1. Introduction
Antisense oligonucleotides (ASOs) are small single-stranded nucleotides that target specific mRNAs by binding to their sense strand through Watson-Crick base pairing, which can be employed to modulate gene expression through various mechanisms [
1]. The therapeutic potential of ASOs was recognized in the 1970s [
2]. However, unmodified ASOs have limited plasma persistence [
3]. ASOs have gone through three generations, with improved stability and binding affinity due to modified sugar moieties, bases, and phosphodiester linkages [
4]. For example, 2′-O-methyl nucleotides (2OMe) and phosphorodiamidate morpholino oligomers (PMOs) are 2nd and 3rd generation ASOs, respectively [
4] .
ASOs modify target mRNA expression through two main mechanisms: RNase H-dependent cleavage and steric block [
5]. RNase H-dependent ASOs, designed as gapmers, bind to the target RNA and trigger cleavage by the endogenous RNase H enzyme, leading to target gene silencing [
6,
7,
8]. Steric blocking ASOs, on the other hand, are often employed to specifically exclude (exon skipping) or retain (exon inclusion) a specific exon(s), leading to alternations in splicing decisions [
2,
9].
Exon skipping, where an ASO causes the exclusion of a specific exon in splicing, has emerged as a promising treatment for genetic diseases, especially muscular dystrophies. US Food and Drug Administration has approved multiple exon-skipping ASO treatments for Duchenne muscular dystrophy (DMD), including eteplirsen, golodirsen, viltolarsen, and casimersen [
10,
11,
12,
13]. Exon skipping has shown promising potential as a treatment option for many genetic diseases beyond DMD. Splicing defects are a common cause of many genetic diseases, and exon skipping can be used to restore proper splicing by skipping over faulty exons. Milasen, a patient-customized n-of-1 ASO drug targeted for a pseudoexon in the CLN7 gene, was recently approved by the FDA for the treatment of Batten’s Disease, demonstrating the potential of exon skipping for personalized medicine [
14,
15]. Exon skipping therapies are also being explored for other genetic diseases such as cystic fibrosis, retinitis pigmentosa, sarcoglycanopathy, dysferlinopathy, fibrodysplasia ossificans progressiva, epidermolysis bullosa, frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17), and cancer, among others [
15,
16,
17,
18,
19,
20,
21,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31,
32,
33,
34].
Despite these promising developments, there are still significant challenges in developing effective exon-skipping therapies. A major hurdle is a difficulty in selecting an optimal sequence for exon skipping, as the efficacy of ASOs is often unpredictable due to numerous factors involved in the exon-skipping process [
35]. Designing effective ASO sequences requires consideration of various criteria [
36], particularly for exon skipping [
37]. Software tools such as eSkip-Finder can aid in this process [
38]. eSkip-Finder (
https://eskip-finder.org) is a web-based tool developed by Chiba et al. that provides a solution for identifying optimal ASO sequences for exon skipping by using machine learning models built from a curated database of publications and patents [
38].
The selection of important features is a crucial step in the tool’s approach, and the eSkip-Finder uses an exhaustive search of subsets of features to identify these critical components. However, due to the high computational cost, the subset size was limited to seven features. To optimize the performance of the models, hyperparameters in the support vector regressor are optimized through a grid search. This optimization process is computationally intensive, requiring a significant amount of computing power, and can take several days to complete.
This paper seeks an alternative solution to reduce the computational cost associated with the eSkip-Finder. Some machine learning algorithms such as decision-tree or random forest have built-in feature ranking capabilities [
39]. Ensemble methods are also proven to have good performance with reasonable computation cost [
40,
41]. We explored their utility in ASO efficacy prediction and demonstrated that a combination of three algorithms, namely random forest, gradient boosting, and XGBoost, through a 3-way voting mechanism can significantly reduce computation time while maintaining or slightly improving the prediction performance. This approach offers a promising solution for reducing computational cost in the ASO efficacy prediction process.
2. Materials and Methods
The datasets used in this study were the same as those used in Chiba et al. [
38]. That is, for PMO, 369 and 57 measurements were used for training and testing and there were 98 and 11 unique ASO sequences in each split without overlapping; for 2OMe, 197 and 31 measurements were used for training and testing and there were 111 and 13 unique ASO sequences in each split without overlapping. As PMO and 2OMe have different chemistry thus different binding affinity, the datasets were handled separately.
For each measurement, there were 32 numerical features calculated via bioinformatics tools as discussed in Chiba et al (such as dose). The categorical feature, Malueka’s category, was excluded from modeling. As reported in [
38]. this feature is not important in determining the ASO efficacy. The feature was specifically linked to dystrophin exons [
42]. Models developed with this feature included will be difficult to generalize to other genes.
The efficacy was measured as a percent in the range 0 to 100. The efficacy is the value to predict, making this a regression problem. All 32 features were inputted into the machine learning models and feature selection was left to the models.
The machine learning libraries included scikit-learn (0.42.2) [
43] and XGBoost (1.6.1) [
44]. The following regressors were used: support vector, random forest, gradient boosting, and XGBoost. The last three were also used to vote by the simple average of the individual predictions. The support vector regressor was included for comparison purpose, as it was used in Chiba et al. All those regressors were built without hyperparameter tuning, i.e., default parameters were used in each regressor (except random seeds). The computation code was developed using Python (3.9.7) on Mac (Quadcore i5, 2 GHz CPU, 16 GB RAM).
Two metrics were used to assess model performances: R2 and mean absolute error (MAE) between true efficacy values and predictions. The models were first assessed on the training data via 10-fold cross-validation. The best model was then selected and applied to the reserved test data. The R2 and MAE on each fold were collected and their mean and standard deviation were further computed to aid the best model selection.
While the random forest, gradient boosting, and XGBoost models were trained, they also collected data to compute the feature importance score. The voting regressor had no feature importance score, however. We therefore used the model-agnostic method, permutation feature importance provided by scikit-learn, to rank the feature importance.
3. Results
The performance metrics for various models using 10-fold cross-validation on the training data are shown in
Table 1. 5-fold and 20-fold cross-validations were also attempted and the results were similar to what was reported here. The data splitting was based on ASOs, i.e., there were no overlapping ASOs in training and validation splits. As can be seen from
Table 1, for both PMO and 2OMe ASOs, the 3-way voting approach gives the largest
R2 and smallest mean absolute error (MAE). We thus chose this approach and applied it to the test datasets. The support vector regressor performed noticeably poorly as there was no hyperparameter optimization in the current study. It shall also be noted that the whole computing took about 10 seconds on a laptop computer.
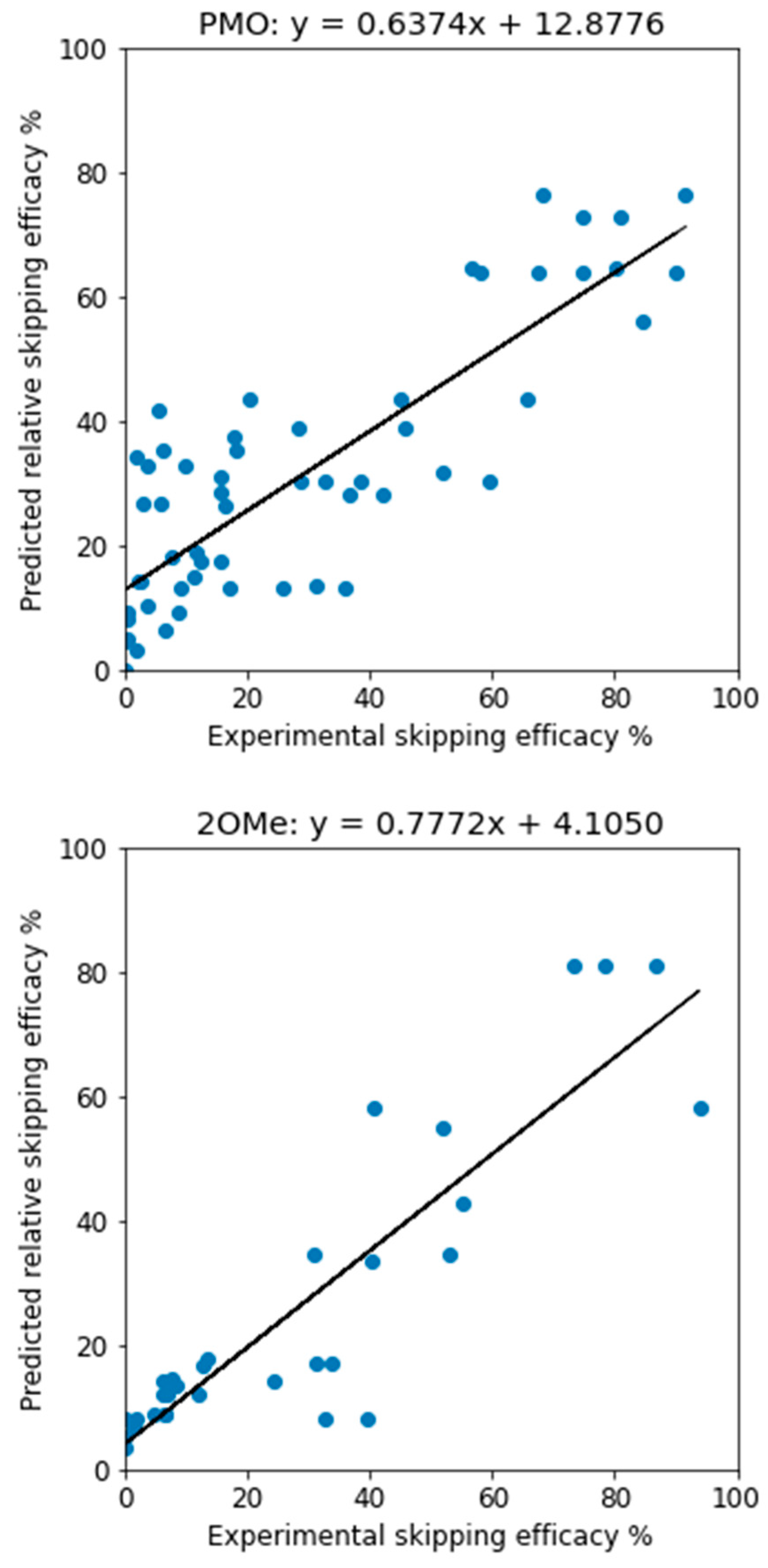
When the 3-way voting models, trained on the training data with all features, were applied to the test data, the predictions were similarly assessed. For PMO, we have
R2 = 0.706 and MAE = 12.25, and for 2OMe,
R2 = 0.795 and MAE = 9.237. The
R2 values are higher than those reported [
38], which were 0.6 and 0.7 respectively. The true efficacy and predicted one have a good linear correlation, as depicted in
Figure 1. It shall be noted that, unlike the support vector regressor which can generate unrealistic, negative efficacy values, the 3-way voting approach will not possibly predict a negative efficacy as long as the input data has no negative efficacy.
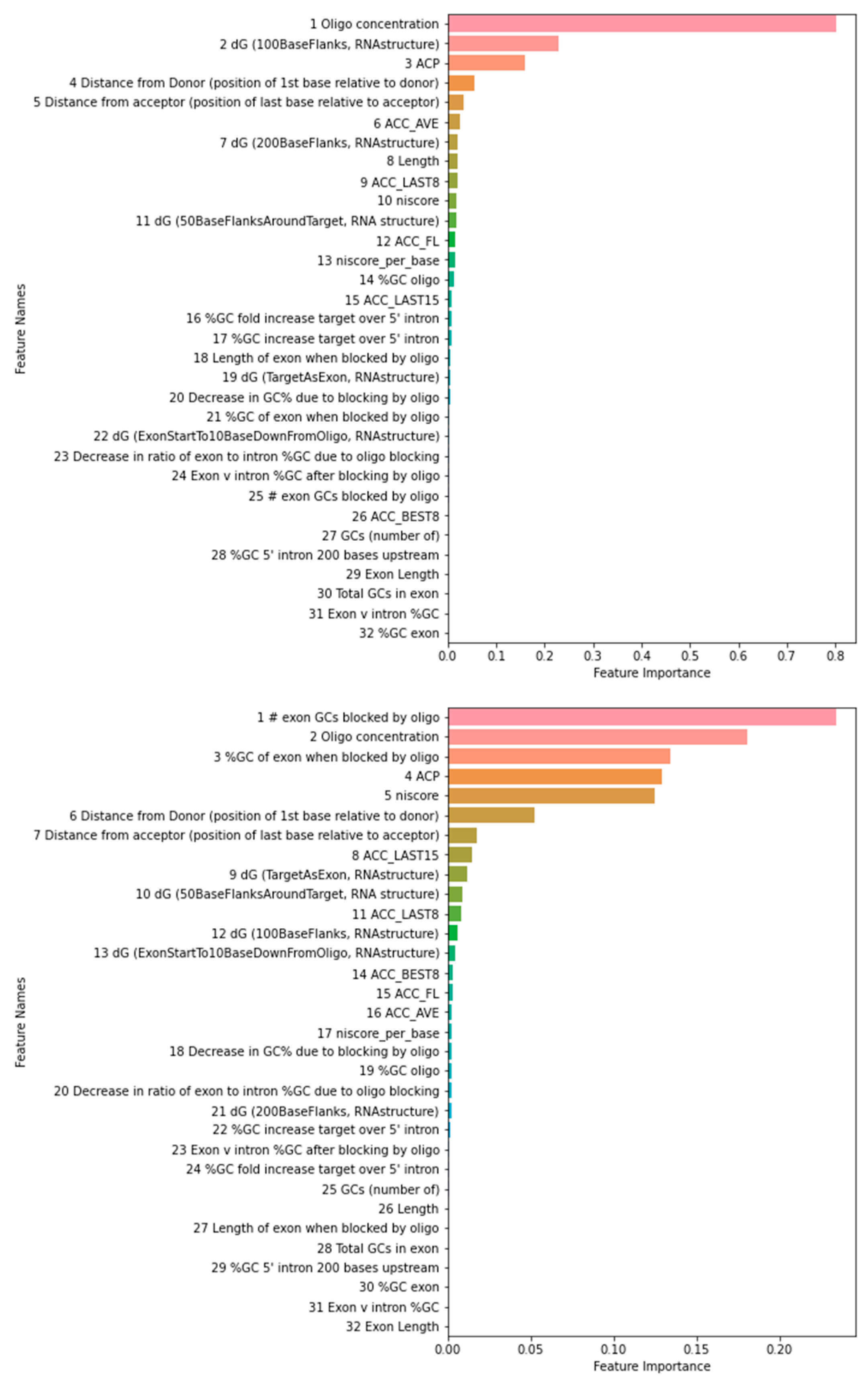
The feature importance ranking using the training data as reported by the 3-way voting is shown in
Figure 2. The rankings using the test data are similar on top-ranked features, suggesting that overfitting is not a concern. Among top 5 and 10 features using training or test dataset, 3 and 8 are common for PMO and 4 and 9 are common for 2OMe. The 4 PMO features used in Chiba et al. here were ranked at 1, 24, 11, and 15. The 6 2OMe features used in Chiba et al. here were ranked at 2, 25, 4, 3, 17, and 11. In both cases, some correlation can be observed. We also noted that some features were strongly correlated, e.g., niscore and niscore_per_base. Niscore_per_base was ranked 17th, but niscore was ranked 5th in our 2OMe model. Therefore, at least some discrepancies can be attributed to the feature correlations. Due to the randomness in the algorithms, the rank order can be slightly different in each run.
To check if the voting approach works for different genes and exons, we applied the trained PMO model to the exon 73 skipping of collagen type VII alpha 1 chain [
9]. The results are summarized in
Table 2. The predictions by the voting approach preserve the ranking order of ASO efficacy experimentally measured. Cautions must be taken when one extends the model to a different application domain, however. As more data is accumulated in databases such as eSkip-Finder, we expect predictive models will be validated rigorously and extended as needed.
4. Discussion
We applied machine learning algorithms with built-in feature selection capabilities to train on and predict exon-skipping PMO and 2OMe ASO efficacy. The model build process requires much less time. Among various algorithms assessed, the voting strategy yielded the best-performing predictors in terms of
R2 and mean absolute error (MAE) between the true and the predicted efficacy.
R2 were 0.706 (PMO) and 0.795 (2OMe), which were slightly higher than were reported [
38]. The MAE was also reported as a reference. This observation on the voting approach was consistent with the general consensus in the machine learning community. Due to the model itself, no negative efficacies are predicted in our approach, whilst the support vector regressor does not have this guarantee. Important features used in our approach were similar to what eSkip-Finder discovered. Features used by Chiba and colleagues were overall ranked high in our voting approach and some differences can be explained by feature correlations. Thus, our modeling approach has similar interpretability.
As mentioned above, the voting approach predicts non-negative efficacies as long as there are no samples with negative efficacies in the training data. However, this can be a drawback, i.e., the approach will not predict any efficacies larger than the highest efficacy in the training data, since decision trees are used essentially in the individual algorithms. This potential limitation can be easily remedied by collecting training samples with large efficacies.
The proposed voting approach has a very short training time. We believe that the same approach might be applicable to developing predictive models for other diseases where ASO efficacy data is available. The voting scheme still relies on engineered features scientists hand-picked. As a possible future extension, one could consider machine learning algorithms in combination with natural language processing techniques, which has been successfully applied to biological sequence analysis [
45].
Author Contributions
Conceptualization, A.Z; software, A.Z.; data analysis, A.Z., S.C. Y.S.; writing and editing, A.Z., S.C, T.Y.; review, all; guidance, T.Y.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used in this study can be accessed from [
46]. No new data were created.
Conflicts of Interest
The authors declare no conflict of interest. T.Y. is a founder and shareholder of OligomicsTx, which aims to commercialize antisense oligonucleotide technology.
References
- Crooke, S.T.; Liang, X.-H.; Baker, B.F.; Crooke, R.M. Antisense technology: A review. Journal of Biological Chemistry 2021, 296.
- Stephenson, M.L.; Zamecnik, P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proceedings of the National Academy of Sciences 1978, 75, 285-288. [CrossRef]
- Chan, J.H.; Lim, S.; Wong, W.F. Antisense oligonucleotides: from design to therapeutic application. Clinical and experimental pharmacology and physiology 2006, 33, 533-540. [CrossRef]
- Quemener, A.M.; Bachelot, L.; Forestier, A.; Donnou-Fournet, E.; Gilot, D.; Galibert, M.D. The powerful world of antisense oligonucleotides: From bench to bedside. Wiley Interdisciplinary Reviews: RNA 2020, 11, e1594. [CrossRef]
- Rinaldi, C.; Wood, M.J. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nature Reviews Neurology 2018, 14, 9-21. [CrossRef]
- Inoue, H.; Hayase, Y.; Iwai, S.; Ohtsuka, E. Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett 1987, 215, 327-330. [CrossRef]
- Lundin, K.E.; Gissberg, O.; Smith, C.E. Oligonucleotide therapies: the past and the present. Human gene therapy 2015, 26, 475-485. [CrossRef]
- Walder, J.A.; Walder, R.Y. Nucleic acid hybridization and amplification method for detection of specific sequences in which a complementary labeled nucleic acid probe is cleaved. Google Patents: 1995.
- Lim, S.R.; Hertel, K.J. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. Journal of Biological Chemistry 2001, 276, 45476-45483.
- Shirley, M. Casimersen: First Approval. Drugs 2021, 81, 875-879. [CrossRef]
- Nelson, S.F.; Miceli, M.C. FDA Approval of Eteplirsen for Muscular Dystrophy. JAMA 2017, 317, 1480. [CrossRef]
- Roshmi, R.R.; Yokota, T. Viltolarsen: From Preclinical Studies to FDA Approval. Methods Mol Biol 2023, 2587, 31-41. [CrossRef]
- Aartsma-Rus, A.; Corey, D.R. The 10th Oligonucleotide Therapy Approved: Golodirsen for Duchenne Muscular Dystrophy. Nucleic Acid Ther 2020, 30, 67-70. [CrossRef]
- Brudvig, J.J.; Weimer, J.M. On the cusp of cures: breakthroughs in Batten disease research. Current Opinion in Neurobiology 2022, 72, 48-54. [CrossRef]
- Kim, J.; Hu, C.; Moufawad El Achkar, C.; Black, L.E.; Douville, J.; Larson, A.; Pendergast, M.K.; Goldkind, S.F.; Lee, E.A.; Kuniholm, A., et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N Engl J Med 2019, 381, 1644-1652. [CrossRef]
- Aartsma-Rus, A.; van Roon-Mom, W.; Lauffer, M.; Siezen, C.; Duijndam, B.; Coenen-de Roo, T.; Schule, R.; Synofzik, M.; Graessner, H. Development of tailored splice switching oligonucleotides for progressive brain disorders in Europe: development, regulation and implementation considerations. RNA 2023, 10.1261/rna.079540.122. [CrossRef]
- Aartsma-Rus, A.; Garanto, A.; van Roon-Mom, W.; McConnell, E.M.; Suslovitch, V.; Yan, W.X.; Watts, J.K.; Yu, T.W. Consensus Guidelines for the Design and In Vitro Preclinical Efficacy Testing N-of-1 Exon Skipping Antisense Oligonucleotides. Nucleic Acid Ther 2023, 33, 17-25. [CrossRef]
- Lemaitre, M.M. Individualized Antisense Oligonucleotide Therapies: How to Approach the Challenge of Manufacturing These Oligos from a Chemistry, Manufacturing, and Control-Regulatory Standpoint. Nucleic Acid Ther 2022, 32, 101-110. [CrossRef]
- Bateman-House, A.; Kearns, L. Individualized Therapeutics Development for Rare Diseases: The Current Ethical Landscape and Policy Responses. Nucleic Acid Ther 2022, 32, 111-117. [CrossRef]
- Hill, S.F.; Meisler, M.H. Antisense Oligonucleotide Therapy for Neurodevelopmental Disorders. Dev Neurosci 2021, 43, 247-252. [CrossRef]
- Amariles, P.; Madrigal-Cadavid, J. Ethical, Economic, Societal, Clinical, and Pharmacology Uncertainties Associated With Milasen and Other Personalized Drugs. Ann Pharmacother 2020, 54, 937-938. [CrossRef]
- Aartsma-Rus, A.; Watts, J.K. The Munich Meeting: Medical Maturation, More Mechanisms, and Milasen. Nucleic Acid Ther 2019, 29, 302-304. [CrossRef]
- Kim, Y.J.; Sivetz, N.; Layne, J.; Voss, D.M.; Yang, L.; Zhang, Q.; Krainer, A.R. Exon-skipping antisense oligonucleotides for cystic fibrosis therapy. Proceedings of the National Academy of Sciences 2022, 119, e2114858118. [CrossRef]
- Covello, G.; Ibrahim, G.H.; Bacchi, N.; Casarosa, S.; Denti, M.A. Exon skipping through chimeric antisense U1 snRNAs to correct retinitis pigmentosa GTPase-regulator (RPGR) splice defect. nucleic acid therapeutics 2022, 32, 333-349. [CrossRef]
- Shi, S.; Cai, J.; de Gorter, D.J.; Sanchez-Duffhues, G.; Kemaladewi, D.U.; Hoogaars, W.M.; Aartsma-Rus, A.; ’t Hoen, P.A.; ten Dijke, P. Antisense-oligonucleotide mediated exon skipping in activin-receptor-like kinase 2: inhibiting the receptor that is overactive in fibrodysplasia ossificans progressiva. PloS one 2013, 8, e69096.
- Rodrigues, M.; Yokota, T. An overview of recent advances and clinical applications of exon skipping and splice modulation for muscular dystrophy and various genetic diseases. Exon Skipping and Inclusion Therapies: Methods and Protocols 2018, 31-55. [CrossRef]
- Isom, L.L.; Knupp, K.G. Dravet syndrome: novel approaches for the most common genetic epilepsy. Neurotherapeutics 2021, 18, 1524-1534. [CrossRef]
- Barthelemy, F.; Blouin, C.; Wein, N.; Mouly, V.; Courrier, S.; Dionnet, E.; Kergourlay, V.; Mathieu, Y.; Garcia, L.; Butler-Browne, G., et al. Exon 32 Skipping of Dysferlin Rescues Membrane Repair in Patients’ Cells. J Neuromuscul Dis 2015, 2, 281-290. [CrossRef]
- Lee, J.J.A.; Maruyama, R.; Duddy, W.; Sakurai, H.; Yokota, T. Identification of Novel Antisense-Mediated Exon Skipping Targets in DYSF for Therapeutic Treatment of Dysferlinopathy. Mol Ther Nucleic Acids 2018, 13, 596-604. [CrossRef]
- Wyatt, E.J.; Demonbreun, A.R.; Kim, E.Y.; Puckelwartz, M.J.; Vo, A.H.; Dellefave-Castillo, L.M.; Gao, Q.Q.; Vainzof, M.; Pavanello, R.C.M.; Zatz, M., et al. Efficient exon skipping of SGCG mutations mediated by phosphorodiamidate morpholino oligomers. JCI Insight 2018, 3. [CrossRef]
- McGrath, J.A.; Ashton, G.H.; Mellerio, J.E.; Salas-Alanis, J.C.; Swensson, O.; McMillan, J.R.; Eady, R.A. Moderation of phenotypic severity in dystrophic and junctional forms of epidermolysis bullosa through in-frame skipping of exons containing non-sense or frameshift mutations. J Invest Dermatol 1999, 113, 314-321. [CrossRef]
- Kalbfuss, B.; Mabon, S.A.; Misteli, T. Correction of alternative splicing of tau in frontotemporal dementia and parkinsonism linked to chromosome 17. J Biol Chem 2001, 276, 42986-42993. [CrossRef]
- Renshaw, J.; Orr, R.M.; Walton, M.I.; Te Poele, R.; Williams, R.D.; Wancewicz, E.V.; Monia, B.P.; Workman, P.; Pritchard-Jones, K. Disruption of WT1 gene expression and exon 5 splicing following cytotoxic drug treatment: antisense down-regulation of exon 5 alters target gene expression and inhibits cell survival. Mol Cancer Ther 2004, 3, 1467-1484. [CrossRef]
- Yu, A.M.; Tu, M.J. Deliver the promise: RNAs as a new class of molecular entities for therapy and vaccination. Pharmacol Ther 2022, 230, 107967. [CrossRef]
- Maruyama, R.; Yokota, T. Tips to Design Effective Splice-Switching Antisense Oligonucleotides for Exon Skipping and Exon Inclusion. Methods Mol Biol 2018, 1828, 79-90. [CrossRef]
- Sciabola, S.; Xi, H.; Cruz, D.; Cao, Q.; Lawrence, C.; Zhang, T.; Rotstein, S.; Hughes, J.D.; Caffrey, D.R.; Stanton, R.V. PFRED: A computational platform for siRNA and antisense oligonucleotides design. PloS one 2021, 16, e0238753. [CrossRef]
- Shimo, T.; Maruyama, R.; Yokota, T. Designing effective antisense oligonucleotides for exon skipping. Duchenne Muscular Dystrophy: Methods and Protocols 2018, 143-155.
- Chiba, S.; Lim, K.R.Q.; Sheri, N.; Anwar, S.; Erkut, E.; Shah, M.N.A.; Aslesh, T.; Woo, S.; Sheikh, O.; Maruyama, R., et al. eSkip-Finder: a machine learning-based web application and database to identify the optimal sequences of antisense oligonucleotides for exon skipping. Nucleic Acids Res 2021, 49, W193-W198. [CrossRef]
- Bishop, C.M.; Nasrabadi, N.M. Pattern recognition and machine learning; Springer: 2006; Vol. 4.
- Chandra, A.; Yao, X. Ensemble learning using multi-objective evolutionary algorithms. Journal of Mathematical Modelling and Algorithms 2006, 5, 417-445. [CrossRef]
- Sahin, E.K. Assessing the predictive capability of ensemble tree methods for landslide susceptibility mapping using XGBoost, gradient boosting machine, and random forest. SN Applied Sciences 2020, 2, 1308. [CrossRef]
- Malueka, R.G.; Takaoka, Y.; Yagi, M.; Awano, H.; Lee, T.; Dwianingsih, E.K.; Nishida, A.; Takeshima, Y.; Matsuo, M. Categorization of 77 dystrophin exons into 5 groups by a decision tree using indexes of splicing regulatory factors as decision markers. BMC Genet 2012, 13, 23. [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: machine learning in Python. J Mach Learn Res.
- Ramraj, S.; Uzir, N.; Sunil, R.; Banerjee, S. Experimenting XGBoost algorithm for prediction and classification of different datasets. International Journal of Control Theory and Applications 2016, 9, 651-662.
- Iuchi, H.; Matsutani, T.; Yamada, K.; Iwano, N.; Sumi, S.; Hosoda, S.; Zhao, S.; Fukunaga, T.; Hamada, M. Representation learning applications in biological sequence analysis. Computational and Structural Biotechnology Journal 2021, 19, 3198-3208. [CrossRef]
- Echigoya, Y.; Mouly, V.; Garcia, L.; Yokota, T.; Duddy, W. In silico screening based on predictive algorithms as a design tool for exon skipping oligonucleotides in Duchenne muscular dystrophy. PLoS One 2015, 10, e0120058. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).