Submitted:
19 March 2023
Posted:
20 March 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection
| Marker | Description | Target Organisms | Forward Primer | Reverse Primer | Reference |
|---|---|---|---|---|---|
| FITS | Fungal rRNA Internal Transcribed Spacer | Fungi | GTCGGTAAAACTCGTGCCAGC | CATAGTGGGGTATCTAATCCCAGTTTG | Miya et al. 2015 |
| 16S | Prokaryotic rRNA small subunit | Bacteria, archaea | GTGYCAGCMGCCGCGGTAA |
GGACTACNVGGGTWTCTAAT | F: 515F and R: 806R, see Caporaso et al., 2012 |
| 18S | Eukaryotic rRNA small subunit | Fungi, algae, protists | GTACACACCGCCCGTC | TGATCCTTCTGCAGGTTCACCTAC | Amaral-Zettler et al. 2009; Euk_1391f and EukBr |
| CO1 | Mitochondrial cytochrome oxidase subunit I | Animals | ATGCGATACTTGGTGTGAAT | GACGCTTCTCCAGACTACAAT | Gu et al. 2013 |
| 12S | Mitochondrial rRNA small subunit | Fish, birds, snakes, insects | GGWACWGGWTGAACWGTWTAYCCYCC | TANACYTCnGGRTGNCCRAARAAYCA | Leray et al. 2013 |
| PITS | Plant rRNA Internal Transcribed Spacer | Plants | GGAAGTAAAAGTCGTAACAAGG | CAAGAGATCCGTTGTTGAAAGTT | F: ITS5, White et al., 1990; R: 5.8S, Epp et al. 2012 |
2.2. DNA Isolation and Amplification
| Marker | Covariate | Factor Levels Tested |
| 16S | LA River Site | Glendale Narrows, Verdugo Wash |
| 16S | River Condition | Soft-Bottom, Concrete |
| 16S | Habitat | Frequently Submerged, Fully Submerged |
| FITS | Habitat | Frequently Submerged, Fully Submerged |
| FITS | LA River Site | Maywood, Arroyo Seco |
2.3. Statistical Approach
2.4. Chi Square Test of Proportions for the 18S Marker
| Kit_Name | LA River Site | Latitude | Longitude | Habitat | River Condition |
|---|---|---|---|---|---|
| K0585_T9 | Arroyo Seco | 34.203154 | -118.166402 | Frequently submerged, intertidal, marsh | soft |
| K0593_C3 | Arroyo Seco | 34.203274 | -118.166417 | Terrestrial, not submerged | soft |
| K0594_E4 | Arroyo Seco | 34.202987 | -118.166335 | Terrestrial, not submerged | soft |
| K0595_B2 | Arroyo Seco | 34.203593 | -118.166448 | Terrestrial, not submerged | soft |
| K0595_L7 | Arroyo Seco | 34.203567 | -118.166415 | Terrestrial, not submerged | soft |
| K0595_T9 | Arroyo Seco | 34.204139 | -118.166314 | Terrestrial, not submerged | soft |
| K0597_M8 | Arroyo Seco | 34.20375 | -118.166481 | Terrestrial, not submerged | soft |
| K0599_L7 | Arroyo Seco | 34.20331 | -118.166408 | Frequently submerged, intertidal, marsh | soft |
| K0526_B2 | Bowtie Parcel | 34.108161 | -118.246186 | Fully submerged | soft |
| K0529_L7 | Bowtie Parcel | 34.108149 | -118.246176 | Fully submerged | soft |
| K0672_C3 | Bowtie Parcel | 34.108433 | -118.246959 | Fully submerged | soft |
| K0672_G5 | Bowtie Parcel | 34.108278 | -118.246926 | Fully submerged | soft |
| K0674_E4 | Bowtie Parcel | 34.108186 | -118.246584 | Fully submerged | soft |
| K0678_E4 | Bowtie Parcel | 34.108131 | -118.246003 | Fully submerged | soft |
| K0679_B2 | Bowtie Parcel | 34.108278 | -118.246341 | Fully submerged | soft |
| K0679_M8 | Bowtie Parcel | 34.108374 | -118.246774 | Fully submerged | soft |
| K0528_A1 | Bull Creek | 34.181558 | -118.497717 | Frequently submerged, intertidal, marsh | soft |
| K0528_E4 | Bull Creek | 34.182029 | -118.49771 | Frequently submerged, intertidal, marsh | soft |
| K0528_K6 | Bull Creek | 34.181975 | -118.497849 | Frequently submerged, intertidal, marsh | soft |
| K0529_K6 | Bull Creek | 34.181652 | -118.497718 | Frequently submerged, intertidal, marsh | soft |
| K0529_T9 | Bull Creek | 34.181651 | -118.497716 | Fully submerged | soft |
| K0530_A1 | Bull Creek | 34.181419 | -118.497763 | Frequently submerged, intertidal, marsh | soft |
| K0530_B2 | Bull Creek | 34.181342 | -118.497657 | Frequently submerged, intertidal, marsh | soft |
| K0530_E4 | Bull Creek | 34.1814 | -118.497865 | Frequently submerged, intertidal, marsh | soft |
| K0528_G5 | Compton Creek | 33.843656 | -118.206466 | Frequently submerged, intertidal, marsh | soft |
| K0528_L7 | Compton Creek | 33.843055 | -118.205667 | Fully submerged | soft |
| K0528_T9 | Compton Creek | 33.843328 | -118.2061 | Frequently submerged, intertidal, marsh | soft |
| K0529_A1 | Compton Creek | 33.843196 | -118.205854 | Frequently submerged, intertidal, marsh | soft |
| K0530_C3 | Compton Creek | 33.843311 | -118.206092 | Frequently submerged, intertidal, marsh | soft |
| K0530_K6 | Compton Creek | 33.842877 | -118.205544 | Frequently submerged, intertidal, marsh | soft |
| K0530_L7 | Compton Creek | 33.842749 | -118.205402 | Fully submerged | soft |
| K0530_M8 | Compton Creek | 33.843196 | -118.205854 | Frequently submerged, intertidal, marsh | soft |
| K0529_C3 | Elysian Valley | 34.083829 | -118.228152 | Fully submerged | concrete |
| K0672_T9 | Elysian Valley | 34.084621 | -118.228071 | Frequently submerged, intertidal, marsh | concrete |
| K0673_A1 | Elysian Valley | 34.084217 | -118.228066 | Frequently submerged, intertidal, marsh | concrete |
| K0673_G5 | Elysian Valley | 34.084227 | -118.228048 | Fully submerged | concrete |
| K0674_G5 | Elysian Valley | 34.08455 | -118.228053 | Fully submerged | concrete |
| K0676_B2 | Elysian Valley | 34.08449 | -118.228157 | Fully submerged | concrete |
| K0676_T9 | Elysian Valley | 34.084721 | -118.228145 | Fully submerged | concrete |
| K0677_A1 | Elysian Valley | 34.084482 | -118.228157 | Frequently submerged, intertidal, marsh | concrete |
| K0593_T9 | Glendale | 34.155282 | -118.275211 | Fully submerged | concrete |
| K0594_L7 | Glendale | 34.15459 | -118.276618 | Fully submerged | concrete |
| K0596_C3 | Glendale | 34.155107 | -118.275459 | Fully submerged | concrete |
| K0596_E4 | Glendale | 34.154774 | -118.27637 | Frequently submerged, intertidal, mars | concrete |
| K0596_L7 | Glendale | 34.154918 | -118.276231 | Fully submerged | concrete |
| K0596_T9 | Glendale | 34.154973 | -118.275799 | Fully submerged | concrete |
| K0597_K6 | Glendale | 34.154997 | -118.275944 | Fully submerged | concrete |
| K0597_L7 | Glendale | 34.155157 | -118.27542 | Fully submerged | concrete |
| K0526_C3 | Glendale Narrows | 34.102813 | -118.242742 | Fully submerged | concrete |
| K0526_G5 | Glendale Narrows | 34.103427 | -118.242642 | Fully submerged | concrete |
| K0529_B2 | Glendale Narrows | 34.103109 | -118.242634 | Fully submerged | soft |
| K0529_G5 | Glendale Narrows | 34.103652 | -118.242686 | Fully submerged | concrete |
| K0529_M8 | Glendale Narrows | 34.103251 | -118.242645 | Fully submerged | concrete |
| K0672_B2 | Glendale Narrows | 34.10274 | -118.242669 | Fully submerged | concrete |
| K0678_B2 | Glendale Narrows | 34.103274 | -118.242544 | Fully submerged | concrete |
| K0678_K6 | Glendale Narrows | 34.103437 | -118.24275 | Fully submerged | concrete |
| K0672_A1 | Long Beach | 33.762909 | -118.202355 | Fully submerged | soft |
| K0674_M8 | Long Beach | 33.762738 | -118.202271 | Fully submerged | concrete |
| K0676_M8 | Long Beach | 33.762683 | -118.202126 | Fully submerged | concrete |
| K0677_B2 | Long Beach | 33.762833 | -118.202418 | Fully submerged | concrete |
| K0677_E4 | Long Beach | 33.762907 | -118.202298 | Fully submerged | concrete |
| K0677_L7 | Long Beach | 33.762841 | -118.20235 | Fully submerged | concrete |
| K0678_L7 | Long Beach | 33.762906 | -118.202305 | Fully submerged | soft |
| K0701_C3 | Long Beach | 33.76269 | -118.202303 | Fully submerged | concrete |
| K0527_A1 | Maywood | 33.986755 | -118.171412 | Frequently submerged, intertidal, marsh | concrete |
| K0527_C3 | Maywood | 33.988033 | -118.172607 | Fully submerged | concrete |
| K0527_E4 | Maywood | 33.987023 | -118.171842 | Fully submerged | concrete |
| K0527_K6 | Maywood | 33.986686 | -118.171342 | Fully submerged | concrete |
| K0527_L7 | Maywood | 33.987668 | -118.172288 | Fully submerged | concrete |
| K0527_T9 | Maywood | 33.986617 | -118.171324 | Fully submerged | concrete |
| K0539_L7 | Maywood | 33.986776 | -118.17165 | Fully submerged | concrete |
| K0593_G5 | Sepulveda Dam | 34.168961 | -118.475292 | Fully submerged | soft |
| K0594_A1 | Sepulveda Dam | 34.168698 | -118.475195 | Fully submerged | soft |
| K0594_T9 | Sepulveda Dam | 34.168961 | -118.475292 | Fully submerged | soft |
| K0595_G5 | Sepulveda Dam | 34.168941 | -118.47461 | Terrestrial, not submerged | soft |
| K0597_T9 | Sepulveda Dam | 34.1688 | -118.475049 | Fully submerged | soft |
| K0599_G5 | Sepulveda Dam | 34.16868 | -118.474846 | Frequently submerged, intertidal, marsh | soft |
| K0599_K6 | Sepulveda Dam | 34.168906 | -118.475125 | Fully submerged | soft |
| K0599_T9 | Sepulveda Dam | 34.168758 | -118.474733 | Rarely submerged, wetland, arroyo | soft |
| K0593_A1 | Tujunga Wash | 34.258032 | -118.386781 | Fully submerged | concrete |
| K0593_E4 | Tujunga Wash | 34.258403 | -118.386614 | Fully submerged | concrete |
| K0595_M8 | Tujunga Wash | 34.257481 | -118.386845 | Fully submerged | concrete |
| K0596_B2 | Tujunga Wash | 34.258667 | -118.386473 | Fully submerged | concrete |
| K0597_E4 | Tujunga Wash | 34.258716 | -118.386376 | Fully submerged | concrete |
| K0599_A1 | Tujunga Wash | 34.258424 | -118.386387 | Fully submerged | concrete |
| K0599_E4 | Tujunga Wash | 34.258395 | -118.386592 | Fully submerged | concrete |
| K0599_M8 | Tujunga Wash | 34.258016 | -118.386744 | Fully submerged | concrete |
| K0593_L7 | Verdugo Wash | 34.203216 | -118.237654 | Fully submerged | soft |
| K0595_A1 | Verdugo Wash | 34.202985 | -118.237755 | Fully submerged | soft |
| K0596_G5 | Verdugo Wash | 34.202611 | -118.237615 | Fully submerged | soft |
Differential Abundance Analysis
- The mean parameter is the expectation value for Kij and is proportional to the actual number of sequence counts for gene i under the experimental condition ρ. The size factor is also accounted for, which is essentially the coverage or sequencing depth of the genetic library for each sample.
- The variance σ2 is the sum of the shot noise and the raw variance.
- The model uses a pooled variance from genes (or taxa) with similar count values to estimate the per gene raw variance.
- m size factors, including 1 for each sample.
- n expression strength parameters qip for each condition ρ. In other words, the expectation values for the abundance of counts for gene or taxon i are proportional to qip.
- The pooled variance parameter simulates the dependence of Vip on the expectation value for the mean, qip, for each condition ρ.
3. Results
| LA RIVER | Taxon Abundance | Assigned Seqs/ Sample | ||||
| Marker | Min | Med | Max | Min | Med | Max |
| FITS | 17 | 211 | 183,729 | 369 | 18,157 | 40,447 |
| 16S | 29 | 181 | 109,927 | 1 | 15,178 | 44,190 |
| 18S | 30 | 168 | 299,045 | 386 | 24,799 | 56,966 |
| COI | 30 | 208 | 153,574 | 14 | 18,555 | 41,257 |
| 12S | 30 | 713 | 31,898 | 0 | 953 | 30,699 |
| PITS | 0 | 265 | 238,793 | 133 | 9,642 | 24,730 |
| LA RIVER | Branch Length NJ | Branch Length UPGMA | NJ vs. UPGMA | ||
| Marker | Mean | Variance | Mean | Variance | Tree Distance |
| FITS | 1,657 | 5,419,114 | 1,585 | 4,124,851 | 8,195 |
| 16S | 620 | 460,349 | 609 | 417,224 | 2,473 |
| 18S | 2,018 | 5,534,355 | 1,978 | 4,278,736 | 10,919 |
| COI | 2,312 | 8,746,132 | 2,114 | 6,010,691 | 9,697 |
| 12S | 634 | 4,710,694 | 1,585 | 4,124,851 | 12,130 |
| PITS | 1,457 | 6,728,373 | 1,351 | 4,241,554 | 8,516 |
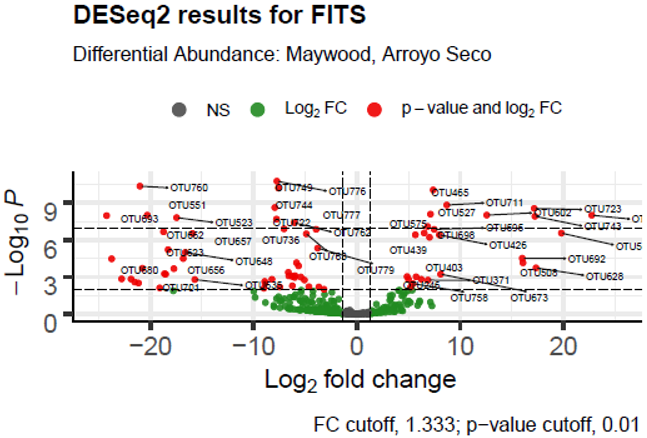
| log2FoldChange | padj | Taxon | Notes | |
| 22.09927 | 3.71E-23 | Prosthecobacter sp. | possible pathogen, anaerobic, tubulin like genes, low nutrient environments | |
| 34.31956 | 1.53E-41 | Dechloromonas sp. | may oxidize benzene | |
| -22.258 | 5.73E-05 | Devosia sp. | Nitrogen fixer | |
| -25.3115 | 1.67E-05 | Bacillus sp. | many beneficial species | |
| 23.78784 | 1.22E-06 | Chromatiaceae (unclassified) | purple sulfur bacteria, use sulfide to fix carbon and generate oxygen | |
| -30.519 | 0.009416 | Sandaracinobacter sp. | metabolism of sulfide to cysteine (or from serine) | |
| 25.68591 | 0.000938 | Chloroflexaceae (unclassified) | green non-sulfur bacteria, many heat-loving anoxygenic photoheterotrophs [29, 30] | |
| -22.3636 | 0.00014 | endosymbiont of Ridgeia piscesae | Gammaproteobacterium, symbiont of a tubeworm | |
| -6.85917 | 4.08E-06 | anaerobic bacterium MO-CFX2 Chloroflexi | ||
| 17.1087 | 4.15E-08 | Rhodocyclales (unclassified) | nitrogen fixing or nitrogen reducing | |
| 33.82601 | 2.58E-14 | Phormidium setchellianum | Potential cause of gastroenteritis, concentrates caused neuro- and hepato-toxicity in mice [31] |
|
| 20.18264 | 0.000268 | Cytophaga xylanolytica | xylan degrading, does well in sulfogenic and methanogenic environments, anaerobic and gliding |
|
| -23.4117 | 0.002659 | Synechococcus sp. | Photolysis of sulfide or water, produces neurotoxins [32] | |
| 11.0032 | 0.000123 | Scenedesmaceae (unclassified) | Green algae, may degrade radioactive materials | |
| 8.245038 | 0.000199 | Flavobacterium sp. | Often associated with plant resistance to pathogens | |
| 7.271474 | 0.005122 | Oscillatoriales cyanobacterium HF1 | Cyanobacterium which may cause illness or death in humans and animals | |
| 10.11933 | 0.001645 | Tetradesmus obliquus | Produces valuable saturated and unsaturated esters, extract has anticancer and antimicrobial effects [33, 34] |
|
| 28.7773 | 1.03E-07 | Microcystis sp. | Cyanobacterium which is toxic to humans [35] | |
| 28.91261 | 5.24E-05 | Rhodocyclaceae bacterium enrichment culture clone Y62 | nitrogen fixing or nitrogen reducing | |
| log2FoldChange | padj | Taxon | Notes | |
| -25.207183 | 3.06E-23 | Oscillatoriales cyanobacterium YACCYB599 | Cyanobacteria which may cause illness or death in humans and animals | |
| -24.66764915 | 4.55E-23 | Chroococcus subviolaceus | Freshwater or high salinity environments, Cyanobacteria which can survive with low O2 [36] | |
| -24.50212313 | 4.55E-23 | Haliea sp. | Marine gamma proteobacterium which tolerates up to12% salinity [37, 38] | |
| 24.49667323 | 3.81E-31 | Halomonas sp. | chloride and saline tolerance | |
| 24.12963073 | 1.43E-27 | Marmoricola sp. | Denitrifying bacteria [39] | |
| 10.00393321 | 8.21E-09 | alpha proteobacterium LS7-MT | Methanol oxidizer, lives in high temperatures [40] | |
| 9.188395232 | 2.37E-18 | Nitrosarchaeum koreense | Aerobic ammonia-oxidizing archaea [41] | |
| -8.382519826 | 0.001244 | Microcystaceae (unclassified) | Common Eutrophic Bloomer, toxin-producing Cyanobacterium | |
| 7.849119335 | 3.12E-07 | Acidobacterium sp. SCGC AAA007-P13 | Potential saprobe | |
| -7.732408042 | 4.32E-08 | Oscillatoriales cyanobacterium IRH12 | Cyanobacterium which may cause illness or death in humans and animals | |
| -7.389766623 | 0.000539 | Roseisolibacter agri | Grows in low oxygen environments [42] | |
| -7.310779292 | 1.03E-07 | Pleurocapsa concharum | Ostracod-dependent Cyanobacterium [43] | |
| 7.242636088 | 5.51E-07 | Devosia sp. | Nitrogen-fixing bacteria | |
| 6.970043209 | 0.001616 | Nitrospira sp. enrichment culture clone LD3 | Nitrifying bacteria nitrite oxidizing bacteria | |
| 6.533527317 | 1.83E-13 | gamma proteobacterium SCGC AAA007-P21 | Uncultivated bacterioplankton | |
| 6.503508981 | 0.001529 | alpha proteobacterium Schreyahn_AOB_Aster_Kultur_5 | Cultured alphaproteobacterium | |
| -6.479686479 | 0.000178 | Chlamydomonadales (unclassified) | Green algae [44] | |
| -6.382235759 | 0.000425 | Chloronema giganteum | Photoautotrophic, anoxygenic green non-sulfur bacteria [91] | |
| -6.230017507 | 0.002384 | Chamaesiphon sp. | Widely distributed Cyanobacterium [45] | |
| 6.02052523 | 0.007591 | Altererythrobacter sp. | Alkaline or salt tolerant aerobic phototroph, anoxygenic [46, 47, 48] | |
| 5.990283542 | 0.000524 | Mycobacteriaceae (unclassified) | Potential human and animal pathogens | |
| 5.737312813 | 2.78E-06 | Acidobacteriaceae (unclassified) | Likely saprobe of plant organic matter | |
| -5.72085055 | 0.009826 | Candidatus Viridilinea mediisalina | Anaerobic phototroph, salt-tolerant and prefers alkaline environments [49] |
|
| -5.56037325 | 2.59E-05 | Veillonellaceae bacterium 6-15 | bacterial vaginosis | |
| -5.548460876 | 0.000699 | Phormidium setchellianum | Cyanobacterium with possible antitumor agents, neuro and hepatotoxicity | |
| -5.531306605 | 0.003193 | Calothrix sp. UAM 374 | Cyanobacterium which grows on plants and hard substrates [89] |
|
| 5.344610141 | 0.0001 | Candidatus Nitrosocosmicus sp. | Aerobic ammonia-oxidizing archaea | |
| -5.019693824 | 0.003193 | Treponema stenostreptum | syphilis relative | |
| -4.952937198 | 0.001067 | Leptolyngbyaceae (unclassified) | Thermophilic and potentially iron-loving Cyanobacterium [50] | |
| -4.934291389 | 0.000964 | Holophagaceae (unclassified) | Anaerobic dweller of freshwater sediments [51] | |
| 4.926495832 | 0.009823 | unidentified eubacterium RB01 (Verrucomicrobia) | ||
| -4.711954167 | 0.002384 | Xanthomonadaceae bacterium | Potential phytopathogens | |
| -4.711366069 | 0.005914 | Leptolyngbya geysericola | Alkaline tolerant non-heteroctic Cyanobacterium, produces calcite on microplastics [52] |
|
| 4.50039412 | 4.71E-06 | Caldilineales bacterium | Thermophilic and anaerobic [53] | |
| -4.35065315 | 0.009823 | Fusibacter sp. enrichment culture | Thiosulfate reducing, potentially halotolerant | |
| -4.16646108 | 0.002439 | Desulfomicrobium sp. | oxidizes sulfide and arsenate in the presence of CO2 and acetate [54], reduces nitrate to ammonium [55] |
|
| -3.874861377 | 0.005914 | Oscillochloridaceae (unclassified) | anoxygenic phototrophic bacteria [29, 56] | |
| -3.695598612 | 0.009826 | Pleurocapsales (unclassified) | Cyanobacterium from calcareous environments | |
| 3.602101991 | 0.002384 | Vicinamibacter silvestris | Polyphosphate accumulating organisms | |
| 2.378738101 | 0.004923 | Firmicutes (unclassified) | High abundance in suburban rivers, negatively correlated with ammonia concentration |
|
| 2.253024076 | 0.008829 | Stenotrophobacter terrae | opportunistic pathogen | |
| 2.126473277 | 0.00044 | Vicinamibacteraceae (unclassified) | Degrades chitin [57] |
|
| 2.033767588 | 0.003193 | Actinobacteria (unclassified) | Many denitrifying bacteria [58, 59] | |
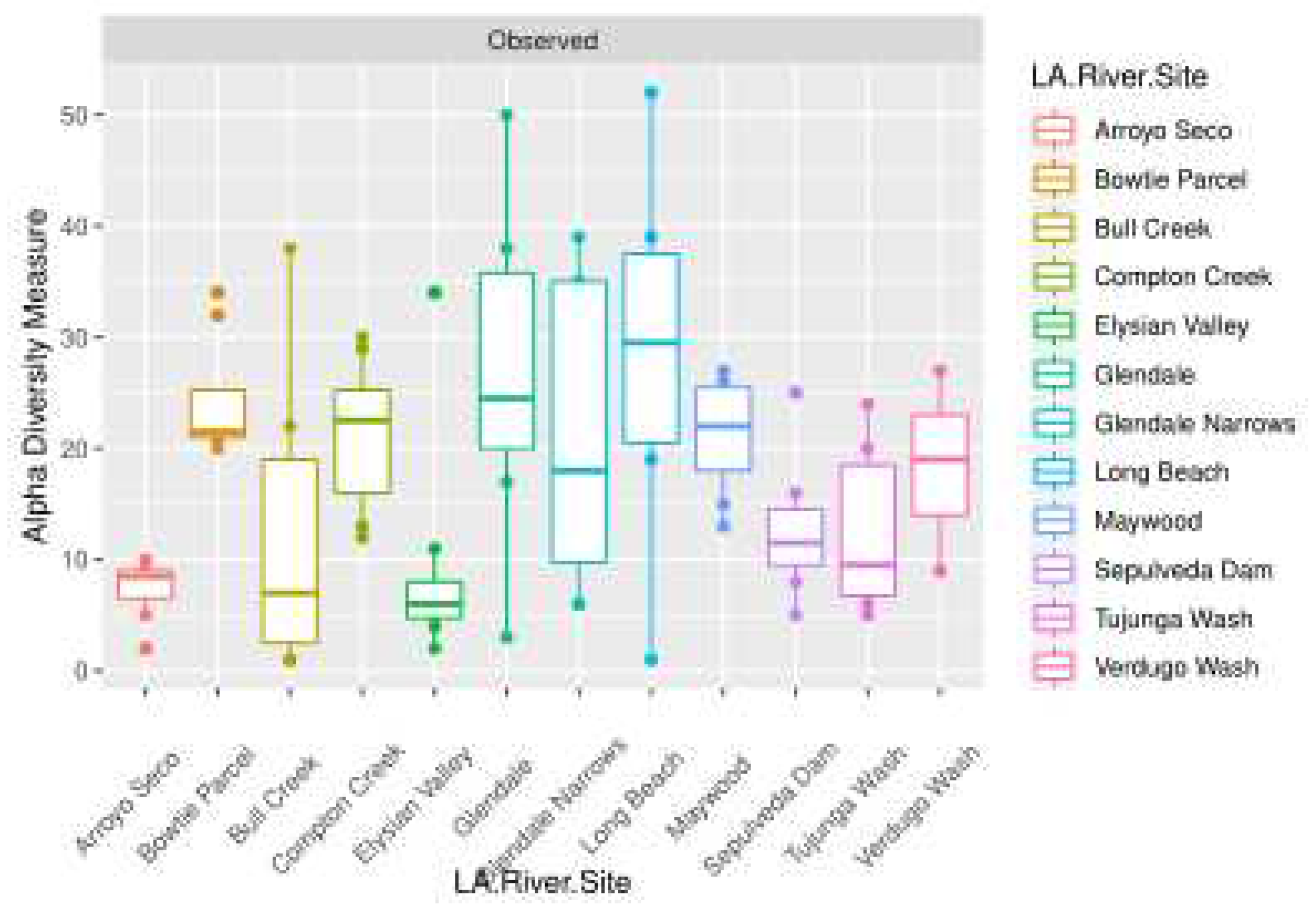
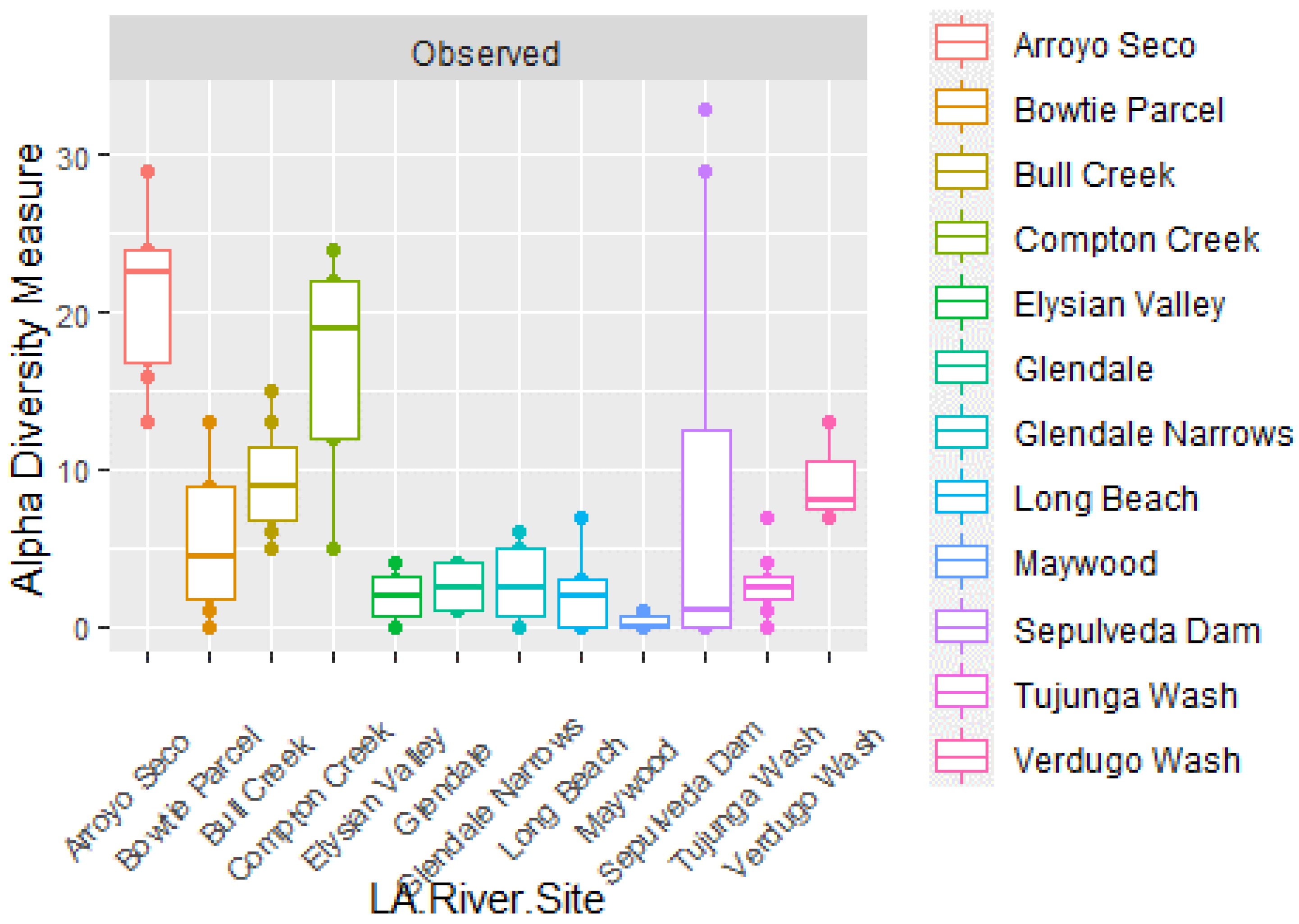
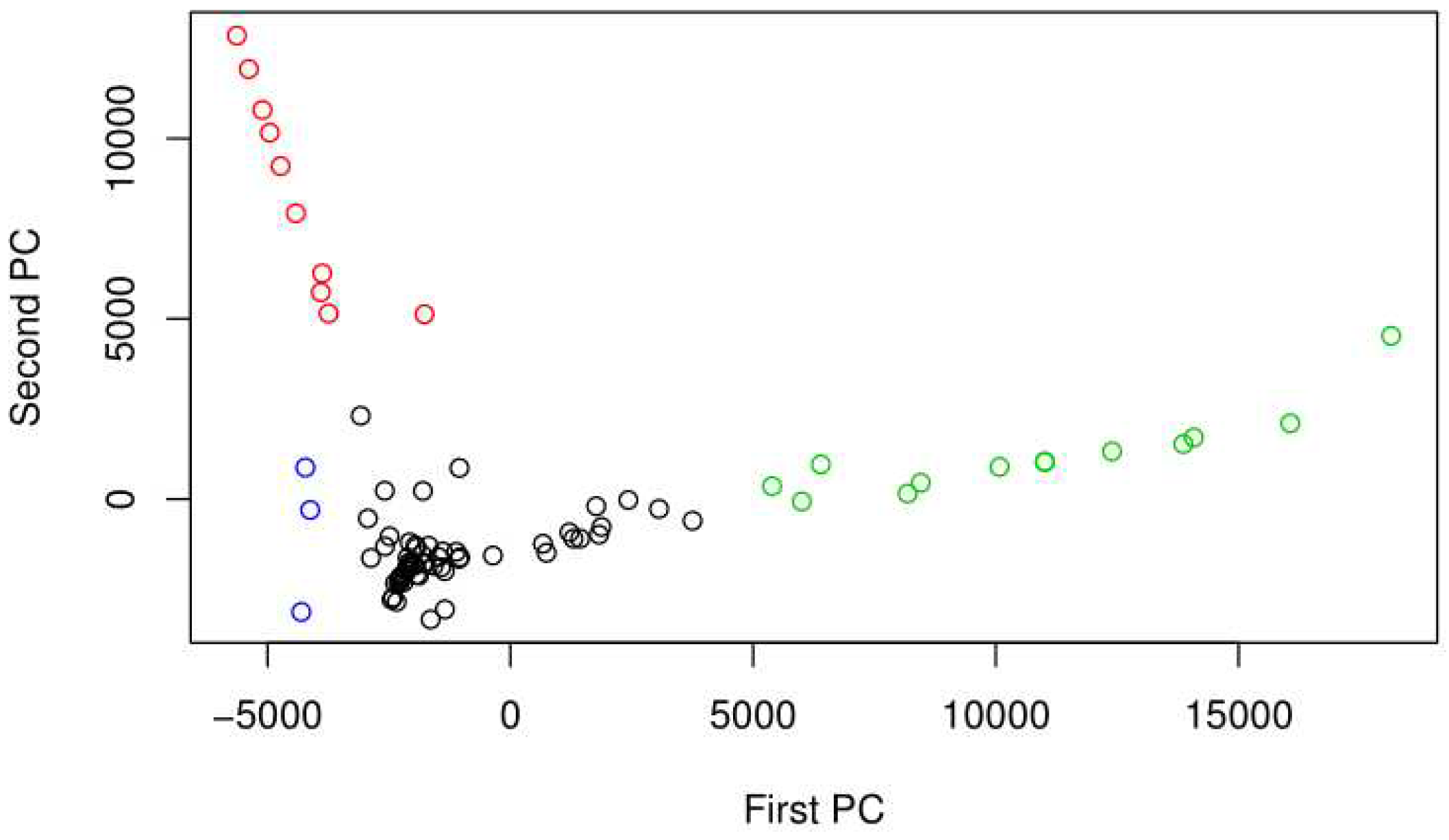
4. Discussion
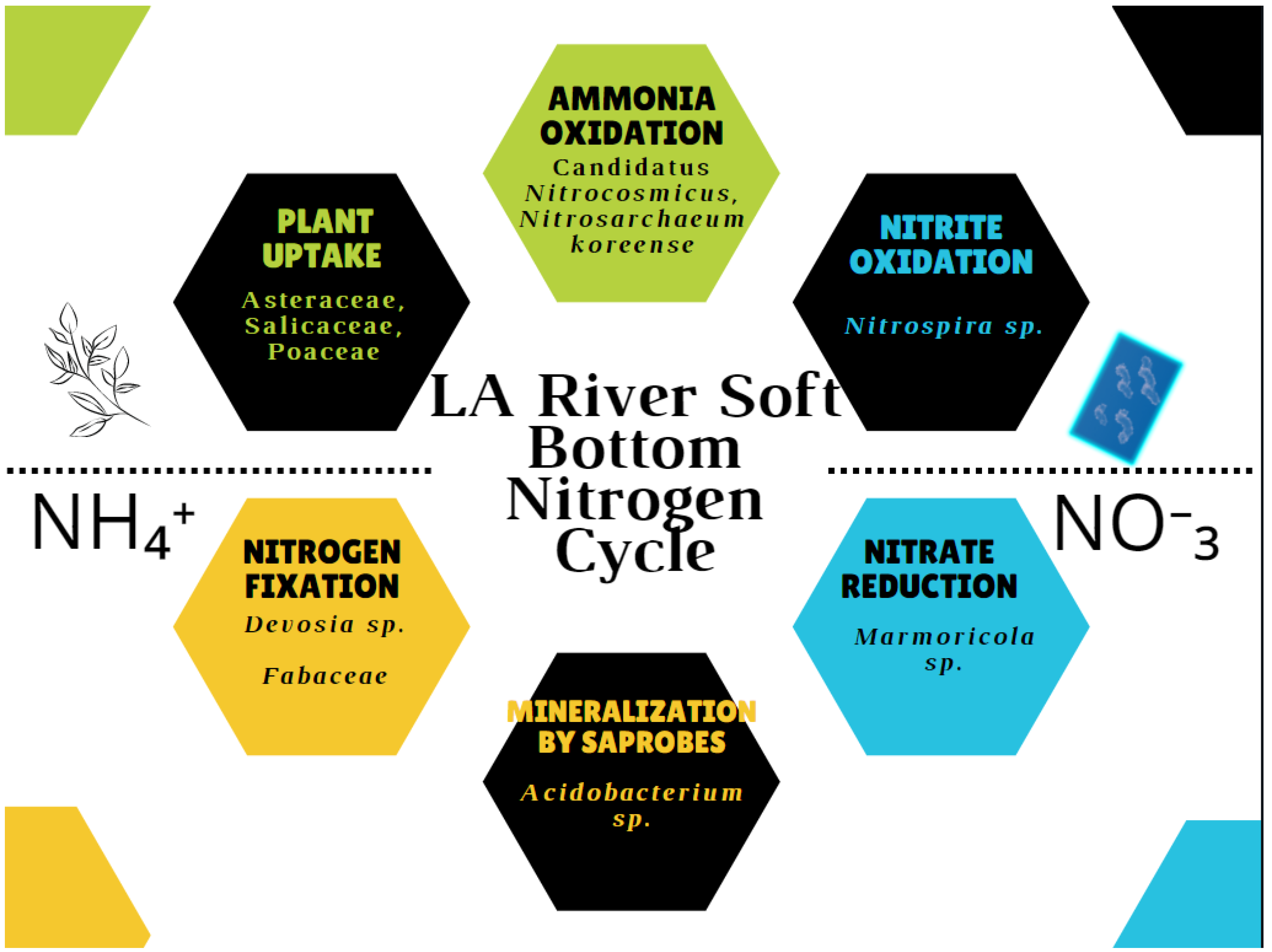
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- St. Louis Federal Reserve. Federal Reserve Economic Data. https://fred.stlouisfed.org/.
- United States Census Bureau. Quick Facts: Los Angeles city, California. https://www.census.gov/quickfacts/losangelescitycalifornia.
- California Environmental Protection Board. Los Angeles River Watershed impaired waters. State Water Resources Control Board - Los Angeles. Retrieved July 6, 2022, from https://www.waterboards.ca.gov/rwqcb4/water_issues/programs/regional_program/Water_Quality_and_Watersheds/los_angeles_river_watershed/303.shtml.
- Protecting Our River. Accessed 10/17/2021. Protectingourriver.org.
- Qiu, Han, Gu, Likun, Sun, Bo, Zhang, Jianyun, Zhang, Miao, He, Shanshan, An, Shuqing, and Leng, Xin. "Metagenomic Analysis Revealed That the Terrestrial Pollutants Override the Effects of Seasonal Variation on Microbiome in River Sediments." Bulletin of Environmental Contamination and Toxicology 105.6 (2020): 892-98. Web.
- Wei, Feifei, Sakata, Kenji, Asakura, Taiga, Date, Yasuhiro, and Kikuchi, Jun. "Systemic Homeostasis in Metabolome, Ionome, and Microbiome of Wild Yellowfin Goby in Estuarine Ecosystem." Scientific Reports 8.1 (2018): 3478-12. Web.
- Garba, Fatima et al. “Deteriorating Water Quality State on the Structural Assemblage of Aquatic Insects in a North-Western Nigerian River.” Water Science 36.1 (2022): 22–31.
- Qiusheng Yuan, Peifang Wang, Chao Wang, Juan Chen, Xun Wang, Sheng Liu, Indicator species and co-occurrence pattern of sediment bacterial community in relation to alkaline copper mine drainage contamination, Ecological Indicators, Volume 120, 2021, 106884, ISSN 1470-160X. [CrossRef]
- Khalilova, E A et al. “Halophilic bacteria of salt lakes and saline soils of the Peri-Caspian lowland (Republic of Daghestan) and their biotechnological potential.” Vavilovskii zhurnal genetiki i selektsii vol. 25,2 (2021): 224-233. [CrossRef]
- Ackerman, D., et al. "Characterization of water quality in the LA River." (2003).
- Desfor, Gene, and Keil, Roger. "Every River Tells a Story: The Don River (Toronto) and the Los Angeles River (Los Angeles) as Articulating Landscapes." Journal of Environmental Policy & Planning 2.1 (2000): 5-23. Web.
- Gandy, Matthew. "Riparian Anomie: Reflections on the Los Angeles River." Landscape Research 31.2 (2006): 135-45. Web.
- Seth J. Wenger, Allison H. Roy, C. Rhett Jackson, Emily S. Bernhardt, Timothy L. Carter, Solange Filoso,et al. "Twenty-six key research questions in urban stream ecology: an assessment of the state of the science," Journal of the North American Benthological Society, 28(4), 1080-1098, (27 October 2009).
- Rachael E. Antwis, Sarah M. Griffiths, Xavier A. Harrison, Paz Aranega-Bou, Andres Arce, Aimee S. Bettridge, Francesca L. Brailsford, et al. Fifty important research questions in microbial ecology, FEMS Microbiology Ecology, Volume 93, Issue 5, May 2017, fix044. [CrossRef]
- Orsi, Jared. Hazardous Metropolis: Flooding and Urban Ecology in Los Angeles, University of California Press, 2004. ProQuest Ebook Central, https://ebookcentral.proquest.com/lib/osu/detail.action?docID=227297.
- Halecki, Wiktor, and Tomasz Stachura. “Evaluation of Soil Hydrophysical Parameters Along a Semiurban Small River; Soil Ecosystem Services for Enhancing Water Retention in Urban and Suburban Green Areas.” Catena (Giessen) 196 (2021): 104910–. Web. Accessed June 28, 2022.
- Behera, Bijay Kumar, Patra, Biswanath, Chakraborty, Hirak Jyoti, Sahu, Parameswar, Rout, Ajaya Kumar, Sarkar, Dhruba Jyoti, Parida, Pranaya Kumar, Raman, Rohan Kumar, Rao, Atmakuri Ramakrishna, Rai, Anil, Das, Basanta Kumar, Jena, Joykrushna, Mohapatra, Trilochan, and Dikhit, Manas Ranjan. "Metagenome Analysis from the Sediment of River Ganga and Yamuna: In Search of Beneficial Microbiome." PloS One 15.10 (2020): E0239594. Web.
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Curd, EE, Gold, Z, Kandlikar, GS, et al. Anacapa Toolkit: An environmental DNA toolkit for processing multilocus metabarcode datasets. Methods Ecol Evol. 2019; 10: 1469– 1475. [CrossRef]
- Borg, I. and Groenen, P. (1997) Modern Multidimensional Scaling. Theory and Applications. Springer.
- Hastie, Tibshirani, and Friedman (2009). The Elements of Statistical Learning (ESL) (2nd edition).
- McMurdie and Holmes (2013) phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE. 8(4):e61217.
- Kandlikar, G.S.; Gold, Z.; Cowen, M.C.; Meyer, R.S.; Freise, A.C.; Kraft, N.J.; Moberg-Parker, J.; Sprague, J.; Kushner, D.J.; Curd, E.E. ranacapa: An R package and Shiny web app to explore environmental DNA data with exploratory statistics and interactive visualizations. F1000Research 2018, 7, 1734. [Google Scholar] [CrossRef] [PubMed]
- Schubert, Erich, and Peter J. Rousseeuw. "Fast and eager k-medoids clustering: O (k) runtime improvement of the PAM, CLARA, and CLARANS algorithms." Information Systems (2021): 101804.
- Lepère C, Domaizon I, Humbert J, Jardillier L, Hugoni M, Debroas D. 2019. Diversity, spatial distribution and activity of fungi in freshwater ecosystems. PeerJ 7:e6247. [CrossRef]
- Anders, S., Huber, W. Differential expression analysis for sequence count data. Genome Biol 11, R106 (2010). [CrossRef]
- Backman, T.W.H and Girke, T. (2016). systemPipeR: NGS Workflow and Report Generation Environment. BMC Bioinformatics, 17: 388.
- Maria Lukarska, Andrés Palencia, Chapter Eleven - Aminoacyl-tRNA synthetases as drug targets, Editor(s): Lluís Ribas de Pouplana, Laurie S. Kaguni, The Enzymes, Academic Press, Volume 48, 2020, Pages 321-350, ISSN 1874-6047, ISBN 9780128202609. [CrossRef]
- Hanada, S. (2014). The Phylum Chloroflexi, the Family Chloroflexaceae, and the Related Phototrophic Families Oscillochloridaceae and Roseiflexaceae. In: Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F. (eds) The Prokaryotes. Springer, Berlin, Heidelberg. [CrossRef]
- John C. Willison, Jean-Pierre Magnin, Chapter Eight - Role and Evolution of Endogenous Plasmids in Photosynthetic Bacteria, Editor(s): J. Thomas Beatty, Advances in Botanical Research, Academic Press, Volume 66, 2013, Pages 227-265, ISSN 0065-2296, ISBN 9780123979230. [CrossRef]
- Teneva, Ivanka et al. “Toxic potential of five freshwater Phormidium species (Cyanoprokaryota).” Toxicon : official journal of the International Society on Toxinology vol. 45,6 (2005): 711-25. [CrossRef]
- “Synechococcus.” Microbewiki, Kenyon College, https://microbewiki.kenyon.edu/index.php/Synechococcus.
- Oliveira, C.Y.B. , Oliveira, C.D.L., Prasad, R., Ong, H.C., Araujo, E.S., Shabnam, N. and Gálvez, A.O. (2021), A multidisciplinary review of Tetradesmus obliquus: a microalga suitable for large-scale biomass production and emerging environmental applications. Rev. Aquacult., 13: 1594-1618. [CrossRef]
- Luisa Gouveia, Jelena Molnar Jazić, Alice Ferreira, Snežana Maletić, Dragoljub Cvetković, Senka Vidović, Jelena Vladić.
- Montclair State University. Microcystis. New Jersey Center For Water Science And Technology. Retrieved July 3, 2022, from https://www.montclair.edu/water-science/freshwater-cyanobacteria-of-new-jersey/visual-guide-to-cyanobacteria-in-new-jersey/coccoid/colonial/microcystis/.
- “Chroococcus.” Microbewiki, Kenyon College, https://microbewiki.kenyon.edu/index.php/Chroococcus#Cell_Structure_and_Metabolism.
- Lucena, Teresa et al. “Haliea mediterranea sp. nov., a marine gammaproteobacterium.” International journal of systematic and evolutionary microbiology vol. 60,Pt 8 (2010): 1844-1848. [CrossRef]
- Yang, L. , Tan, Z., Wang, D. et al. Species identification through mitochondrial rRNA genetic analysis. Sci Rep 4, 4089 (2014). [CrossRef]
- Kasozi N, Kaiser H, Wilhelmi B. Metabarcoding Analysis of Bacterial Communities Associated with Media Grow Bed Zones in an Aquaponic System. Int J Microbiol. 2020 Oct 1;2020:8884070. [CrossRef] [PubMed]
- Islam T, Hernández M, Gessesse A, Murrell JC, Øvreås L. A Novel Moderately Thermophilic Facultative Methylotroph within the Class Alphaproteobacteria. Microorganisms. 2021 Feb 25;9(3):477. [CrossRef] [PubMed]
- Jung, Man-Young et al. “Nitrosarchaeum koreense gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon member of the phylum Thaumarchaeota isolated from agricultural soil.” International journal of systematic and evolutionary microbiology vol. 68,10 (2018): 3084-3095. [CrossRef]
- Pascual, Javier et al. “Roseisolibacter agri gen. nov., sp. nov., a novel slow-growing member of the under-represented phylum Gemmatimonadetes.” International journal of systematic and evolutionary microbiology vol. 68,4 (2018): 1028-1036. [CrossRef]
- “Pleurocapsa.” Microbewiki, Kenyon College, microbewiki.kenyon.edu/index.php/Pleurocapsa.
- Hu Y, Xing W, Song H, Zhu H, Liu G and Hu Z (2019) Evolutionary Analysis of Unicellular Species in Chlamydomonadales Through Chloroplast Genome Comparison With the Colonial Volvocine Algae. Front. Microbiol. 10:1351. [CrossRef]
- Kurmayer, R., Christiansen, G., Holzinger, A. et al. Single colony genetic analysis of epilithic stream algae of the genus Chamaesiphon spp.. Hydrobiologia 811, 61–75 (2018). [CrossRef]
- Dahal, Ram Hari, and Jaisoo Kim. “Altererythrobacter Fulvus Sp. Nov., a Novel Alkalitolerant Alphaproteobacterium Isolated from Forest Soil.” International journal of systematic and evolutionary microbiology 68.5 (2018): 1502–1508. Web.
- Li, Hui-Ping et al. “Altererythrobacter rhizovicinus sp. nov., isolated from rhizosphere soil of Haloxylon ammodendron.” International journal of systematic and evolutionary microbiology vol. 70,1 (2020): 680-686. [CrossRef]
- Fidalgo, Cátia et al. “Altererythrobacter halimionae sp. nov. and Altererythrobacter endophyticus sp. nov., two endophytes from the salt marsh plant Halimione portulacoides.” International journal of systematic and evolutionary microbiology vol. 67,8 (2017): 3057-3062. [CrossRef]
- Gaisin, Vasil A et al. “'Candidatus Viridilinea mediisalina', a novel phototrophic Chloroflexi bacterium from a Siberian soda lake.” FEMS microbiology letters vol. 366,5 (2019): fnz043. [CrossRef]
- Brown, Igor, Tringe, Susannah G., Ivanova, Natalia, Goodwin, Lynne, Shapiro, Nicole, Alcorta, Jaime, Pan, Donald, Chistoserdov, Andrei, Sarkisova, Svetlana, Woyke, Tanja, and Maresca, ed., Julia A. High-Quality Draft Genome Sequence of the Siderophilic and Thermophilic Leptolyngbyaceae Cyanobacterium JSC-12. United States: 2021. Web. [CrossRef]
- Fukunaga, Y., Ichikawa, N. (2014). The Class Holophagaceae . In: Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F. (eds) The Prokaryotes. Springer, Berlin, Heidelberg. [CrossRef]
- Mateos Cárdenas, Alicia et al. “Microplastics in the Freshwater Environment.” Reference Module in Earth Systems and Environmental Sciences 2021. Web.
- Yamada, Takeshi, et al. “Anaerolinea Thermolimosa Sp. Nov., Levilinea Saccharolytica Gen. Nov., Sp. Nov. and Leptolinea Tardivitalis Gen. Nov., Sp. Nov., Novel Filamentous Anaerobes, and Description of the New Classes Anaerolineae Classis Nov. and CALDILINEAE Classis Nov. in the Bacterial Phylum Chloroflexi.” International Journal of Systematic and Evolutionary Microbiology, vol. 56, no. 6, 2006, pp. 1331–1340. [CrossRef]
- Rabus, Ralf et al. “A Post-Genomic View of the Ecophysiology, Catabolism and Biotechnological Relevance of Sulphate-Reducing Prokaryotes.” Advances in microbial physiology 66 (2015): 55–. Print.
- Zhang, Tong et al. “Investigation of Dissimilatory Nitrate Reduction to Ammonium (DNRA) in Urban River Network Along the Huangpu River, China: Rates, Abundances, and Microbial Communities.” Environmental science and pollution research international 29.16 (2021): 23823–23833.
- Keppen, O I et al. “Proposal of Oscillochloridaceae fam. nov. on the basis of a phylogenetic analysis of the filamentous anoxygenic phototrophic bacteria, and emended description of Oscillochloris and Oscillochloris trichoides in comparison with further new isolates.” International journal of systematic and evolutionary microbiology vol. 50 Pt 4 (2000): 1529-1537. [CrossRef]
- Huber, Katharina J, and Jörg Overmann. “Vicinamibacteraceae Fam. Nov., the First Described Family Within the Subdivision 6 Acidobacteria.” International journal of systematic and evolutionary microbiology 68.7 (2018): 2331–2334. Web.
- Zhang, Haihan et al. “Nitrate reduction by the aerobic denitrifying actinomycete Streptomyces sp. XD-11-6-2: Performance, metabolic activity, and micro-polluted water treatment.” Bioresource technology vol. 326 (2021): 124779. [CrossRef]
- St. Clair, Savanah, et al. "Analysis of the soil microbiome of a Los Angeles urban farm." Applied and Environmental Soil Science 2020 (2020): 1-16.
- WoRMS Editorial Board (2021). World Register of Marine Species. Available from http://www.marinespecies.org at VLIZ. Accessed 2021-04-20. [CrossRef]
- ITIS Standard Report: Cyprididae. Retrieved Mar 2021, from the Integrated Taxonomic Information System on-line database, http://www.itis.gov.
- Myers, P., R. Espinosa, C. S. Parr, T. Jones, G. S. Hammond, and T. A. Dewey. 2021. The Animal Diversity Web (online). Accessed at https://animaldiversity.org, 6/1/2021.
- Smith, R.J. , Martens K. (2000) The ontogeny of the cypridid ostracod Eucypris virens (Jurine, 1820) (Crustacea, Ostracoda). In: Horne D.J., Martens K. (eds) Evolutionary Biology and Ecology of Ostracoda. Developments in Hydrobiology, vol 148. Springer, Dordrecht. [CrossRef]
- USDA, NRCS. 2021. The PLANTS Database (http://plants.usda.gov, 20 April 2021). National Plant Data Team, Greensboro, NC 27401-4901 USA.
- White Alder (U.S. National PARK SERVICE). Last updated Jan 31, 2020. https://www.nps.gov/articles/white-alder.htm.
- Gu, Qiyan et al. “Pollution Profile of Waterborne Bacterial and Fungal Community in Urban Rivers of Pearl River Estuary: Microbial Safety Assessment.” Journal of freshwater ecology 36.1 (2021): 305–322.
- Lin, Xianbiao et al. “Bacterial Community Shifts Driven by Nitrogen Pollution in River Sediments of a Highly Urbanized City.” International journal of environmental research and public health 16.20 (2019): 3794–.
- Wang, Peng et al. “Characteristics of Aquatic Bacterial Community and the Influencing Factors in an Urban River.” The Science of the total environment 569-570 (2016): 382–389.
- Luisa W Hugerth et al. “Metagenome-Assembled Genomes Uncover a Global Brackish Microbiome.” bioRxiv (2015).
- Iepure, Sanda et al. “Response of Microcrustacean Communities from the Surface—groundwater Interface to Water Contamination in Urban River System of the Jarama Basin (central Spain).” Environmental science and pollution research international 20.8 (2013): 5813–5826.
- Garcia, X, and D Pargament. “Reusing Wastewater to Cope with Water Scarcity: Economic, Social and Environmental Considerations for Decision-Making.” Resources, conservation and recycling 101 (2015): 154–166.
- Francis, Robert A. “Positioning Urban Rivers Within Urban Ecology.” Urban ecosystems 15.2 (2012): 285–291.
- Li, Kexin et al. “Microbial Abundance and Diversity Investigations Along Rivers: Current Knowledge and Future Directions.” Wiley interdisciplinary reviews. Water 8.5 (2021): e1547.
- Pini, Andrea K, and Pamela Geddes. “Fungi Are Capable of Mycoremediation of River Water Contaminated by E. Coli.” Water, air, and soil pollution 231.2 (2020).
- Cecchi, Grazia et al. “From Waste to Resource: Mycoremediation of Contaminated Marine Sediments in the SEDITERRA Project.” Journal of soils and sediments 20.6 (2019): 2653–2663.
- Cai, Xianlei et al. “Properties of Ammonia-oxidising Bacteria and Archaea in a Hypereutrophic Urban River Network.” Freshwater biology 67.2 (2022): 250–262. Web.
- J.P. Megonigal, M.E. Hines, P.T. Visscher, 8.08 - Anaerobic Metabolism: Linkages to Trace Gases and Aerobic Processes, Editor(s): Heinrich D. Holland, Karl K. Turekian, Treatise on Geochemistry, Pergamon, 2003, Pages 317-424, ISBN 9780080437514. [CrossRef]
- Wen, X.; Chen, F.; Lin, Y.; Zhu, H.; Yuan, F.; Kuang, D.; Jia, Z.; Yuan, Z. Microbial Indicators and Their Use for Monitoring Drinking Water Quality—A Review. Sustainability 2020, 12, 2249. [Google Scholar] [CrossRef]
- Herve, Vincent et al. “Aquatic Urban Ecology at the Scale of a Capital: Community Structure and Interactions in Street Gutters.” The ISME Journal 12.1 (2018): 253–266.
- Stal, L.J. (2007) Cyanobacteria. In: Seckbach J. (eds) Algae and Cyanobacteria in Extreme Environments. Cellular Origin, Life in Extreme Habitats and Astrobiology, vol 11. Springer, Dordrecht. [CrossRef]
- Bláha, Luděk et al. “Toxins produced in cyanobacterial water blooms - toxicity and risks.” Interdisciplinary toxicology vol. 2,2 (2009): 36-41. [CrossRef]
- Wang, Kai, Mandy Razzano, and Xiaozhen Mou. “Cyanobacterial Blooms Alter the Relative Importance of Neutral and Selective Processes in Assembling Freshwater Bacterioplankton Community.” The Science of the total environment 706 (2020): 135724–135724.
- Caro-Borrero, Angela, and Javier Carmona-Jiménez. “The Use of Macroinvertebrates and Algae as Indicators of Riparian Ecosystem Services in the Mexican Basin: a Morpho-Functional Approach.” Urban ecosystems 22.6 (2019): 1187–1200.
- “Assessment of Aquatic Life Use Needs for the Los Angeles River.” Los Angeles River Environmental Flows Project, Southern California Coastal Water Research Project, ftp.sccwrp.org/pub/download/DOCUMENTS/TechnicalReports/1154_LARiverAquaticLifeUses.pdf.
- Levi, Peter S., and Peter B. McIntyre. “Ecosystem Responses to Channel Restoration Decline with Stream Size in Urban River Networks.” Ecological applications 30.5 (2020): e02107–n/a.
- North Dakota State University Extension. Prevent Cyanobacteria Blooms. Aug 2020. https://www.ag.ndsu.edu/news/newsreleases/2020/aug-10-2020/prevent-cyanobacteria-blooms.
- Liu, Huan et al. “The Distribution Variance of Airborne Microorganisms in Urban and Rural Environments.” Environmental pollution (1987) 247 (2019): 898–906.
- US EPA. Control Measures for Cyanobacterial HABs in Surface Water. https://www.epa.gov/cyanohabs/control-measures-cyanobacterial-habs-surface-water.
- “Calothrix (Cyanobacteria).” Phycokey - Calothrix, University of New Hampshire Center for Freshwater Biology, cfb.unh.edu/phycokey/Choices/Cyanobacteria/cyano_filaments/cyano_unbranched_fil/tapered_filaments/CALOTHRIX/Calothrix_key.htm#:~:text=Habitat%3A,in%20the%20marine%20intertidal%20zone.
- Blighe K, Rana S, Lewis M (2021). EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R package version 1.10.0, https://github.com/kevinblighe/EnhancedVolcano.
- Gorlenko, Vladimir & Pierson, Beverly. (2015). Chloronema. 10.1002/9781118960608.gbm00381.
- Matsuoka S, Sugiyama Y, Sato H, Katano I, Harada K, Doi H (2019) Spatial structure of fungal DNA assemblages revealed with eDNA metabarcoding in a forest river network in western Japan. Metabarcoding and Metagenomics 3: e36335. [CrossRef]
- D. Baragaño, G. Ratié, C. Sierra, V. Chrastný, M. Komárek, J.R. Gallego, Multiple pollution sources unravelled by environmental forensics techniques and multivariate statistics, Journal of Hazardous Materials,Volume 424, Part B, 2022, 127413, ISSN 0304-3894. [CrossRef]
- UC CalEDNA DNA DATA SERVICE — CALeDNA (ucedna.com).
- Badotti, F., de Oliveira, F.S., Garcia, C.F. et al. Effectiveness of ITS and sub-regions as DNA barcode markers for the identification of Basidiomycota (Fungi). BMC Microbiol 17, 42 (2017). [CrossRef]
- Gilbert, J. (2015) Metagenomics, Metadata, and Meta-analysis. In: Nelson K.E. (eds) Encyclopedia of Metagenomics. Springer, Boston, MA. https://doi-org.ezproxy.proxy.library.oregonstate.edu/10.1007/978-1-4899-7478-5_17.
- Tran, Q. , Pham, DT. & Phan, V. Using 16S rRNA gene as marker to detect unknown bacteria in microbial communities. BMC Bioinformatics 18, 499 (2017). [CrossRef]
- Kisand, Veljo, Talas, Liisi, Kisand, Anu, Stivrins, Normunds, Reitalu, Triin, Alliksaar, Tiiu, Vassiljev, Jüri, Liiv, Merlin, Heinsalu, Atko, Seppä, Heikki, and Veski, Siim. "From Microbial Eukaryotes to Metazoan Vertebrates: Wide Spectrum Paleo-diversity in Sedimentary Ancient DNA over the Last ~14,500 years." Geobiology 16.6 (2018): 628-39. Web.
- Rach, Jessica, Bergmann, Tjard, Paknia, Omid, DeSalle, Rob, Schierwater, Bernd, and Hadrys, Heike. "The Marker Choice: Unexpected Resolving Power of an Unexplored CO1 Region for Layered DNA Barcoding Approaches." PloS One 12.4 (2017): E0174842. Web.
- Pumariño L, Alomar O, Agustí N. Development of specific ITS markers for plant DNA identification within herbivorous insects. Bull Entomol Res. 2011 Jun;101(3):271-6. [CrossRef] [PubMed]
- Green approach for the valorization of microalgae Tetradesmus obliquus, Sustainable Chemistry and Pharmacy, Volume 24, 2021, 100556, ISSN 2352-5541. [CrossRef]
- Shoun H, Kano M, Baba I, Takaya N, Matsuo M. Denitrification by actinomycetes and purification of dissimilatory nitrite reductase and azurin from Streptomyces thioluteus. J Bacteriol. 1998 Sep;180(17):4413-5. [CrossRef] [PubMed]
- Amaral-Zettler, L.A., McCliment, E.A., Ducklow, H.W. and Huse, S.M., 2009. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PloS one, 4: p.e6372.
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., et al. - Ultrahigh-throughput microbial community analysis on the Illumina HiSeq and MiSeqplatforms. The ISME Journal 6 (2012) 1621–1624. [CrossRef] [PubMed]
- Epp, L.S., Boessenkool, S., Bellemain, E.P., Haile, J., Esposito, A., Riaz, T., Erseus, C., Gusarov, V.I., Edwards, M.E., Johnsen, A. and Stenøien, H.K., 2012. New environmental metabarcodes for analysing soil DNA: potential for studying past and present ecosystems. Molecular Ecology, 21: pp.1821-1833.
- Gu, W., Song, J., Cao, Y., Sun, Q., Yao, H., Wu, Q., Chao, J., Zhou, J., Xue, W. and Duan, J., 2013. Application of the ITS2 region for barcoding medicinal plants of Selaginellaceae in Pteridophyta. PloS one, 8: p.e67818.
- Leray, M., Yang, J.Y., Meyer, C.P., Mills, S.C., Agudelo, N., Ranwez, V., Boehm, J.T. and Machida, R.J., 2013. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Frontiers in Zoology, 10 p.34.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





