Submitted:
08 April 2023
Posted:
10 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
“The whole case here rests upon the demonstration that codon-amino acid pairing interactions exist and that the codon assignments in some way reflect these interactions… all-or-none specificities are not required for such interactions to determine the form of the codon catalog, either a general form or one specified down to the very last detail. All that is required here is that a sufficient number of slight preferences be shown.”.Carl Woese, 1969
2. Methods
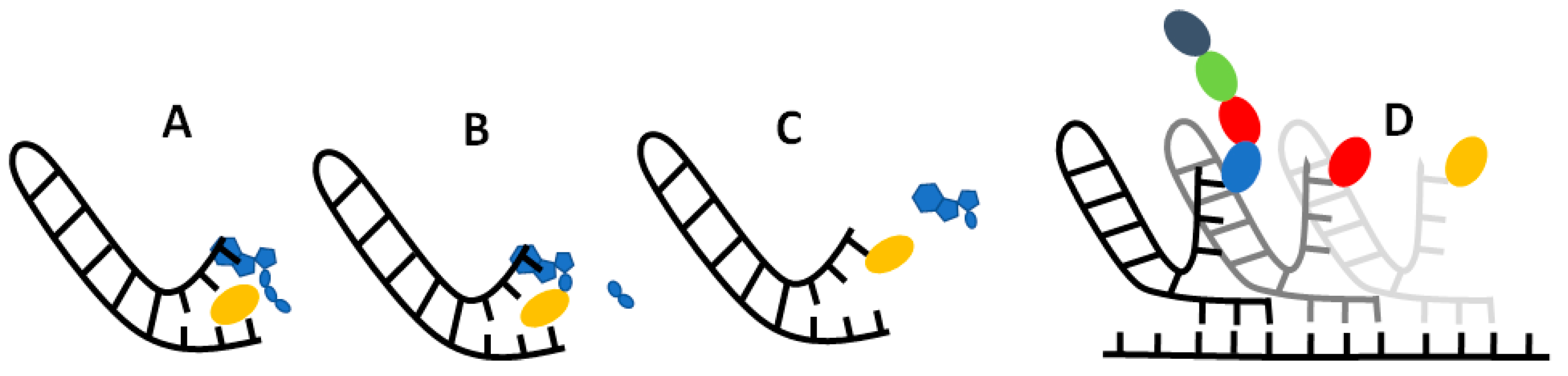
2.1. Molecular dynamics pipeline
2.2. Hydrophobicity trends
2.3. Rings
2.4. NMR
3. Results
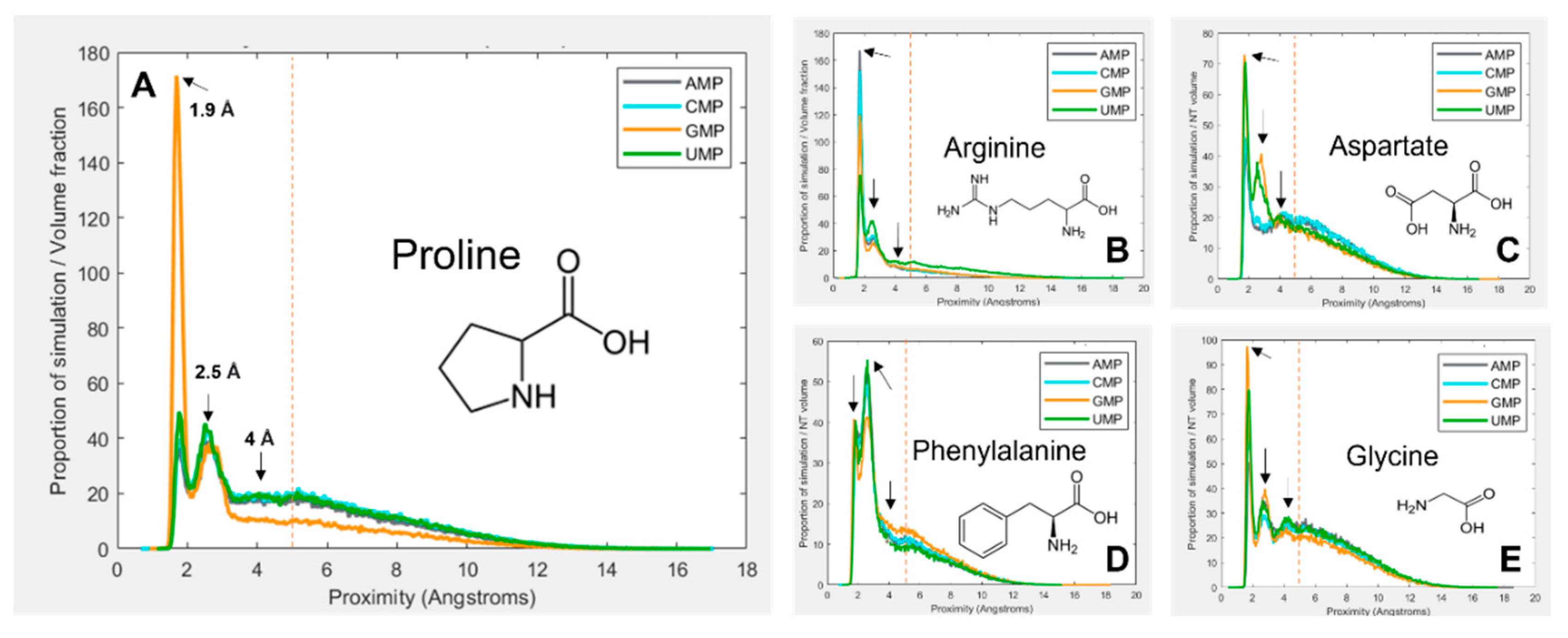
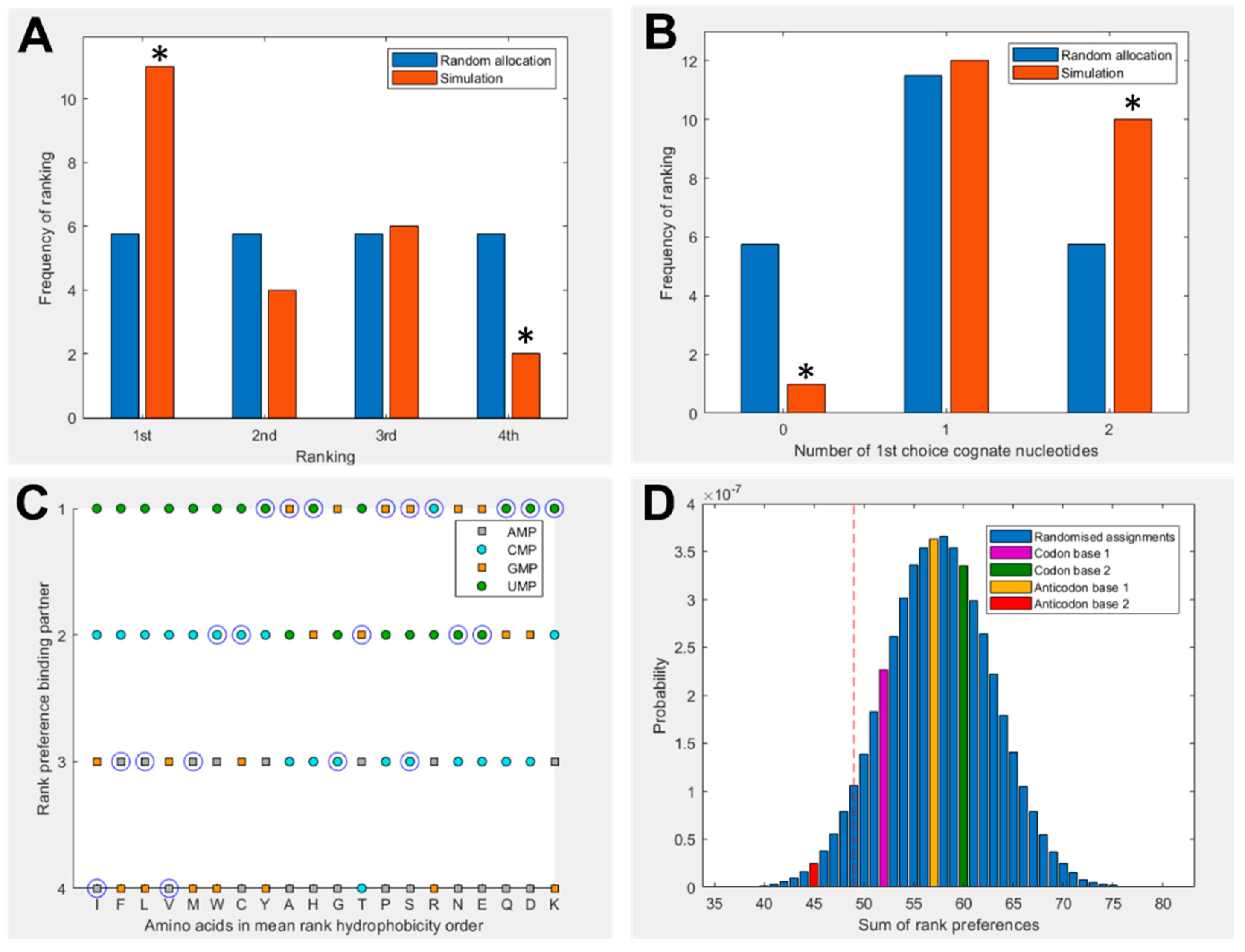
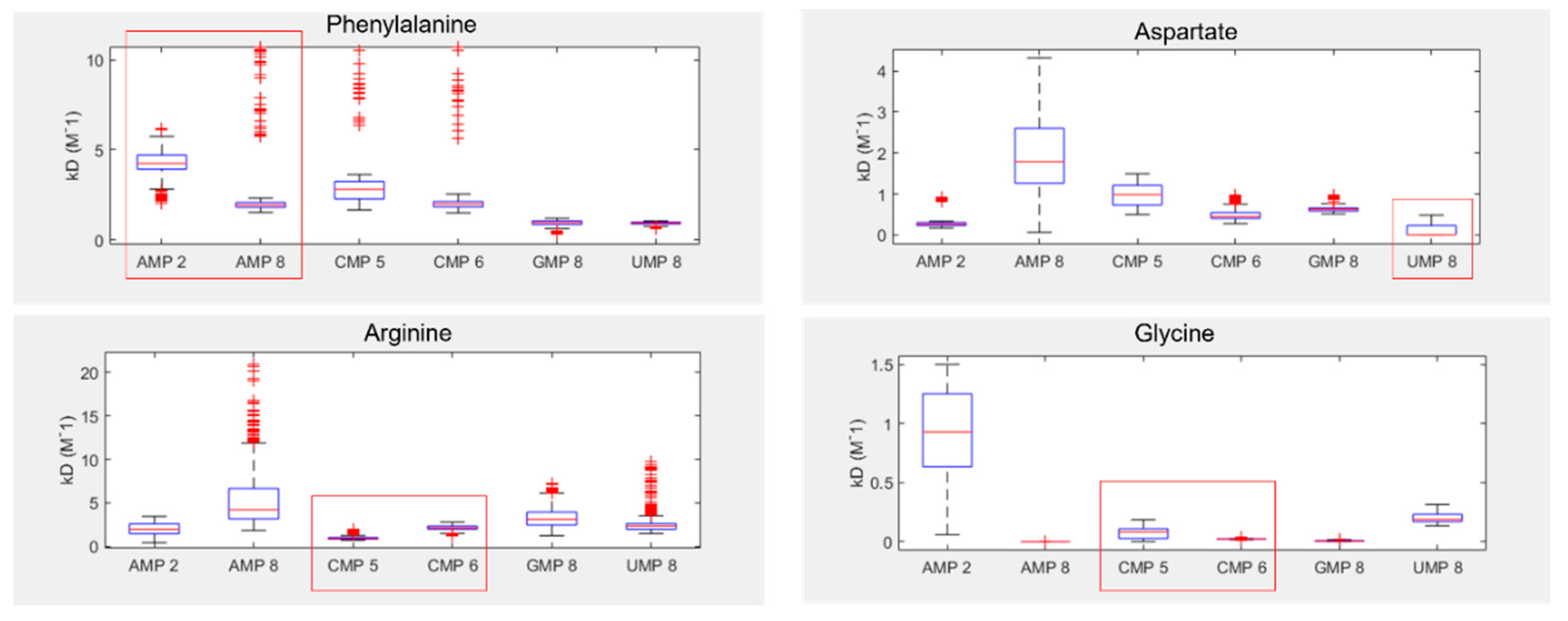
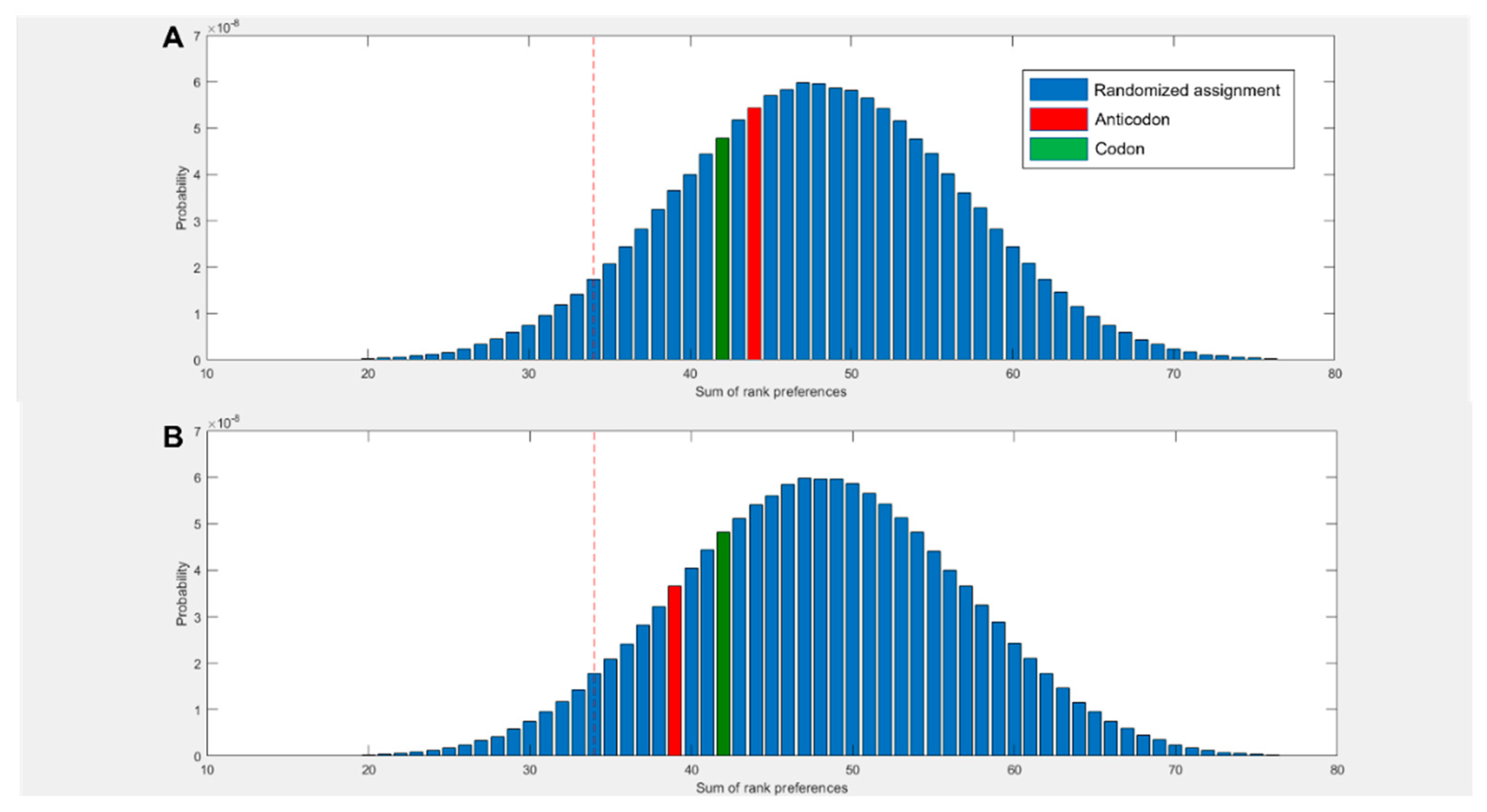
3.1. Amino acids prefer cognate nucleotides
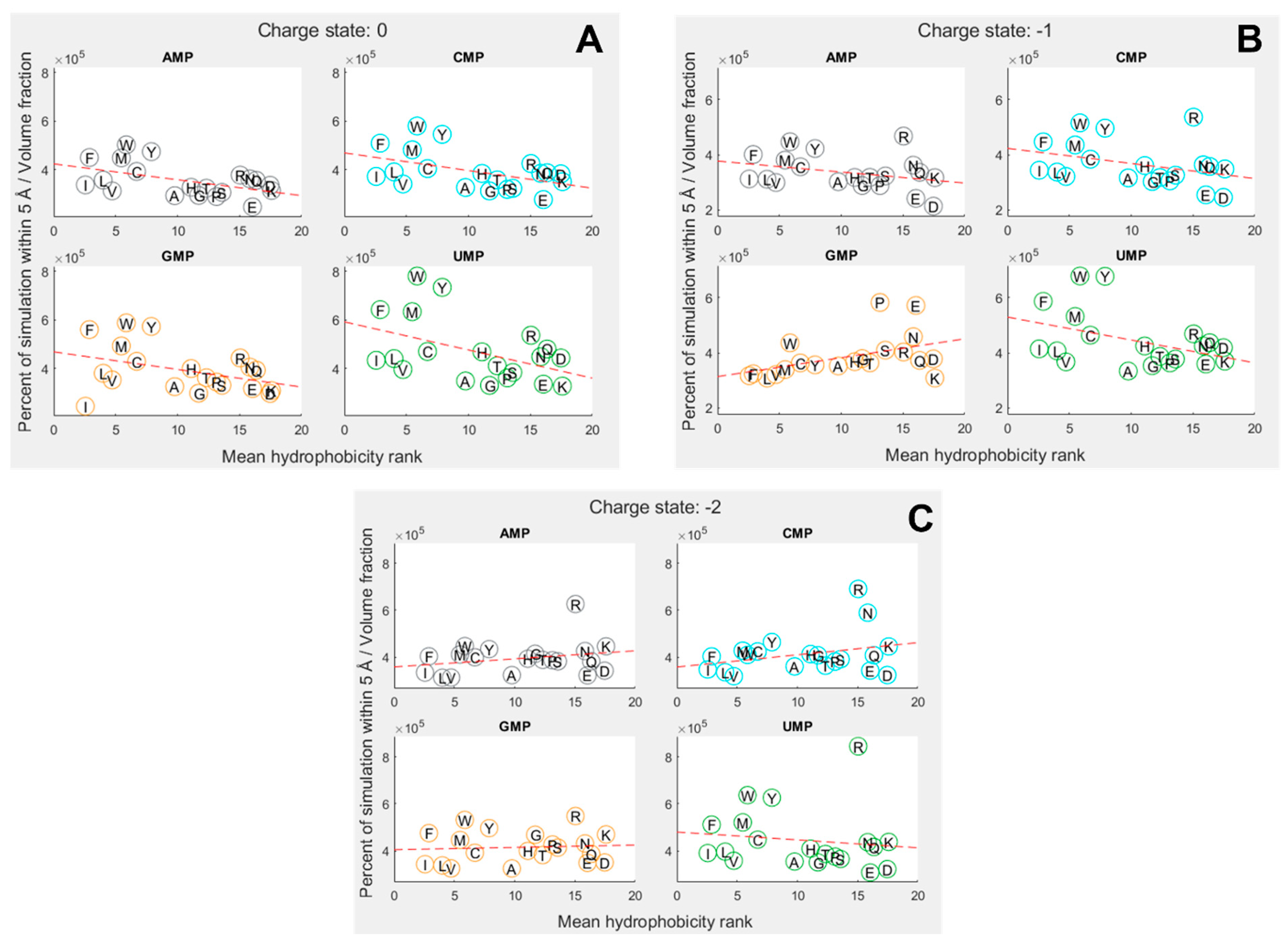
3.2. Hydrophobicity plays a role in binding
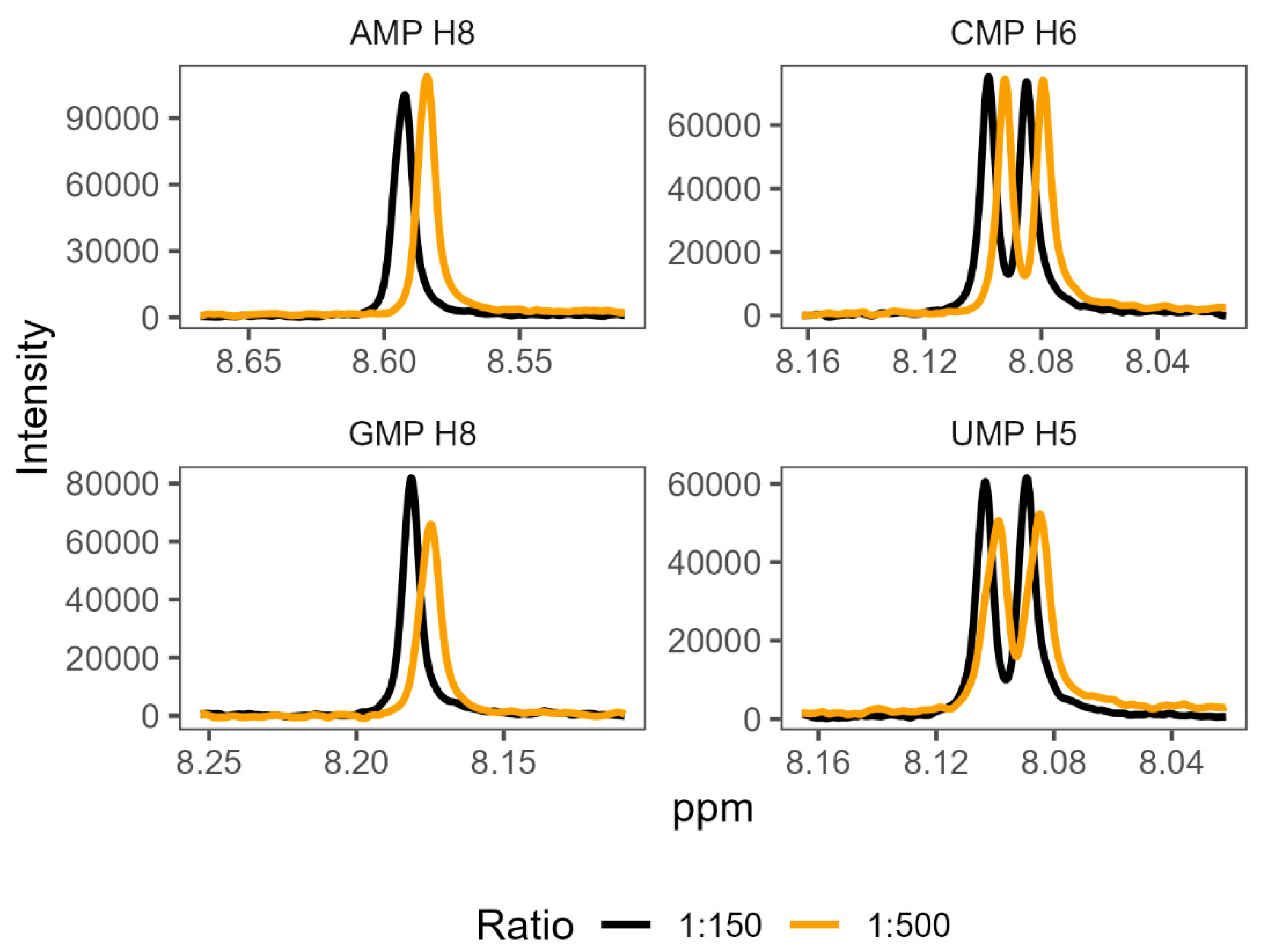
3.3. NMR corroborates binding interactions
3.4. Dinucleotides also show affinity for their cognate amino acids:
4. Discussion
Supplementary Materials
Acknowledgements
References
- Woese, C.R. Order in the Genetic Code. Proc Natl Acad Sci U S A 1965, 54, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Eck, R.V. Genetic Code: Emergence of a Symmetrical Pattern. Science 1963, 140, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Nirenberg, M.W.; Jones, O.W.; Leder, P.; Clark, B.F.C.; Sly, W.S.; Pestka, S. On the Coding of Genetic Information. Cold Spring Harb Symp Quant Biol 1963, 28, 549–557. [Google Scholar] [CrossRef]
- Taylor, F.J.R.; Coates, D. The Code within the Codons. Biosystems 1989, 22, 177–187. [Google Scholar] [CrossRef]
- Copley, S.D.; Smith, E.; Morowitz, H.J. A Mechanism for the Association of Amino Acids with Their Codons and the Origin of the Genetic Code. Proceedings of the National Academy of Sciences of the United States of America 2005, 102, 4442–4447. [Google Scholar] [CrossRef]
- Wong, J.T. A Co-Evolution Theory of the Genetic Code. Proceedings of the National Academy of Sciences of the United States of America 1975, 72, 1909–1912. [Google Scholar] [CrossRef]
- Di Giulio, M. An Autotrophic Origin for the Coded Amino Acids Is Concordant with the Coevolution Theory of the Genetic Code. J Mol Evol 2016, 83, 93–96. [Google Scholar] [CrossRef]
- Trinquier, G.; Sanejouand, Y.H. Which Effective Property of Amino Acids Is Best Preserved by the Genetic Code? Protein Engineering Design and Selection 1998, 11, 153–169. [Google Scholar] [CrossRef]
- Woese, C.R.; Dugre, D.H.; Saxinger, W.C.; Dugre, S.A. The Molecular Basis for the Genetic Code. Proc Natl Acad Sci U S A 1966, 55, 966–974. [Google Scholar] [CrossRef]
- Fontecilla-Camps, J.C. The Stereochemical Basis of the Genetic Code and the (Mostly) Autotrophic Origin of Life. Life (Basel) 2014, 4, 1013–1025. [Google Scholar] [CrossRef]
- Woese, C. Models for the Evolution of Codon Assignments. J Mol Biol 1969, 43, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H. On Protein Synthesis. Symp Soc Exp Biol 1958, 12, 138–163. [Google Scholar] [PubMed]
- Crick, F.H. The Origin of the Genetic Code. Journal of molecular biology 1968, 38, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H.C. As quoted by Hoagland, M., in The Nucleic Acids, ed. E. Chargaff and J. Davidson (New York: Academic Press, 1960), vol. 3; Crick, F.H.C., J. Griffith, and L. Orgel, these PROCEEDINGS, 43, 416.
- Weber, A.L.; Lacey, J.C. Genetic Code Correlations: Amino Acids and Their Anticodon Nucleotides. Journal of Molecular Evolution 1978, 11, 199–210. [Google Scholar] [CrossRef]
- Lacey, J.C.; Pruitt, K.M. Origin of the Genetic Code. Nature 1969, 223, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Lacey, J.C.; Mullins, D.W. Experimental Studies Related to the Origin of the Genetic Code and the Process of Protein Synthesis--a Review. Orig Life 1983, 13, 3–42. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M. Specific Aminoacylation of C4N Hairpin RNAs with the Cognate Aminoacyl-Adenylates in the Presence of a Dipeptide: Origin of the Genetic Code. Journal of Biochemistry 1995, 117, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M. Molecular Basis for the Genetic Code. J Mol Evol 1982, 18, 297–303. [Google Scholar] [CrossRef]
- Yarus, M.; Widmann, J.J.; Knight, R. RNA–Amino Acid Binding: A Stereochemical Era for the Genetic Code. Journal of Molecular Evolution 2009, 69, 406–429. [Google Scholar] [CrossRef]
- Yarus, M.; Caporaso, J.G.; Knight, R. ORIGINS OF THE GENETIC CODE: The Escaped Triplet Theory. Annual Review of Biochemistry 2005, 74, 179–198. [Google Scholar] [CrossRef]
- Hobish, M.K.; Wickramasinghe, N.S.M.D.; Ponnamperuma, C. Direct Interaction between Amino Acids and Nucleotides as a Possible Physicochemical Basis for the Origin of the Genetic Code. Advances in Space Research 1995, 15, 365–382. [Google Scholar] [CrossRef]
- Moghadam, S.A.; Preto, J.; Klobukowski, M.; Tuszynski, J.A. Testing Amino Acid-Codon Affinity Hypothesis Using Molecular Docking. BioSystems 2020, 198. [Google Scholar] [CrossRef]
- Koonin, E.V.; Novozhilov, A.S. Origin and Evolution of the Genetic Code: The Universal Enigma. IUBMB Life 2009, 61, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Di Giulio, M. Arguments against the Stereochemical Theory of the Origin of the Genetic Code. Biosystems 2022, 221, 104750. [Google Scholar] [CrossRef] [PubMed]
- Haig, D.; Hurst, L.D. A Quantitative Measure of Error Minimization in the Genetic Code. J Mol Evol 1991, 33, 412–417. [Google Scholar] [CrossRef]
- Sonneborn, T.M. (1965). In Evolving Genes and Proteins, ed. by V. Bryson & H. Vogel,p. 377. New York: Academic Press.
- Barbieri, M. Evolution of the Genetic Code: The Ambiguity-Reduction Theory. Biosystems 2019, 185, 104024. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R. On the Evolution of the Genetic Code. Proc Natl Acad Sci U S A 1965, 54, 1546–1552. [Google Scholar] [CrossRef]
- Delarue, M. An Asymmetric Underlying Rule in the Assignment of Codons: Possible Clue to a Quick Early Evolution of the Genetic Code via Successive Binary Choices. RNA 2007, 13, 161–169. [Google Scholar] [CrossRef]
- Woese, C.R. The Fundamental Nature of the Genetic Code: Prebiotic Interactions between Polynucleotides and Polyamino Acids or Their Derivatives. Proceedings of the National Academy of Sciences of the United States of America 1968, 59, 110–117. [Google Scholar] [CrossRef]
- Orgel, L.E. Evolution of the Genetic Apparatus. J Mol Biol 1968, 38, 381–393. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-Splicing RNA: Autoexcision and Autocyclization of the Ribosomal RNA Intervening Sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA Moiety of Ribonuclease P Is the Catalytic Subunit of the Enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R. The RNA Worlds in Context. Cold Spring Harbor Perspectives in Biology 2012, 4, a006742–a006742. [Google Scholar] [CrossRef]
- Gilbert, W. Origin of Life: The RNA World. Nature 1986, 319, 618–618. [Google Scholar] [CrossRef]
- Leslie, E.O. Prebiotic Chemistry and the Origin of the RNA World. Critical Reviews in Biochemistry and Molecular Biology 2004, 39, 99–123. [Google Scholar] [CrossRef] [PubMed]
- Joyce, G.F. and Orgel, L.E. Prospects for understanding the origin of the RNA world. Cold Spring Harbor Monograph Series 1993, 24, 1. [Google Scholar]
- Bose, T.; Fridkin, G.; Davidovich, C.; Krupkin, M.; Dinger, N.; Falkovich, A.H.; Peleg, Y.; Agmon, I.; Bashan, A.; Yonath, A. Origin of Life: Protoribosome Forms Peptide Bonds and Links RNA and Protein Dominated Worlds. Nucleic Acids Research 2022, 50, 1815–1828. [Google Scholar] [CrossRef] [PubMed]
- Joyce, G.F. Evolution in an RNA World. Cold Spring Harb Symp Quant Biol 2009, 74, 17–23. [Google Scholar] [CrossRef]
- Orgel, L.E. Some Consequences of the RNA World Hypothesis. Orig Life Evol Biosph 2003, 33, 211–218. [Google Scholar] [CrossRef]
- Spiegelman, S.; Haruna, I.; Holland, I.B.; Beaudreau, G.; Mills, D. The Synthesis of a Self-Propagating and Infectious Nucleic Acid with a Purified Enzyme. Proceedings of the National Academy of Sciences of the United States of America 1965, 54, 919–927. [Google Scholar] [CrossRef]
- Szathmáry, E.; Demeter, L. Group Selection of Early Replicators and the Origin of Life. J Theor Biol 1987, 128, 463–486. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, S.; Kun, Á.; Ryckelynck, M.; Coldren, F.; Szilágyi, A.; Jossinet, F.; Rick, C.; Nghe, P.; Szathmáry, E.; Griffiths, A.D. Transient Compartmentalization of RNA Replicators Prevents Extinction Due to Parasites. Science 2016, 354, 1293–1296. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M. Hypercycles and the Origin of Life. Nature 1979, 280, 445–446. [Google Scholar] [CrossRef] [PubMed]
- Eigen, M. Selforganization of Matter and the Evolution of Biological Macromolecules. Naturwissenschaften 1971, 58, 465–523. [Google Scholar] [CrossRef] [PubMed]
- Salditt, A.; Keil, L.M.R.; Horning, D.P.; Mast, C.B.; Joyce, G.F.; Braun, D. Thermal Habitat for RNA Amplification and Accumulation. Phys. Rev. Lett. 2020, 125, 048104. [Google Scholar] [CrossRef] [PubMed]
- Joyce, G.F. The RNA world: Life before DNA and protein. Extraterrestrials: where are they 1995, 2, 139–151. [Google Scholar]
- Ralser, M. The RNA World and the Origin of Metabolic Enzymes. Biochem Soc Trans 2014, 42, 985–988. [Google Scholar] [CrossRef]
- Ralser, M. An Appeal to Magic? The Discovery of a Non-Enzymatic Metabolism and Its Role in the Origins of Life. Biochem J 2018, 475, 2577–2592. [Google Scholar] [CrossRef]
- Harrison, S.A.; Lane, N. Life as a Guide to Prebiotic Nucleotide Synthesis. Nature Communications 2018, 9, 1–4. [Google Scholar] [CrossRef]
- Lazcano, A.; Miller, S.L. On the Origin of Metabolic Pathways. J Mol Evol 1999, 49, 424–431. [Google Scholar] [CrossRef]
- Horowitz, N.H. On the Evolution of Biochemical Syntheses. Proc Natl Acad Sci U S A 1945, 31, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, N.H. The evolution of biochemical synthesis—Retrospect and prospect. In Evolving Genes and Proteins; Bryson, V., Vogel, H.J., Eds.; Academic Press: New York, NY, USA, 1965; pp. 15–23. [Google Scholar]
- Walker, S.I.; Davies, P.C.W. The Algorithmic Origins of Life. Journal of The Royal Society Interface 2013, 10, 20120869. [Google Scholar] [CrossRef] [PubMed]
- Amend, J.P.; McCollom, T.M. Energetics of Biomolecule Synthesis on Early Earth. In Chemical Evolution II: From the Origins of Life to Modern Society; ACS Symposium Series; American Chemical Society, 2009; Vol. 1025, pp 63–94. [CrossRef]
- Amend, J.P.; LaRowe, D.E.; McCollom, T.M.; Shock, E.L. The Energetics of Organic Synthesis inside and Outside the Cell. Philosophical Transactions of the Royal Society B: Biological Sciences 2013, 368, 20120255. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, J.L.E.; Xavier, J.C.; Vieira, A. d. N.; Pereira, D.P.H.; Leidner, J.; Sousa, F.L.; Kleinermanns, K.; Preiner, M.; Martin, W.F. Energy at Origins: Favorable Thermodynamics of Biosynthetic Reactions in the Last Universal Common Ancestor (LUCA). Frontiers in Microbiology 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Nunes Palmeira, R.; Colnaghi, M.; Harrison, S.A.; Pomiankowski, A.; Lane, N. The Limits of Metabolic Heredity in Protocells. Proceedings of the Royal Society B: Biological Sciences 2022, 289, 20221469. [Google Scholar] [CrossRef]
- Orgel, L.E. Self-Organizing Biochemical Cycles. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 12503–12507. [Google Scholar] [CrossRef] [PubMed]
- Preiner, M.; Igarashi, K.; Muchowska, K.B.; Yu, M.; Varma, S.J.; Kleinermanns, K.; Nobu, M.K.; Kamagata, Y.; Tüysüz, H.; Moran, J.; et al. A Hydrogen-Dependent Geochemical Analogue of Primordial Carbon and Energy Metabolism. Nat Ecol Evol 2020, 4, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Beyazay, T.; Belthle, K.S.; Farès, C.; Preiner, M.; Moran, J.; Martin, W.F.; Tüysüz, H. Ambient Temperature CO2 Fixation to Pyruvate and Subsequently to Citramalate over Iron and Nickel Nanoparticles. Nat Commun 2023, 14, 570. [Google Scholar] [CrossRef]
- Hudson, R.; de Graaf, R.; Strandoo Rodin, M.; Ohno, A.; Lane, N.; McGlynn, S.E.; Yamada, Y.M.A.; Nakamura, R.; Barge, L.M.; Braun, D.; et al. CO2 Reduction Driven by a PH Gradient. Proceedings of the National Academy of Sciences 2020, 117, 22873–22879. [Google Scholar] [CrossRef]
- Varma, S.J.; Muchowska, K.B.; Chatelain, P.; Moran, J. Native Iron Reduces CO2 to Intermediates and End-Products of the Acetyl-CoA Pathway. Nat Ecol Evol 2018, 2, 1019–1024. [Google Scholar] [CrossRef]
- Muchowska, K.B.; Varma, S.J.; Chevallot-Beroux, E.; Lethuillier-Karl, L.; Li, G.; Moran, J. Metals Promote Sequences of the Reverse Krebs Cycle. Nat Ecol Evol 2017, 1, 1716–1721. [Google Scholar] [CrossRef] [PubMed]
- Muchowska, K.B.; Varma, S.J.; Moran, J. Synthesis and Breakdown of Universal Metabolic Precursors Promoted by Iron. Nature 2019, 569, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Barge, L.M.; Flores, E.; Baum, M.M.; VanderVelde, D.G.; Russell, M.J. Redox and PH Gradients Drive Amino Acid Synthesis in Iron Oxyhydroxide Mineral Systems. Proceedings of the National Academy of Sciences 2019, 116. [Google Scholar] [CrossRef] [PubMed]
- Huber, C.; Wächtershäuser, G. Primordial Reductive Amination Revisited. Tetrahedron Letters 2003, 44, 1695–1697. [Google Scholar] [CrossRef]
- Mayer, R.J.; Moran, J. Quantifying Reductive Amination in Nonenzymatic Amino Acid Synthesis. Angewandte Chemie International Edition 2022, 61, e202212237. [Google Scholar] [CrossRef] [PubMed]
- Whicher, A.; Camprubi, E.; Pinna, S.; Herschy, B.; Lane, N. Acetyl Phosphate as a Primordial Energy Currency at the Origin of Life. Origins of Life and Evolution of Biospheres 2018, 48, 159–179. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.A.; Turchyn, A.V.; Ralser, M. Non-Enzymatic Glycolysis and Pentose Phosphate Pathway-like Reactions in a Plausible Archean Ocean. Mol Syst Biol 2014, 10, 725. [Google Scholar] [CrossRef]
- Messner, C.B.; Driscoll, P.C.; Piedrafita, G.; De Volder, M.F.L.; Ralser, M. Nonenzymatic Gluconeogenesis-like Formation of Fructose 1,6-Bisphosphate in Ice. Proceedings of the National Academy of Sciences 2017, 114, 7403–7407. [Google Scholar] [CrossRef]
- Piedrafita, G.; Varma, S.J.; Castro, C.; Messner, C.B.; Szyrwiel, L.; Griffin, J.L.; Ralser, M. Cysteine and Iron Accelerate the Formation of Ribose-5-Phosphate, Providing Insights into the Evolutionary Origins of the Metabolic Network Structure. PLOS Biology 2021, 19, e3001468. [Google Scholar] [CrossRef]
- Camprubi, E.; Harrison, S.A.; Jordan, S.F.; Bonnel, J.; Pinna, S.; Lane, N. Do Soluble Phosphates Direct the Formose Reaction towards Pentose Sugars? Astrobiology 2022, 22, 981–991. [Google Scholar] [CrossRef]
- Yi, J.; Kaur, H.; Kazöne, W.; Rauscher, S.A.; Gravillier, L.-A.; Muchowska, K.B.; Moran, J. A Nonenzymatic Analog of Pyrimidine Nucleobase Biosynthesis. Angewandte Chemie International Edition 2022, 61, e202117211. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.A.; Palmeira, R.N.; Halpern, A.; Lane, N. A Biophysical Basis for the Emergence of the Genetic Code in Protocells. Biochimica et biophysica acta. Bioenergetics 2022, 1863, 148597. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. Journal of Cheminformatics 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. Journal of Computational Chemistry 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, J.; Jo, S.; Brooks, C.L.; Lee, H.S.; Im, W. CHARMM-GUI Ligand Reader and Modeler for CHARMM Force Field Generation of Small Molecules. Journal of Computational Chemistry 2017, 38, 1879–1886. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. Journal of Chemical Theory and Computation 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable Molecular Dynamics on CPU and GPU Architectures with NAMD. The Journal of Chemical Physics 2020, 153, 044130. [Google Scholar] [CrossRef]
- Matsunaga, Y. MDToolbox 1.0. https://mdtoolbox.readthedocs.io/en/latest/introduction.html (accessed 2022-02-21).
- Tien, M.Z.; Meyer, A.G.; Sydykova, D.K.; Spielman, S.J.; Wilke, C.O. Maximum Allowed Solvent Accessibilites of Residues in Proteins. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Williamson, M.P. Using Chemical Shift Perturbation to Characterise Ligand Binding. Prog Nucl Magn Reson Spectrosc 2013, 73, 1–16. [Google Scholar] [CrossRef]
- Lacey, J.C.; Mullins, D.W.; Khaled, M.A. The Case for the Anticode. Origins of Life 1984, 14, 505–511. [Google Scholar] [CrossRef]
- Krigbaum, W.R.; Komoriya, A. Local Interactions as a Structure Determinant for Protein Molecules: II. Biochim Biophys Acta 1979, 576, 204–248. [Google Scholar] [CrossRef] [PubMed]
- Sweet, R.M.; Eisenberg, D. Correlation of Sequence Hydrophobicities Measures Similarity in Three-Dimensional Protein Structure. J Mol Biol 1983, 171, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database (Oxford) 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Ostell, J.; Pruitt, K.D.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res 2019, 47, D94–D99. [Google Scholar] [CrossRef] [PubMed]
- The Merck Index Online. The Merck Index Online - chemicals, drugs and biologicals. https://www.rsc.org/merck-index (accessed 2023-02-23).
- Riniker, S. Fixed-Charge Atomistic Force Fields for Molecular Dynamics Simulations in the Condensed Phase: An Overview. J. Chem. Inf. Model. 2018, 58, 565–578. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yan, J.; Zhang, H.; Xu, Z.; Zhang, J.Z.H. An Electrostatic Energy-Based Charge Model for Molecular Dynamics Simulation. J. Chem. Phys. 2021, 154, 134107. [Google Scholar] [CrossRef] [PubMed]
- Condon, D.E.; Kennedy, S.D.; Mort, B.C.; Kierzek, R.; Yildirim, I.; Turner, D.H. Stacking in RNA: NMR of Four Tetramers Benchmark Molecular Dynamics. Journal of Chemical Theory and Computation 2015, 11, 2729–2742. [Google Scholar] [CrossRef]
- Johnson, D.B.F.; Wang, L. Imprints of the Genetic Code in the Ribosome. Proceedings of the National Academy of Sciences of the United States of America 2010, 107, 8298–8303. [Google Scholar] [CrossRef]
- Pinna, S.; Kunz, C.; Halpern, A.; Harrison, S.A.; Jordan, S.F.; Ward, J.; Werner, F.; Lane, N. A Prebiotic Basis for ATP as the Universal Energy Currency. PLOS Biology 2022, 20, e3001437. [Google Scholar] [CrossRef]
- Sosunov, V.; Zorov, S.; Sosunova, E.; Nikolaev, A.; Zakeyeva, I.; Bass, I.; Goldfarb, A.; Nikiforov, V.; Severinov, K.; Mustaev, A. The Involvement of the Aspartate Triad of the Active Center in All Catalytic Activities of Multisubunit RNA Polymerase. Nucleic Acids Res 2005, 33, 4202–4211. [Google Scholar] [CrossRef]
- Unciuleac, M.-C.; Goldgur, Y.; Shuman, S. Two-Metal versus One-Metal Mechanisms of Lysine Adenylylation by ATP-Dependent and NAD+-Dependent Polynucleotide Ligases. Proc Natl Acad Sci U S A 2017, 114, 2592–2597. [Google Scholar] [CrossRef] [PubMed]
- Hopfield, J.J. Origin of the Genetic Code: A Testable Hypothesis Based on TRNA Structure, Sequence, and Kinetic Proofreading. Proc. Natl. Acad. Sci. U.S.A. 1978, 75, 4334–4338. [Google Scholar] [CrossRef] [PubMed]
- Betat, H.; Mörl, M. The CCA-Adding Enzyme: A Central Scrutinizer in TRNA Quality Control. Bioessays 2015, 37, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Francklyn, C.; Carter, C.W., Jr. Aminoacylating Urzymes Challenge the RNA World Hypothesis. The Journal of biological chemistry 2013, 288, 26856–26863. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.W.; Wills, P.R. Hierarchical Groove Discrimination by Class I and II Aminoacyl-TRNA Synthetases Reveals a Palimpsest of the Operational RNA Code in the TRNA Acceptor-Stem Bases. Nucleic Acids Research 2018, 46, 9667–9683. [Google Scholar] [CrossRef]
- Davidovich, C.; Belousoff, M.; Bashan, A.; Yonath, A. The Evolving Ribosome: From Non-Coded Peptide Bond Formation to Sophisticated Translation Machinery. Research in Microbiology 2009, 160, 487–492. [Google Scholar] [CrossRef]
- Farias, S.T.; Rêgo, T.G.; José, M.V. Origin and Evolution of the Peptidyl Transferase Center from Proto-TRNAs. FEBS open bio 2014, 4, 175–178. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




