennema, O.R. Fennema’s food chemistry, 4th Eds., 2007, CRC Press.
- He, S.; Franco, C.; Zhang, W. Process optimisation and physicochemical characterisation of enzymatic hydrolysates of proteins from co-products of Atlantic salmon (Salmo salar) and yellowtail kingfish (Seriola lalandi). Int. J. Food Sci. Technol. 2012, 47, 2397–2404. [Google Scholar] [CrossRef]
- Saha, B.C.; Hayashi, K. Debittering of protein hydrolyzates. Biotechnol. Adv. 2001, 19, 355–370. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.J.; Unklesbay, N.; Hsieh, F.H.; Clarke, A.D. Hydrophobicity of bitter peptides from soy protein hydrolysates. J. Agric. Food Chem 2004, 52, 5895–5901. [Google Scholar] [CrossRef]
- Sun, X.D. Enzymatic hydrolysis of soy proteins and the hydrolysates utilisation. Int. J. Food Sci. Technol. 2011, 46, 2447–2459. [Google Scholar] [CrossRef]
- Benjakul, S.; Yarnpakdee, S.; Senphan, T.; Halldorsdottir, S.M.; Kristinsson, H.G. Fish protein hydrolysates: Production, bioactivities, and applications. In: Antioxidants and Functional Components in Aquatic Foods. 2014, John Wiley & Sons, Ltd, Chichester, UK, pp 237–281.
- Xu, Q.; Hong, H.; Yu, W.; Jiang, X.; Yan, X.; Wu, J. Sodium chloride suppresses the bitterness of protein hydrolysates by decreasing hydrophobic interactions. J. Food Sci. 2019, 84, 86–91. [Google Scholar] [CrossRef]
- Gong, M.; Mohan, A.; Gibson, A.; Udenigwe, C.C. Mechanisms of plastein formation, and prospective food and nutraceutical applications of the peptide aggregates. Biotechnol. Rep. 2015, 5, 63–69. [Google Scholar] [CrossRef]
- Aspevik, T.; Totland, C.; Lea, P.; Oterhals, Å. Sensory and surface-active properties of protein hydrolysates based on Atlantic salmon (Salmo salar) by-products. Process Biochem. 2016, 51, 1006–1014. [Google Scholar] [CrossRef]
- Shahidi, F.; Zhong, Y. Bioactive peptides. J. AOAC Int. 2008, 91, 914–931. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, C.G.; Tagliazucchi, D.; Babini, E.; Sefora, R.G.; Taneyo, S.D.L.; Gianotti, A. Bioactive peptides from vegetable food matrices: Research trends and novel biotechnologies for synthesis and recovery. J. Funct. Foods 2016, 27, 549–569. [Google Scholar] [CrossRef]
- Daliri, E.B.M.; Oh, D.H.; Lee, B.H. Bioactive peptides. Foods 2017, 6, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Ryan, J.T.; Ross, R.P.; Bolton, D.; Fitzgerald, G.F.; Stanton, C. Bioactive peptides from muscle sources: Meat and fish. Nutrients 2011, 3, 765–791. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, H.; Xing, R.; Li, P. Characterization, preparation, and purification of marine bioactive peptides. Biomed. Res. Int. 2017, 2017, 9746720. [Google Scholar] [CrossRef] [PubMed]
- Phadke, G.G.; Rathod, N.B.; Ozogul, F.; Elavarasan, K.; Karthikeyan, M.; Shin, K.-H.; Kim, S.-K. Exploiting of secondary raw materials from fish processing industry as a source of bioactive peptide-rich protein hydrolysates. Mar. Drugs 2021, 19, 480. [Google Scholar] [CrossRef]
- Ahn, C.B.; Jeon, Y.J.; Kim, Y.T.; Je, J.Y. AngiotensinI converting enzyme (ACE) inhibitory peptides from salmon byproduct protein hydrolysate by Alcalase hydrolysis. Process Biochem. 2012, 47, 2240–2245. [Google Scholar] [CrossRef]
- Gu, R.Z.; Li, C.Y.; Liu, W.Y.; Yi, W.X.; Cai, M.Y. Angiotensin I-converting enzyme inhibitory activity of low-molecular-weight peptides from Atlantic salmon (Salmo salar L.) skin. Food Res. Int 2011, 44, 1536–1540. [Google Scholar] [CrossRef]
- Ahn, C.B.; Cho, Y.S.; Je, J.Y. Purification and anti-inflammatory action of tripeptide from salmon pectoral fin byproduct protein hydrolysate. Food Chem. 2015, 168, 151–156. [Google Scholar] [CrossRef]
- Li-Chan, E.C.Y.; Hunag, S.L.; Jao, C.L.; Ho, K.P.; Hsu, K.C. Peptides derived from Atlantic salmon skin gelatin as dipeptidyl-peptidase IV inhibitors. J. Agric. Food Chem. 2012, 60, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Teng, X.; Shang, J.; Wang, D.; Liu, N. Identification of novel DPP–IV inhibitory peptides from Atlantic salmon (Salmo salar) skin. Food Res. Int. 2020, 133, 109161. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, F.; Han, F.; Wang, H. Purification and characterization of antioxidative peptides from salmon protamine hydrolysate. J. Food Biochem. 2008, 32, 654–671. [Google Scholar] [CrossRef]
- Darewicz, M.; Borawska, J.; Vegarud, G.E.; Minkiewicz, P.; Iwaniak, A. Angiotensin I-converting enzyme (ACE) inhibitory activity and ace inhibitory peptides of salmon (Salmo salar) protein hydrolysates obtained by human and porcine gastrointestinal enzymes. Int. J. Mol. Sci. 2014, 15, 14077–14101. [Google Scholar] [CrossRef] [PubMed]
- Picot, L.; Bordenave, S.; Didelot, S. Antiproliferative activity of fish protein hydrolysates on human breast cancer cell lines. Process Biochem. 2006, 41, 1217–1222. [Google Scholar] [CrossRef]
- Ahn, C.B.; Je, J.Y.; Cho, Y.S. Antioxidant and anti-inflammatory peptide fraction from salmon byproduct protein hydrolysates by peptic hydrolysis. Food Res. Int. 2012, 49, 92–98. [Google Scholar] [CrossRef]
- Girgih, A.T.; Udenigwe, C.C.; Hasan, F.M.; Gill, T.A.; Aluko, R.E. Antioxidant properties of Salmon (Salmo salar) protein hydrolysate and peptide fractions isolated by reverse-phase HPLC. Food Res. Int. 2013, 52, 315–322. [Google Scholar] [CrossRef]
- De La Fuente, B.; Pallarés, N.; Berrada, H.; Barba, F.J. Salmon (Salmo salar) side streams as a bioresource to obtain potential antioxidant peptides after applying pressurized liquid extraction (PLE). Mar. Drugs 2021, 19, 323. [Google Scholar] [CrossRef]
- Hanachi, A.; Bianchi, A.; Kahn, C.J.F.; Velot, E.; Arab-Tehrany, E.; Cakir-Kiefer, C.; Linder, M. Encapsulation of salmon peptides in marine liposomes: Physico-chemical properties, antiradical activities and biocompatibility assays. Mar. Drugs 2022, 20, 249. [Google Scholar] [CrossRef]
- Rajendran, S.R.C.K.; Mohan, A.; Khiari, Z.; Udenigwe, C.C.; Mason, B. Yield, physicochemical, and antioxidant properties of Atlantic salmon visceral hydrolysate: Comparison of lactic acid bacterial fermentation with Flavourzyme proteolysis and formic acid treatment. J. Food Process. Preserv. 2018, 42, e13620. [Google Scholar] [CrossRef]
- Wang, K.; Siddanakoppalu, P.N.; Ahmed, I.; Pavase, T.R.; Lin, H.; Li, Z. Purification and identification of anti-allergic peptide from Atlantic salmon (Salmo salar) byproduct enzymatic hydrolysates. J. Funct. Foods 2020, 72, 104084. [Google Scholar] [CrossRef]
- Agyei, D.; Ongkudon, C.M.; Wei, C.Y.; Chan, A.S.; Danquah, M.K. Bioprocess challenges to the isolation and purification of bioactive peptides. Food Bioprod. Process. 2016, 98, 244–256. [Google Scholar] [CrossRef]
- Senadheera, T.R.L.; Hossain, A.; Dave, D.; Shahidi, F. In Silico analysis of bioactive peptides produced from underutilized sea cucumber by-products—A bioinformatics approach. Mar. Drugs 2022, 20, 610. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Cheng, S.; Lu, W.; Du, M. Advancement and prospects of bioinformatics analysis for studying bioactive peptides from food-derived protein: Sequence, structure, and functions. TRAC - Trends Analyt. Chem 2018, 105, 7–17. [Google Scholar] [CrossRef]
- Vilas Boas, L.C.P.; Campos, M.L.; Berlanda, R.L.A.; de Carvalho Neves, N.; Franco, O.L. Antiviral peptides as promising therapeutic drugs. Cell. Mol. Life Sci. 2019, 76, 3525–3542. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-Y.; Hsieh, C.-H.; Hung, C.-C.; Jao, C.-L.; Lin, P.-Y.; Hsieh, Y.-L.; Hsu, K.-C. A study to evaluate the potential of an in silico approach for predicting dipeptidyl peptidase-IV inhibitory activity in vitro of protein hydrolysates. Food Chem. 2017, 234, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, I.M.E.; Li-Chan, E.C.Y. Evaluation of the potential of dietary proteins as precursors of dipeptidyl peptidase (DPP)-IV inhibitors by an in silico approach. J. Funct. Foods 2012, 4, 403–422. [Google Scholar] [CrossRef]
- Tichý, M.; Hanzlíková, I.; Rucki, M.; Pokorná, A.; Uzlová, R.; Tumová, J. Acute toxicity of binary mixtures: Alternative methods, QSAR and mechanisms. Interdiscip. Toxicol. 2008, 1, 15–17. [Google Scholar] [CrossRef]
- Gad, S.C. Encyclopedia of Toxicology, 2014, Third Edition. Elsevier, pp 1–9.
- Nongonierma, A.B.; Fitzgerald, R.J. Learnings from quantitative structure-activity relationship (QSAR) studies with respect to food protein-derived bioactive peptides: A review. RSC Adv. 2016, 6, 75400–75413. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Strategies for the discovery and identification of food protein-derived biologically active peptides. Trends Food Sci. Technol. 2017, 69, 289–305. [Google Scholar] [CrossRef]
- Peter, S.C.; Dhanjal, J.K.; Malik, V.; Radhakrishnan, N.; Jayakanthan, M.; Sundar, D.; Sundar, D. Quantitative structure-activity relationship (QSAR): Modeling approaches to biological applications. In: Encyclopedia of Bioinformatics and Computational Biology: ABC of Bioinformatics. 2018, Elsevier, pp 661–676.
- Lemes, A.C.; Sala, L.; Ores, J.D.C.; Braga, A.R.C.; Egea, M.B.; Fernandes, K.F. A review of the latest advances in encrypted bioactive peptides from protein-richwaste. Int. J. Mol. Sci. 2016, 17, 950. [Google Scholar] [CrossRef] [PubMed]
- Agyei, D.; Danquah, M.K. Industrial-scale manufacturing of pharmaceutical-grade bioactive peptides. Biotechnol Adv. 2011, 29, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Udenigwe, C.C.; Aluko, R.E. Food protein-derived bioactive peptides: Production, processing, and potential health benefits. J. Food Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef] [PubMed]
- Bazinet, L.; Firdaous, L. Separation of bioactive peptides by membrane processes: Technologies and devices. Recent Pat. Biotechnol. 2013, 7, 9–27. [Google Scholar] [CrossRef] [PubMed]
- Hakami, M.W.; Alkhudhiri, A.; Al-Batty, S.; Zacharof, M.P.; Maddy, J.; Hilal, N. Ceramic microfiltration membranes in wastewater treatment: Filtration behavior, fouling and prevention. Membranes 2020, 10, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Muro, C.; Riera, F.; Fernandez, A. (2013) Advancements in the fractionation of milk biopeptides by means of membrane processes. In: Bioactive Food Peptides in Health and Disease. 2013, InTech.
- Suwal, S.; Ketnawa, S.; Liceaga, A.M.; Huang, J.Y. Electro-membrane fractionation of antioxidant peptides from protein hydrolysates of rainbow trout (Oncorhynchus mykiss) byproducts. Innov. Food Sci. Emerg. Technol. 2018, 45, 122–131. [Google Scholar] [CrossRef]
- Doyen, A.; Beaulieu, L.; Saucier, L.; Pouliot, Y.; Bazinet, L. Demonstration of in vitro anticancer properties of peptide fractions from a snow crab by-products hydrolysate after separation by electrodialysis with ultrafiltration membranes. Sep. Purif. Technol. 2011, 78, 321–329. [Google Scholar] [CrossRef]
- Roblet, C.; Akhtar, M.J.; Mikhaylin, S.; Pilon, G.; Gill, T.; Marette, A.; Bazinet, L. Enhancement of glucose uptake in muscular cell by peptide fractions separated by electrodialysis with filtration membrane from salmon frame protein hydrolysate. J. Funct. Foods 2016, 22, 337–346. [Google Scholar] [CrossRef]
- Perez Espitia, P.J.; de Fátima Ferreira Soares, N.; dos Reis Coimbra, J.S.; de Andrade, N.J.; Souza Cruz, R.; Alves Medeiros, E.A. Bioactive peptides: Synthesis, properties, and applications in the packaging and preservation of food. Compr. Rev. Food Sci. Food Saf. 2012, 11, 187–204. [Google Scholar] [CrossRef]
- Ahn, C.B.; Kim, J.G.; Je, J.Y. Purification and antioxidant properties of octapeptide from salmon byproduct protein hydrolysate by gastrointestinal digestion. Food Chem. 2014, 147, 78–83. [Google Scholar] [CrossRef]
- Health Canada. Licensed natural health products database (LNHPD). In: Online query Phoslax. 2021, https://health-products.canada.ca/lnhpd-bdpsnh/info.do?licence=80000689#fn1-rf. Accessed 20 Apr 2021.
- Health Canada Questions and Answers about Preparing Submissions for Food Health Claims. In: Heal. Canada. 2015, https://www.canada.ca/en/health-canada/services/food-nutrition/legislation-guidelines/guidance-documents/questions-answers-about-preparing-submissions-food-health-claims-2015.html. Accessed 20 Apr 2021.
- Bureau of Nutritional Sciences. Guidance Document for Preparing a Submission for Food Health Claims; Health Canada, 2009; pp. 1–45. [Google Scholar]

| Sample | Moisture | Protein | Lipid | Ash | References |
|---|---|---|---|---|---|
| Gut | 50-60.45 | 10.38-13.8 | 22.12-30 | 1.8-1.94 | [33,34,35] |
| Head | 62-63.35 | 11.31-14 | 17-21.86 | 3.51-4.4 | |
| Frame/ trimmings | 55-57.18 | 16.36-19 | 22.65-27 | 3.64-6 | |
| Fillet | 68.80-75 | 18.7-20.90 | 11.30-21.3 | 1.80-2.2 | |
| Whole fish | 64.50-75 | 16.7-17.90 | 17.30-22.12 | 1.88-2.2 |
| Enzymes | Salmon Part | pH | Time (min) | Temperature (°C) | Enzyme Concentration | Protein Yield (%) | Reference |
|---|---|---|---|---|---|---|---|
| Alcalase | Muscle | 7.5 | 180 | 40 | 7.5% | 86.92-88.39 | [9] |
| Flavourzyme | 78.95-84.26 | ||||||
| Corolase PN-L | 72.88-79.32 | ||||||
| Corolase 7089 | 82.41-86.48 | ||||||
| Endogenous | 71.67-79.12 | ||||||
| Alcalase | Head | 8 | 120 | 55 | 5.5% | 71.00 | [59] |
| Alcalase | Skin | 8.39 | 120 | 55 | 2.5% | 77.03 | [60] |
| Neutrase Alcalase Flavourzyme and Protamex | Skin | 6.7-8.0 | 240 | 53 | 10000 U/g | 45.11 | [61] |
| Endogenous | Viscera | N/A | 60 and 120 | 52 | 0.1% | 77.40 | [16] |
| Protamex +Endogenous | Viscera, head and frame | 82.80 | |||||
| Papain + Bromelain + Endogenous | Viscera, head and frame | 0.05+0.05% | 86.0 | ||||
| Protamex + Endogenous | Viscera | 0.1% | 75.50 | ||||
| Papain plus Bromelain + Endogenous | Viscera | 76.60 | |||||
| Papain plus Bromelain | Viscera | 69.90 | |||||
| Corolase PP | Backbone | N/A | 120 | 50 | 0.1% | 11.50 | [62] |
| Corolase 7089 | 0.1% | 9.30 | |||||
| Protamex | 0.1% | 9.40 | |||||
| Bromelain 400 GDU/g + Papain 100TU/mg | 0.05+0.05% | 11.60 | |||||
| Protex 6L | 0.1% | 8.50 | |||||
| Seabzyme L200 | 0.1% | 9.90 | |||||
| Trypsin | 0.1% | 12.10 | |||||
| Alcalase | Head and backbone | 6.5 | 120 | 50 | 5.5 - 88 U/g | 64.70 | [63] |
| Promod 671L | 66.00 | ||||||
| Protex 7L | 62.20 | ||||||
| Protamex | Frame | 6.5 | 60 | 50-56 | 10-90 AU/kg | 43.00-61.00 | [64] |
| Alcalase | Head, trimming, and frame | 8.98 | 180 | 64.2 | 0.2% |
84 |
[65] |
| Alcalase and papain | Frame | 8.0 | 240 | 85 | 1-3% | 79.2-82.01 | [66] |
| Bromelain BR1200 and FoodPro PNL | Backbone and head | N/A | 60 | 50 | 10 U/g |
89.3-92.1 |
[67] |
| Amino acids | Hydrolysates (mg/100 g) | Skin (g/100 g) |
Trimmings (g/100 g) | Viscera+head+frame (mg/g) | Viscera (mg/g) |
||
| Pepsin | Pepsin + pancreatin | Thermolysin | |||||
| Aspartic acid | 0.4 ± 0.2 | 11.6 ± 1.1 | 12.4 ± 0.5 | 5.72 | 7.31 | 77.8 ± 0.8 | 77.0 ± 0.1 |
| Threonine | 2.9 ± 0.2 | 6.0 ± 0.3 | 17.8 ± 0.8 | 2.03 | 3.23 | 39.3 ± 0.1 | 42.4 ± 0.3 |
| Serine | 4.8 ± 0.2 | 6.4 ± 0.3 | 21.8 ± 0.6 | 3.92 | 3.19 | 39.2 ± 0.2 | 41.2 ± 0.9 |
| Glutamic acid | 3.1 ± 0.1 | 29.0 ± 1.2 | 54.2 ± 5.4 | 9.05 | 11.07 | 113.1 ± 1.3 | 109.6 ± 1.9 |
| Glycine | 13.7 ± 0.8 | 15.2 ± 1.0 | 33.0 ± 2.7 | 22.6 | 7.07 | 85.8 ± 1.4 | 66.5 ± 0.0 |
| Alanine | 24.7 ± 1.5 | 35.5 ± 0.9 | 95.0 ± 5.3 | 7.81 | 5.22 | 57.0 ± 0.0 | 53.4 ± 2.0 |
| Valine | 2.5 ± 0.1 | 15.2 ± 0.3 | 18.4 ± 1.7 | 1.71 | 3.83 | 42.6 ± 0.2 | 45.6 ± 0.1 |
| Methionine | 1.3 ± 0.1 | 11.8 ± 0.4 | 31.9 ± 1.6 | 2.46 | 2.66 | 24.3 ± 0.1 | 24.4 ± 0.8 |
| Isoleucine | 1.2 ± 0.1 | 21.9 ± 2.2 | – | 1.05 | 2.98 | 36.0 ± 0.2 | 37.4 ± 0.0 |
| Leucine | 2.4 ± 0.1 | 84.3 ± 1.4 | 69.8 ± 2.3 | 1.75 | 4.97 | 57.9 ± 0.4 | 62.6 ± 0.3 |
| Tyrosine | 1.9 ± 0.1 | 76.9 ± 4.3 | 11.4 ± 0.5 | 0.35 | 1.76 | 18.9 ± 0.5 | 22.2 ± 0.2 |
| Phenylalanine | 3.1 ± 0.3 | 76.3 ± 5.3 | 137.0 ± 4.0 | 1.71 | 2.53 | 31.2 ± 0.2 | 33.9 ± 0.3 |
| Histidine | 16.4 ± 0.9 | 21.0 ± 0.8 | 62.4 ± 2.7 | 1.33 | 3.88 | 26.3 ± 0.2 | 23.2 ± 0.7 |
| Arginine | 1.3 ± 0.6 | 196 ± 14 | – | 7.8 | 5.04 | 56.2 ± 0.3 | 50.8 ± 0.7 |
| Tryptophan | – | 18.3 ± 0.5 | – | 8.2 ± 0.0 | 10.4 ± 0.0 | ||
| Lysine | – | – | – | 2.93 | 6.4 | 71.2 ± 0.6 | 69.4 ± 0.4 |
| Proline | – | – | – | 10.63 | 3.98 | 49.2 ± 0.5 | 47.0 ± 0.3 |
| Cysteine | 0.49 | 0.94 | 9.4 ± 0.0 | 12.4 ± 0.0 | |||
| Reference | [84] | [85] | [16] | ||||
| Peptide/AA | Activities | Reference | ||||
|---|---|---|---|---|---|---|
| ACE IC50 | DPP-IV IC50 | ORAC (µmol TE/µmol peptide) |
AIA (mM) | HRSA (µg/mL) | ||
| GPAV | 415.91±2.65 (µM) | 245.58 ± 7.15 (µM) | 9.51 ± 1.40 | [17] | ||
| VP | 1215.15 ± 83.14 (µM) | 758.15 ± 12.45 (µM) | 19.45 ± 2.15 | |||
| VC | 134.45 ± 5.15 (µM) | 5413.45 ± 62.15 (µM) | 3.45 ± 0.78 | |||
| YP | 5.21 ± 0.94 (µM) | 7564.02 ± 42.45 (µM) | 17.18 ± 1.54 | |||
| FF | 59.15 ± 0.54 (µM) | 546.84 ± 8.15 (µM) | 8.47 ± 1.05 | |||
| PP | 1912.46 ± 63.15 (µM) | 4343.48 ± 29.78 (µM) | 12.48 ± 1.47 | |||
| W | 135.48 ± 5.48 (µM) | 438.15 ± 10.97 (µM) | 4.31 ± 0.19 | |||
| F | 125.15 ± 2.64 (µM) | 295.15 ± 6.45 (µM) | 4.59 ± 0.89 | |||
| Y | 182.84 ± 0.52 (µM) | 75.15 ± 0.84 (µM) | 2.74 ± 0.65 | |||
| GF | 178.14 ± 24.51 (µM) | 1547.15 ± 34.15 (µM) | 19.74 ± 1.01 | [18] | ||
| GPVA | 445.61 ± 6.94 (µM) | 264.74 ± 1.59 (µM) | 9.48 ± 0.94 | |||
| GGPAGPAV | 673.16 ± 15.03 (µM) | 8139.11 ± 134.68 (µM) | 5.47 ± 0.94 | |||
| R | 98.04 ± 0.15 (µM) | 110.44 ± 0.47 (µM) | 4.71 ± 0.09 | |||
| PAY | 0.75 | [111] | ||||
| VWDPPKFD | 9.12 ± 1.07 (µM) | [109] | ||||
| FEDYVPLSCF | 13.72 ± 2.54 (µM) | |||||
| FNVPLYE | 6.79 ± 0.78 (µM) | |||||
| AP | 0.060 ± 0.001 mg/mL | [110] | ||||
| VR | 0.332 ± 0.005 mg/mL | |||||
| GAAGR | ||||||
| AGPS | ||||||
| VDGK | ||||||
| RER | ||||||
| LN | ||||||
| VTGK | ||||||
| GHAGE | ||||||
| VGGK | ||||||
| GHGR | ||||||
| GAPE | 49.6 (µM) | [112] | ||||
| GPGA | 41.9 (µM) | |||||
| YYGYTGAFR | 1.21 mg/mL | [113] | ||||
| LDKVFR | 0.10 mg/mL | |||||
| VLATSGPG | 0.18 mg/mL | |||||
| PA | 91.3 | [114] | ||||
| IVY | 0.48 (µM) | [115] | ||||
| VW | 1.4 (µM) | |||||
| IY | 2.1 (µM) | |||||
| IW | 4.7 (µM) | |||||
| VY | 7.1 (µM) | |||||
| TVY | 15 (µM) | |||||
| VFPS | 0.46 (µM) | |||||
| VTVNPYKWLP | 5.5 (µM) | |||||
| IWHHT | 5.8 (µM) | |||||
| YALPHA | 9.8 (µM) | |||||
| ALPHA | 10 (µM) | |||||
| ACE: Angiotensin-I converting enzyme inhibition; DPP-IV: Dipeptidyl-peptidase IV (DPP-IV)-inhibitory activity; ORAC: Oxygen radical absorbance capacity; TE: Trolox equivalent; AIA: Anti-Inflammatory Activity; HRSA: Hydrogen Radical Scavenging Activity | ||||||
| Databases | Name | Webpage | Purpose | Reference |
|---|---|---|---|---|
| Protein/peptide sequence databases | BIOPEP | http://www.uwm.edu.pl/biochemia/index.php/en/biopep | Protein and peptide database | [119,123,124,125,126,] |
| PepBank | http://pepbank.mgh.harvard.edu/ | Peptide database | ||
| BioPD | http://biopd.bjmu.edu.cn/ | Peptide database | ||
| SwePep | http://www.swepep.org/ | Sweden peptide database | ||
| EROP-Moscow | http://erop.inbi.ras.ru/ | Peptide database | ||
| MilkAMP | http://milkampdb.org/home.php | Dairy antimicrobial peptide database | ||
| UniProtKB | http://www.uniprot.org/ | Primary sequence and structural information for proteins | ||
| NCBI Protein database | http://www.ncbi.nlm.nih.gov | Basic sequence information for proteins | ||
| PeptideDB | http://www.peptides.be/ | Peptide database | ||
| AVPdb | http://crdd.osdd.net/servers/avpdb/index.php | Database of anti-viral peptides | ||
| CAMPR3 | http://www.camp.bicnirrh.res.in/ | Collection of anti-microbial peptides | ||
| HIPdb | http://crdd.osdd.net/servers/hipdb/index.php | Database of HIV inhibiting peptides | ||
| BACTIBASE | http://bactibase.hammamilab.org/main.php | Database of bacteriocins | ||
| APD | https://wangapd3.com/main.php | Antimicrobial peptide database | ||
| http://aps.unmc.edu/AP/ | ||||
| BitterDB | http://bitterdb.agri.huji.ac.il/dbbitter.php | Database for bitter peptides | ||
| In silico digestion Tools | BIOPEP | http://www.uwm.edu.pl/biochemia/index.php/en/biopep | Tools for peptide predication | |
| PeptideCutter | http://web.expasy.org/peptidecutter/ | Predict peptide cleavage sites | ||
| POPS | http://pops.csse.monash.edu.au/pops-cgi/index.php | Tool for modelling and predicting protease specificity | ||
| http://www.mybiosoftware.com/pops-2-1-1-prediction-protease-specificity.html | ||||
| NeuroPred | http://stagbeetle.animal.uiuc.edu/cgi-bin/neuropred.py | Prediction of cleavage sites and mass from neuropeptide precursors | ||
| CAMPSign | http://www.campsign.bicnirrh.res.in/ | Identification of AMPs using AMP family signatures | ||
| Enzyme Predictor | http://bioware.ucd.ie/~enzpred/Enzpred.php | Evaluation tool to determine the cleavage ability of enzymes | ||
| Potential bioactivity prediction | PeptideRanker | http://bioware.ucd.ie/ ∼compass/biowareweb/ | Pediction of bioactive peptides | |
| BIOPEP | http://www.uwm.edu.pl/biochemia/index.php/en/biopep | Pediction of bioactive peptides | ||
| AntiBP2 | http://www.imtech.res.in/raghava/antibp2/ | Antibacterial peptide prediction tool | ||
| AVPpred | http://crdd.osdd.net/servers/avppred/submit.php | Collection and prediction of antiviral peptides | ||
| AVP-IC50Pred | http://crdd.osdd.net/servers/ic50avp/index.html | Prediction of peptide antiviral activity in terms of IC50 | ||
| Toxicity/allergenicity prediction | ||||
| ToxinPred | http://crdd.osdd.net/raghava/toxinpred/ | Predicting the toxicity of peptides | ||
| BIOPEP | http://www.uwm.edu.pl/biochemia/index.php/en/biopep | Allergenic protein database | ||
| AlgPred | http://crdd.osdd.net/raghava/algpred/ | Predicting allergenic proteins and peptides | ||
| Allerdictor | https://openebench.bsc.es/tool/allerdictor | Allergen prediction tool | ||
| EPIMHC | http://bio.med.ucm.es/epimhc/ | Database of Naturally Processed MHC-restricted Peptide Ligands and Epitopes for Customized Computational Vaccinology | ||
| Physicochemical characteristics prediction | Expasy-Compute pI/Mw | http://web.expasy.org/compute_pi/ | A tool to compute the theoretical pI (isoelectric point) and MW (mol weight) | |
| ProtParam | http://web.expasy.org/protparam/ | A tool to compute the grand average of hydropathicity (GRAVY) and Instability index | ||
| Peptide Structure Prediction Server | PepDraw | http://www.tulane.edu/~biochem/WW/PepDraw/ | A tool to compute net charge and hydrophobicity | |
| Pep-Fold | https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD/ | Predicting peptide structures from amino acid sequences. | ||
| PEPstrMOD | http://osddlinux.osdd.net/raghava/pepstrmod/ | Predicts the tertiary structure of small peptides with sequence length varying between 7 to 25 residues | ||
| I-TASSER | https://zhanglab.ccmb.med.umich.edu/I-TASSER/ | Protein structure prediction and structure-based function annotation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





