Introduction
The evolution of multicellularity is the premier example of an evolutionary transition in individuality, which involves the integration of lower-level individuals (cells in this case) into a new kind of higher-level individual, the multicellular organism. As an evolutionary transition in individuality, the evolution of multicellularity is especially interesting because it occurred repeatedly throughout the tree of life (in both prokaryotes and eukaryotes) and at vastly different points in the history of life (Grosberg & Strathmann, 2007; Herron et al., 2022). Other examples of evolutionary transitions in individuality are the integration of networks of cooperating genes into the genome of the first cell, the origin of the eukaryotic cell, and the evolution of eusocial insect societies (Buss, 1987; Maynard Smith & Szathmáry, 1995; Michod, 1999).
Multicellular phenotypes can be either stable/obligate or facultative (i.e., in response to environmental cues, as a part of the life cycle) and involve clonal (e.g., animals, plants, algae, fungi) or non-clonal (e.g., social amoebae, myxobacteria) groups, with or without cell specialization (Bonner, 1998; Herron et al., 2022). Multicellularity can also evolve in the lab and can be studied using experimental evolution (Boraas et al., 1998; Herron et al., 2019; Ratcliff et al., 2012, 2013; Rose et al., 2020).
For billions of years, life on earth was comprised of solely single-celled individuals. At different points in the history of life, cells formed groups and these cell-groups evolved into stable, integrated multicellular individuals with heritable variation in fitness at the group level. But how do groups of cells evolve into a new kind of individual? We have previously suggested that the transition of a group of cells into a multicellular individual requires the reorganization of fitness from being a property of single cells to being a property of the multicellular group (Michod & Nedelcu, 2003; Nedelcu & Michod, 2003).
To envision the reorganization of fitness during the transition to multicellular individuality we need to understand the general properties of fitness, its components, and how these fitness components trade-off with one another. The fitness of any evolutionary individual involves two basic components: reproduction and survival. High overall fitness requires a balance between these components; in the simplest mathematical model, fitness is taken as the product of survival and reproduction (Michod, 2006b). But survival and reproduction often trade-off with one another: investment in one fitness component tends to detract from the other.
During the transition from unicellular to multicellular individuals, the activities associated with reproduction and survival, initially expressed in a way that enhances the fitness of individual cells, become reorganized among the cells of the multicellular group. Ultimately, cells differentially express one or the other component (i.e., cells become specialized) to increase the fitness of the new multicellular individual. This division of labor between cells in the multicellular group reorganizes the two fitness components, reproduction and survival, from the cell-level to the group level. Once the fitness components are compartmentalized between cells specialized in reproduction (germ) and survival (soma), the multicellular group becomes a multicellular individual; that is, it is indivisible in the sense that the specialized cells lost their own individuality and can no longer both survive and reproduce outside of the context of the group (Michod, 2006a, 2006b; Michod & Nedelcu, 2003).
How might fitness become reorganized during an evolutionary transition in individuality? In particular, what is the genetic basis underlying the reorganization of fitness resulting in cell specialization in fitness components during the evolution of multicellularity? Understanding the relative contribution of the two fitness components throughout the life of all individuals can provide a mechanistic framework. Many life-history traits control resource contribution to survival and reproduction as the individual proceeds through its life cycle. Furthermore, in times of stress, or when resources are limited, overall fitness is compromised, and individuals must commit to survival and delay reproduction until conditions improve. Genes that control investment in fitness components in times of stress are examples of life history genes. All organisms must possess life history genes, including genes that decrease reproduction in times of stress. Our hypothesis is that genes that turn off reproduction in times of stress in unicellular organisms have been co-opted during the evolution of multicellularity to produce non-reproductive somatic cells in cell groups (Nedelcu, 2009; Nedelcu & Michod, 2006).
Consistent with our hypothesis, current evidence indicates that de novo gene evolution and gene family expansion played a minor role in the evolution of multicellularity and cellular differentiation in both the animal and green algal/plant lineages. Instead, gene co-option (often involving gene duplication followed by diversification) appears to have been a major contributor (Brunet & King, 2017; Olson & Nedelcu, 2016). Specifically, many genes that are involved in traits associated with multicellularity (and that have initially been thought to have evolved during the evolution of multicellularity) have in fact been co-opted from genes present in their unicellular ancestors, where they played roles associated with the unicellular lifestyle. Such a scenario can be especially envisioned for the evolution of soma and germ – the two components that embody the full transition to multicellular individuality.
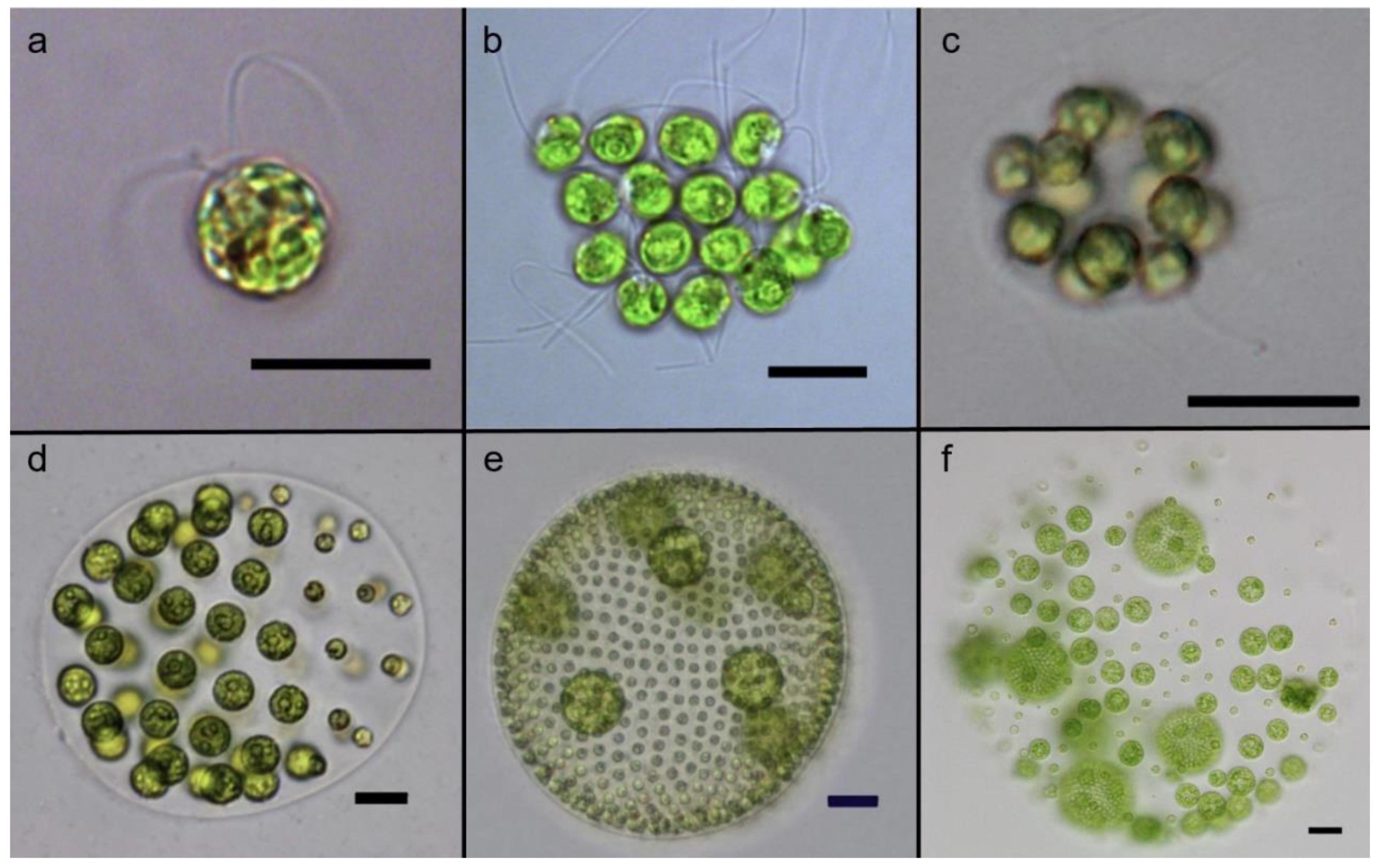
Studies into such gene co-option events are challenging in many multicellular lineages due to the ancient origin of multicellularity and lack of closely related unicellular and simple multicellular relatives. For example, metazoans evolved multicellularity 574-852 million years ago (Sharpe et al., 2014) and lineages resembling transitionary ancestors are not known. However, a group of green algae in the order Volvocales provides an ideal model system for studying the transition from unicellular to differentiated multicellularity. This group – known as the volvocine algae – evolved multicellularity relatively recently (~240 million years ago) and contains extant relatives that span a range of complexity from unicellularity, to undifferentiated multicellularity, to differentiated multicellularity (Hanschen et al., 2017; Herron et al., 2009; Umen, 2020) (
Figure 1). Furthermore, consistent with our hypothesis, the evolution of somatic cell differentiation in this lineage involved the co-option of a stress-induced life history gene that belongs to the
regA-like gene family (König & Nedelcu, 2020; Michod & Nedelcu, 2003; Nedelcu & Michod, 2006), which is the subject of this review. As this gene family is specific to the volvocine green algae and not found in other lineages, our hope is that the research reviewed here may stimulate efforts to identify similar genes in other lineages. We predict that such work would reveal additional examples of life history genes that have been co-opted for fitness reorganization and the evolution of soma during the transition to multicellular individuality in other groups.
The volvocine model system
The volvocine algae are a group of freshwater haploid bi-flagellated chlorophyte green algae that reproduce asexually in optimal environments, but can undergo rounds of sexual reproduction under stressful conditions (Coleman, 2012). This group has been developed as a model system for the evolution of multicellularity and cellular differentiation because its species span a range of morphological and developmental traits from single-celled organisms (e.g., Chlamydomonas), to multicellular forms without cell specialization (e.g, Gonium, Eudorina), to multicellular organisms with complex embryonic development and germ-soma differentiation (i.e., Volvox) (Herron & Michod, 2008; Kirk, 2005; Umen, 2020).
The multicellular volvocine species are included in three families: the Tetrabaenaceae, Goniaceae, and Volvocaceae. In addition, within the Volvocaceae family, two sub-clades have been defined: the “Eudorina Group” and the Euvolvox (or section Volvox) (Nozaki et al., 2000). The Tetrabaenaceae contains two species, Tetrabaena socialis and Basichlamys sacculifera. These are the simplest multicellular volvocine algae with four Chlamydomonas-like cells arranged like a four-leaf clover (Arakaki et al., 2013; Nozaki et al., 1996). The polyphyletic Goniaceae includes several species in two genera, Gonium and Astrephomene. Gonium species have 8 – 16 Chlamydomonas-like cells arranged as a flat plate. Whereas Astrephomene species are 32 or 64-celled spheroidal colonies with 2 to 4 sterile somatic cells in the posterior of the colony (Coleman, 2012; Hanschen et al., 2016; Yamashita et al., 2021). The Volvocaceae is the largest and most diverse family of volvocine green algae with many polyphyletic genera. Algae in the genera Eudorina, Pandorina, Volvulina, Yamagishiella and Colemanosphaera all have spheroidal body plans with between 16 – 64 cells (cell numbers vary between genera) with no germ-soma cellular differentiation under standard growth conditions. Species in the Pleodorina genus have 32 to 128 cells with specialized somatic cells in the anterior portion of the colony, except for one species, Pleodorina sphaerica, that has somatic cells distributed in both the anterior and posterior of the colony (Coleman, 2012; Nozaki et al., 2017). Finally, species in the genus Volvox are the largest and most complex members of the Volvocaceae, with several hundred to several thousand cells and two distinct cell types, specialized germ and specialized soma (Coleman, 2012).
The multicellular volvocine algae evolved from unicellular ancestors related to species in the
Chlamydomonas and
Vitreochlamys genera. Historically, analyses based on single or small sets of genes, indicated that multicellularity has arisen only once in the volvocine green algae clade (Herron et al., 2009). However, a recent phylotranscriptomic analysis that more than quadrupled the number of single-copy nuclear genes used for phylogenetic reconstruction suggests that (i) multicellularity possibly evolved twice in this group, and (ii) the Goniaceae family is not monophyletic (Lindsey et al., 2021). While this conclusion requires additional confirmation, we adopt this new phylogenetic hypothesis as the evolutionary framework for this review. The current tree topology (
Figure 2) also implies that cellular differentiation evolved four to six times independently in the volvocine green algae, which is consistent with past analyses (Grochau-Wright et al., 2017; Herron & Michod, 2008).
The species
Volvox carteri forma
nagariensis has served as the primary model organism for studying the developmental and genetic mechanisms that underlie cellular differentiation (Kirk, 1998, 2001; Umen, 2020). An asexual
V. carteri individual contains 1,000-2,000 small
Chlamydomonas-like flagellated somatic cells and up to 16 large unflagellated germ cells known as gonidia (
Figure 1). The somatic cells are terminally differentiated and have no cell division potential. The lack of cell division ensures that the motility of the individual is maintained because flagellar activity is compromised during cell division in volvocine algae due to the so-called “flagellation constraint” and the presence of a rigid cell wall (Koufopanou, 1994). Juvenile
V. carteri develop from gonidia, which grow to up to ~1000 times the volume of somatic cells before they start dividing. Development of a juvenile
V. carteri begins with a series of five symmetric divisions resulting in a 32-celled embryo. Then, during the sixth division cycle, the 16 cells in the anterior of the embryo divide asymmetrically with one daughter cell inheriting a larger volume of cytoplasm than the other. These large cells go through two additional asymmetric divisions then cease dividing, while all other cells go through a series of 11 to 12 symmetric divisions. At the end of cleavage, the embryo contains ~2,000 cells, most of which are small soma-initial cells, except the 16 large germ-cell initials generated through asymmetric division. At this stage, the embryo is effectively inside-out relative to the adult organization, with the flagella of the somatic cells pointing inward. To gain the adult configuration, the embryo goes through an inversion process.
Cytodifferentiation occurs just after inversion; all cells <8 µm will terminally differentiate into somatic cells while larger cells will become germ (Kirk, 2001). Cell size has been shown to be sufficient for determining cell fate in V. carteri (Kirk et al., 1993) though this is not the case in all Volvox species (Grochau-Wright, 2019; Grochau-Wright et al., 2021; Ransick, 1991, 1993). In V. carteri, a gene known as regA is turned on in the small cells, which results in the suppression of germ cell development and the differentiation of somatic cells. On the other hand, a set of lag genes are specifically induced in the large cells, which suppresses somatic cell development and initiates the differentiation of germ cells (Kirk, 2001). Currently, the presence of lag genes has only been established through mutant phenotypes and linkage mapping while the actual genes themselves are unknown. Below, we describe characteristics of the regA gene in V. carteri.
regA gene structure and function
Early investigations into cellular differentiation in
V. carteri identified a class of mutants called “somatic regenerators” in which somatic cells first develop normally but then dedifferentiate and become reproductive (Sessoms & Huskey, 1973; Starr, 1970) (
Figure 1f). Linkage analysis found that all such regenerator mutants map to a single locus
1, which was named
regA (from “
regenerator”) (Huskey et al., 1979; Huskey & Griffin, 1979; Kirk, 1998). However, it is worth noting that the annotation “RegA” or “Reg genes” has been used multiple times independently in other species to refer to many different types of genes. Such similarities in name are due to historical and linguistic coincidence rather than any shared function or homology. In this review, we are strictly discussing the
regA gene and its gene family that is restricted to volvocine algae.
regA is known as the master regulatory gene that controls somatic cell development in V. carteri (D. L. Kirk, 1998, 2001; M. M. Kirk et al., 1999). M. M. Kirk et al. (1999) used transposon tagging to identify the regA gene and went on to determine that the RegA protein is localized in the nuclei of somatic cells. This finding, the amino acid composition, and the presence of a DNA-binding SAND domain in the RegA protein (Duncan et al., 2006) helped establish the current working model that RegA acts as a transcriptional repressor of genes needed for gonidial development (Kirk, 2001).
In V. carteri f. nagariensis, expression of regA in somatic-progenitor cells begins during development shortly after inversion and persists throughout the rest of the life-cycle exclusively in somatic cells (Harryman, 2012; M. M. Kirk et al., 1999; Klein et al., 2017; Matt & Umen, 2018). A long-standing hypothesis is that regA suppresses the expression of nuclear encoded chloroplast proteins required for chloroplast biogenesis and turn-over (Choi et al., 1996; Meissner et al., 1999; Tam & Kirk, 1991). These negative effects on chloroplast would reflect in the inability of the somatic cells to photosynthesize, grow and divide. However, Matt & Umen (2018) cast some doubt on this idea. They used whole transcriptome analysis to compare the expression profiles of germ cells and somatic cells. While photosynthetic genes were expressed at around two-fold higher levels in germ cells, photosynthetic genes were nevertheless highly abundant in somatic cells as well. Matt & Umen (2018) propose that both germ cells and somatic cells maintain active photosynthesis, but germ cells are specialized in anabolic processes like starch, fatty acid, and amino acid biosynthesis while somatic cells breakdown starch and lipids to provide substrates needed to synthesize ECM glycoproteins. So, while it remains plausible that regA down-regulates photosynthetic genes, it is also plausible that regA down-regulates other genes related to germ cell growth like starch synthesis.
The structure of the
regA gene has been well described for
V. carteri and serves as the basic template for the gene structure of many other homologs in the
regA gene family
– referred to as the VARL (
Volvocine
Algae
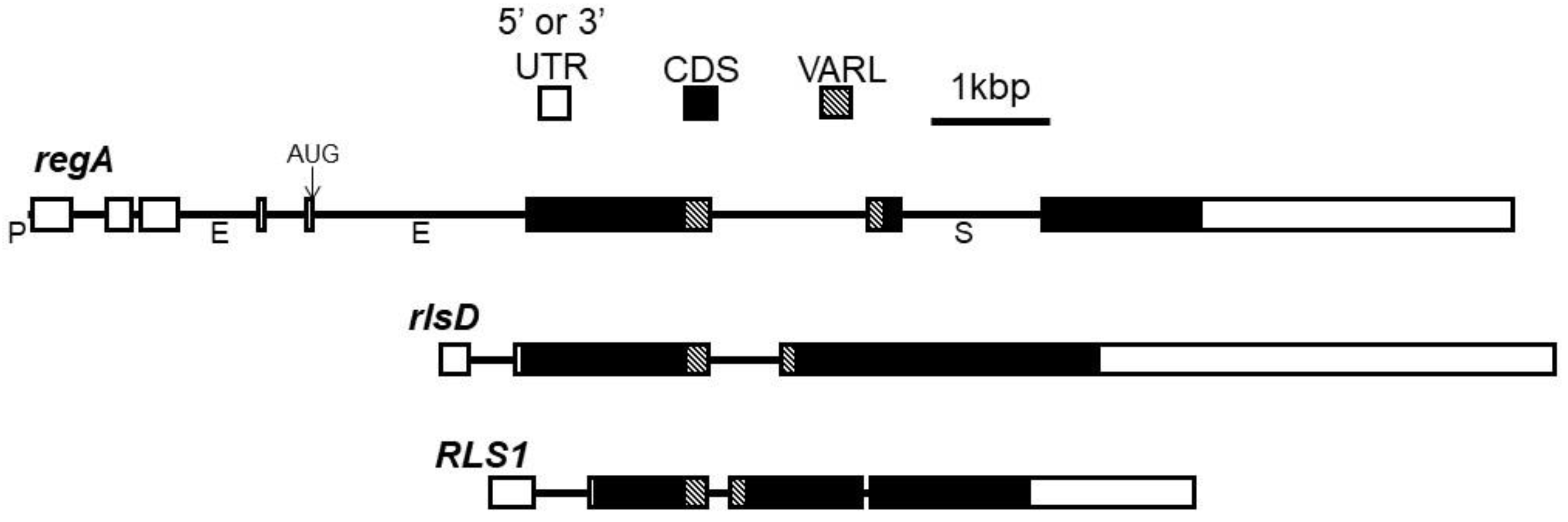
regA-Like) family. The minimal promoter of
regA consists of only 42 nt found directly upstream of the transcription start site with a plausible TATA box with the sequence TAATTGA beginning at -28 and an initiator region with the sequence CACTCAT beginning -1 relative to the transcription start site (Stark et al., 2001). The transcriptional unit of
regA is 12,477 nt-long and contains seven introns and eight exons; after introns are spliced out, the mature
regA mRNA is 6,725 nt-long and consists of a 940 nt 5’UTR (exons 1 – 5), a 3,147 nt coding region (exons 5 – 8), and a 2,638 nt 3’UTR with a UGUAA polyadenylation signal (Kirk et al., 1999) (
Figure 3).
However, a splice variant that retains intron 7 (1,194 bp) is expressed at low levels in V. carteri f. nagariensis as well. The donor splice site of intron 7 is GC instead of the typical GU, which may explain the variation in splicing. Remarkably, intron 7 encodes an ORF in the same frame as the rest of the regA coding region and therefore is likely to be translated, resulting in two different RegA protein products. However, experiments using modified regA transformation constructs to alter the splicing and translation of intron 7 have demonstrated that the presence or absence of intron 7 splicing has no detectable effect on phenotypic rescue of regenerator mutants, despite retention of intron 7 adding nearly 400 more amino acid residues to the RegA protein (Stark et al., 2001). Interestingly, the homologous region to intron 7 is not spliced out in the closely related V. carteri f. kawasakiensis, and protein-level homology has been described within the intron 7 region across a wide variety of volvocine algae species (Duncan et al., 2006; Grochau-Wright et al., 2017). Thus, it appears likely that splicing out intron 7 is a quirk specific to V. carteri f. nagariensis while homologous regions are exonic in other species.
In addition to the promoter, the differential transcription of regA is regulated by two enhancers found in introns 3 and 5, and a silencer found in intron 7 (Stark et al., 2001). Eight possible AUG start codons are found in the 5’UTR of the mature regA mRNA, which are thought to be bypassed by a ribosome shunting mechanism so that translation begins at the 9th AUG sequence of the mRNA (Babinger et al., 2006).
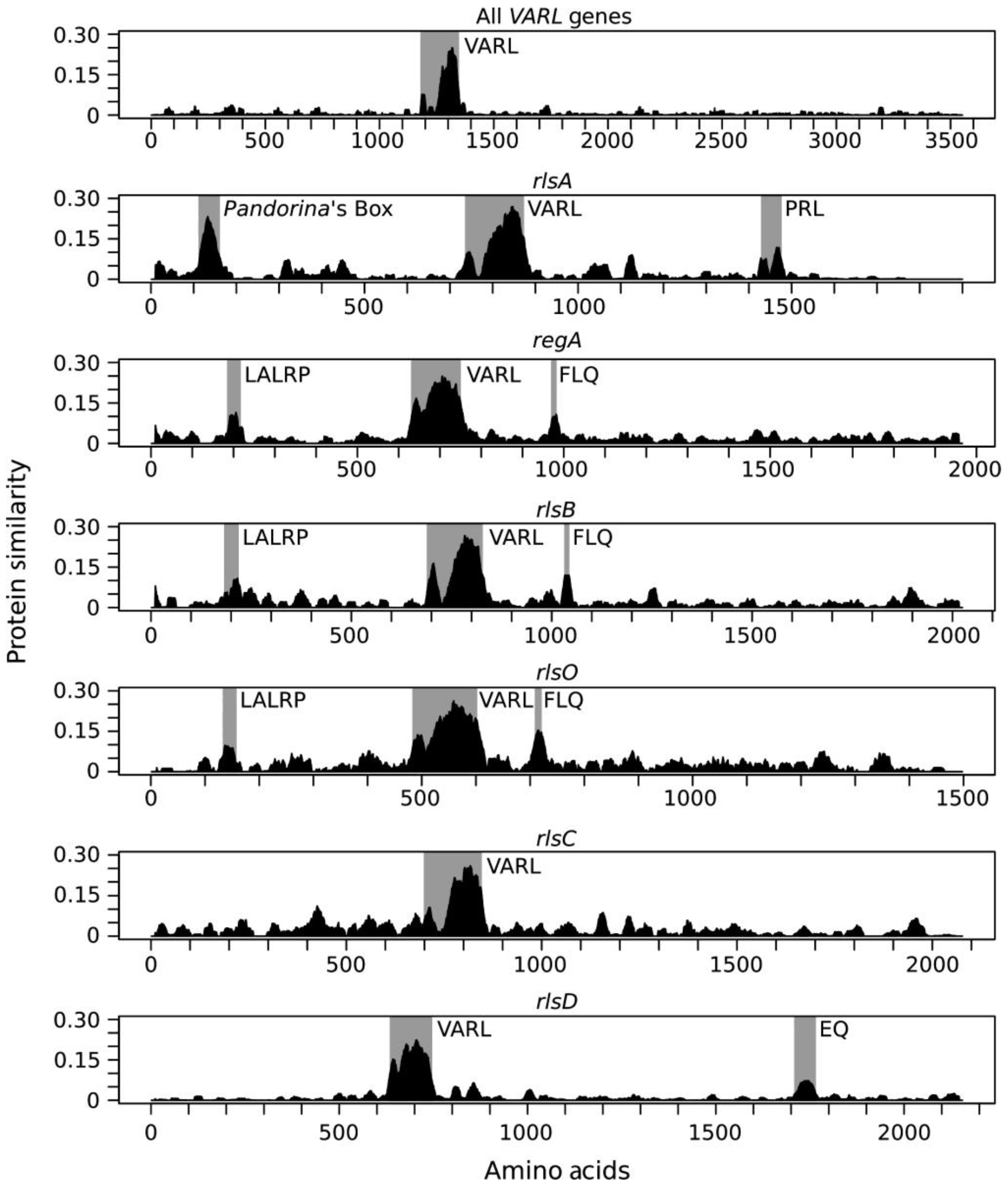
Following translation, the predicted RegA protein is 1,049 amino acids long without the inclusion of intron 7 or 1,447 with intron 7; and contains a high proportion of glutamine, alanine and proline residues (M. M. Kirk et al., 1999; Stark et al., 2001). A key structural region within the RegA protein is the VARL domain, which is the distinguishing feature of the VARL gene family (Duncan et al., 2006, 2007) (
Figure 4). The VARL domain is located between amino acids 444 – 558 in the RegA of
V. carteri f.
nagariensis and is composed of a highly conserved core VARL region (sites 484 – 558), a short but highly conserved N-terminal extension region (sites 444 – 455) and a less conserved linker region between these two (Duncan et al., 2006, 2007; Grochau-Wright et al., 2017). In addition, two short motifs of high amino acid conservation have been identified that are shared across the predicted RegA proteins of numerous volvocine algae species: a “LALRP” motif upstream of the VARL domain and an “FLQ” motif found within the intron 7 region downstream of the VARL domain (Grochau-Wright et al., 2017) (
Figure 5).
The core VARL domain appears to encode a DNA-binding SAND domain (Duncan et al., 2006). The SAND domain (IPR000770/PF01342) – named after Sp100, AIRE-1, NucP41/75, DEAF-1, is a DNA-binding domain found in animal and plant proteins that function in chromatin-dependent transcriptional control or bind specific DNA sequences (e.g., (Bottomley et al., 2001)). SAND-containing proteins are involved in multiple distinct processes, both general and lineage/tissue-specific. However, most of the known roles of SAND-containing proteins are related to multicellular development – including cell differentiation, cell proliferation, tissue homeostasis and organ formation. For instance, DEAF-1 (Deformed epidermal autoregulatory factor-1) is required for embryonic development in the fly Drosophila melanogaster (Veraksa et al., 2002) and also regulates breast epithelial cell differentiation in mammals (Barker et al., 2008). GMEB (Glucocorticoid Modulatory Element Binding) controls neural apoptosis in the nematode Caenorhabditis elegans (Nakagawa, 2008). Spe44 (Speckled protein 44 kDa) is a master switch of germ cell fate in C. elegans and like the mammalian AIRE (Autoimmune regulator 1) plays a role in sperm cell differentiation (Kulkarni et al., 2012; Radhakrishnan et al., 2016; Schaller et al., 2008). In land plants, SAND domains are associated with ATX (Arabidopsis homolog of trithorax) and ULTRAPETALA (ULT) proteins. Both protein types are involved in cell proliferation, cell differentiation, and tissue patterning. For instance, ATX1 in Arabidopsis thaliana is required for root, leaf and floral development through its histone methyltransferase activity (Chen et al., 2017); whereas ULT is a negative regulator that influences shoot and floral meristem size through controlling cell accumulation (Carles, 2005; Carles & Fletcher, 2009; Fletcher, 2001).
Evolution of the VARL gene family
The VARL gene family is defined by the presence of a homologous VARL domain within the predicted protein (note that volvocine algae possess additional SAND-containing proteins outside the VARL family; see next section and
Figure 6). Although all VARL genes contain the VARL domain, the sequence level conservation outside of the VARL domain is very low. Thus, entire gene sequences cannot be aligned and used for phylogenetic analyses. The VARL domain itself is very short (~86 amino acids) and not highly conserved, such that its utility for inferring evolutionary relationships among the members of the VARL gene family is also limited. Nevertheless, information from gene synteny, sequence signatures outside of the VARL domain, and the location of conserved introns can help draw more robust conclusions regarding the evolution of the VARL family. We summarize the available data below but direct readers to Grochau-Wright et al., (2017) (in particular Supplementary Table 3) for more detailed information.
Based on currently available whole genome sequence data, the VARL gene family contains 12 members in C. reinhardtii (Duncan et al., 2007), 8 in Gonium pectorale (Hanschen et al., 2016) and Tetrabaena socialis (Featherston et al., 2018), 6 in Astrephomene gubernaculifera (Yamashita et al., 2021), and 14 in V. carteri (Duncan et al., 2007). With the exception of regA orthologs (when present), all other regA homologs are known as regA-like sequences: annotated as RLS1-12 in Chlamydomonas and Goniaceae or rlsA-O in Volvocaceae.
To date,
regA orthologs have been found in every member of the Volvocaceae family that was investigated (including species without somatic cells) but appear to be absent in volvocine species outside of the Volvocaceae (Duncan et al., 2007; Featherston et al., 2018; Grochau-Wright et al., 2017, 2021; Hanschen et al., 2014, 2016; Yamashita et al., 2021). In all species with a
regA ortholog for which complete genome sequence is available, the gene is found in a syntenic gene cluster of 4 – 5 paralogs of closely related VARL genes called the
regA cluster (
Figure 2 and
Figure 5). The first gene in the
regA gene cluster is the
rlsA gene which has a unique highly conserved ~40 amino acid protein motif upstream of the VARL domain called “Pandorina’s Box” and a second short conserved motif, called the “PRL” motif after its conserved sequence, downstream of the VARL domain (Grochau-Wright et al., 2017). Downstream of
rlsA is
regA followed by
rlsB. Then, some species (i.e.,
Pandorina morum,
Platydorina caudata, and
Yamagishiella unicocca) have an additional paralog called
rlsO. It appears that
rlsO is found only in species outside of the “Eudorina-group” of the Volvocaceae but further investigation is needed to confirm this hypothesis. These three
regA cluster genes (
regA,
rlsB, and
rlsO) all contain two short conserved regions called the “LALRP” and “FLQ” motifs, found upstream and downstream of the VARL domain, respectively. However, instead of
rlsO,
Volvox ferrisii has a different gene in the same location called
rlsN, which is unique among all VARL genes because it has two VARL domains instead of one (Hanschen et al., 2014). The relationship between
rlsO and
rlsN is not clear, though it seems plausible that
rlsO underwent a domain duplication event to give rise to
rlsN in
V. ferrisii. Finally, the last VARL gene in the
regA cluster is
rlsC, whose orthologs do not appear to share any strongly conserved regions outside the VARL domain (Grochau-Wright et al., 2017).
C. reinhardtii and other volvocine algae outside the Volvocaceae lack orthologs of any of the regA cluster genes. The closest homolog to the regA cluster genes found in these species is RLS1. This gene is an ortholog of the Volvocaceaen rlsD, which is the closest rls paralog of the regA cluster. Duncan et al. (2007) proposed that in the common ancestor of the lineages leading to V. carteri and C. reinhardtii a VARL gene underwent duplication to give rise to two paralogs they referred to as “proto-RLS1/rlsD” and “proto-regA”. Following the separation of the C. reinhardtii and V. carteri lineages they proposed that proto-regA was lost from C. reinhardtii but underwent additional duplication in V. carteri’s lineage to give rise to the regA cluster. Meanwhile, proto-RLS1/rlsD was retained in both lineages and evolved into modern day RLS1 and rlsD. However, the addition of more regA sequences from a variety of species and lack of a proto-regA candidate in Gonium pectorale, Tetrabaena socialis, and Astrephomene gubernaculifera favor a different model in which the ancestral RLS1/rlsD underwent a series of tandem duplications to form the regA gene cluster at the origin of the Volvocaceae (Grochau-Wright et al., 2017; Hanschen et al., 2014).
Finally, several lineage-specific details about the
regA cluster and
rlsD are relevant to further understanding the evolution of this gene family in different volvocine species. First, in all species for which sufficient data is available, the
rlsD gene is found near the
regA gene cluster, except for
V. carteri (
Figure 2). This supports the idea that
RLS1/rlsD duplicated to give rise to the
regA cluster; but, later, in the
V. carteri lineage,
rlsD translocated away from the
regA cluster (Duncan et al., 2007; Grochau-Wright et al., 2017). Second, the
regA cluster is inverted with respect to nearby syntenic markers in
Platydorina caudata although the
regA cluster genes themselves are in the same order relative to each other. Third, the
regA,
rlsB, and
rlsO genes of
Y. unicocca are more similar to each other than they are to their homologs in other species suggesting two of these genes were lost and were later replaced via duplication of the remaining gene (thus, these three genes in
Y. unicocca are not orthologs of the similarly labeled genes in the other species) (Grochau-Wright et al., 2017). It is possible that similar mechanisms of complex evolution have happened within the
regA cluster in other species as well, but phylogenomic analysis supports the orthology of
regA cluster genes within Eudorina-group species (Grochau-Wright et al., 2021).
Synthesizing the information above, we propose the following scenario for the evolution of the
regA gene family (
Figure 2). The VARL gene family comprising several paralogs including
RLS1/rlsD was already present in the common ancestor of all volvocine green algae.
RLS1/rlsD underwent one or more duplication events in the common ancestor of the Volvocaceae family to give rise to a five-gene
regA gene cluster comprised of
rlsA, regA, rlsB, rlsO, and
rlsC. After the lineage leading to
V. ferrisii diverged from the rest of the Volvocaceae, its
rlsO gene gained a second VARL domain, and evolved into
rlsN. Whereas, the common ancestor of the Eudorina group lost
rlsO. In addition,
Y. unicocca lost two internal
regA cluster genes (
regA,
rlsB, or
rlsO) but restored the 5 gene cluster through gene duplication; and the
regA cluster of
P. caudata became inverted relative to nearby syntenic markers (
Figure 2).
SAND domain-containing sequences beyond volvocine algae
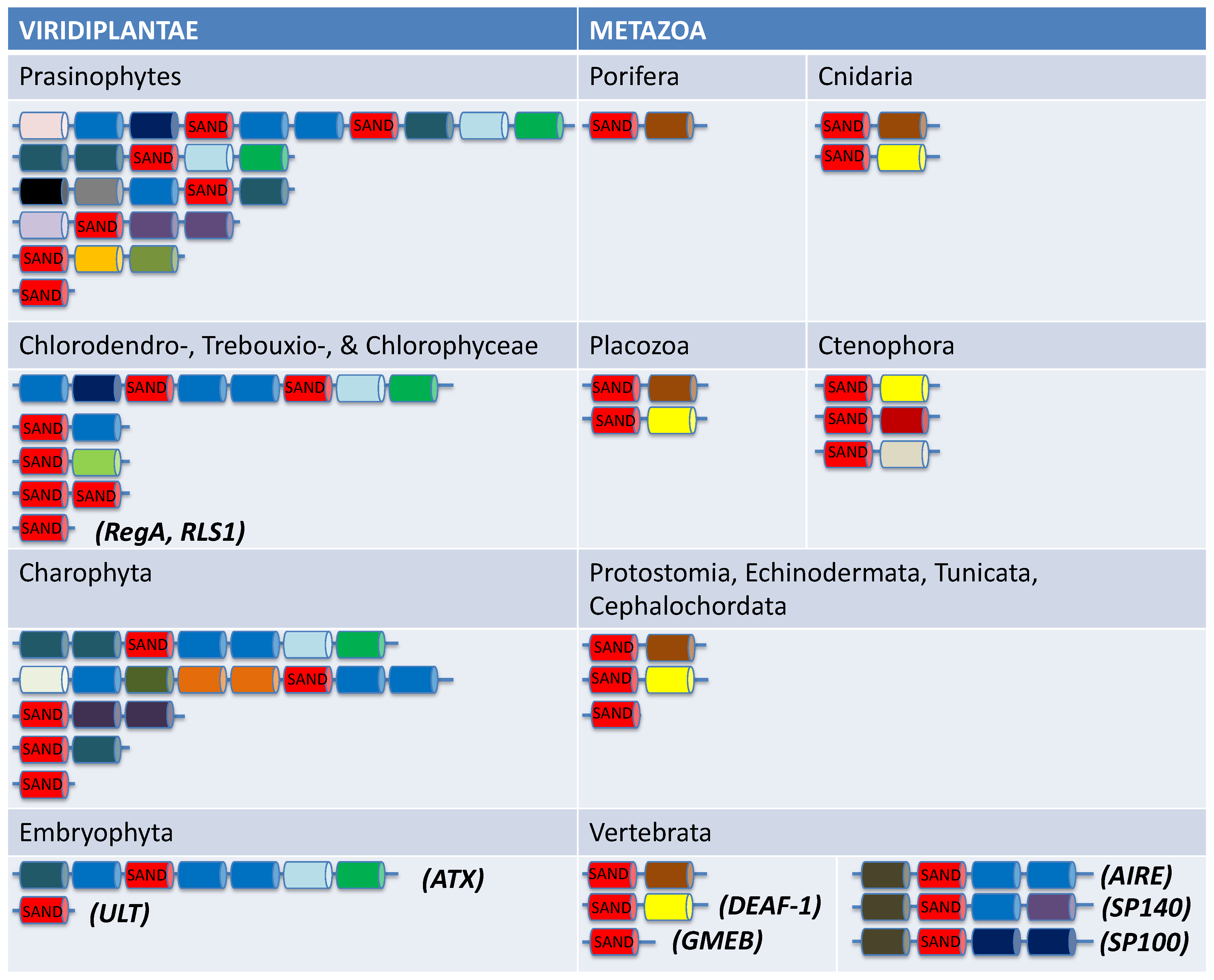
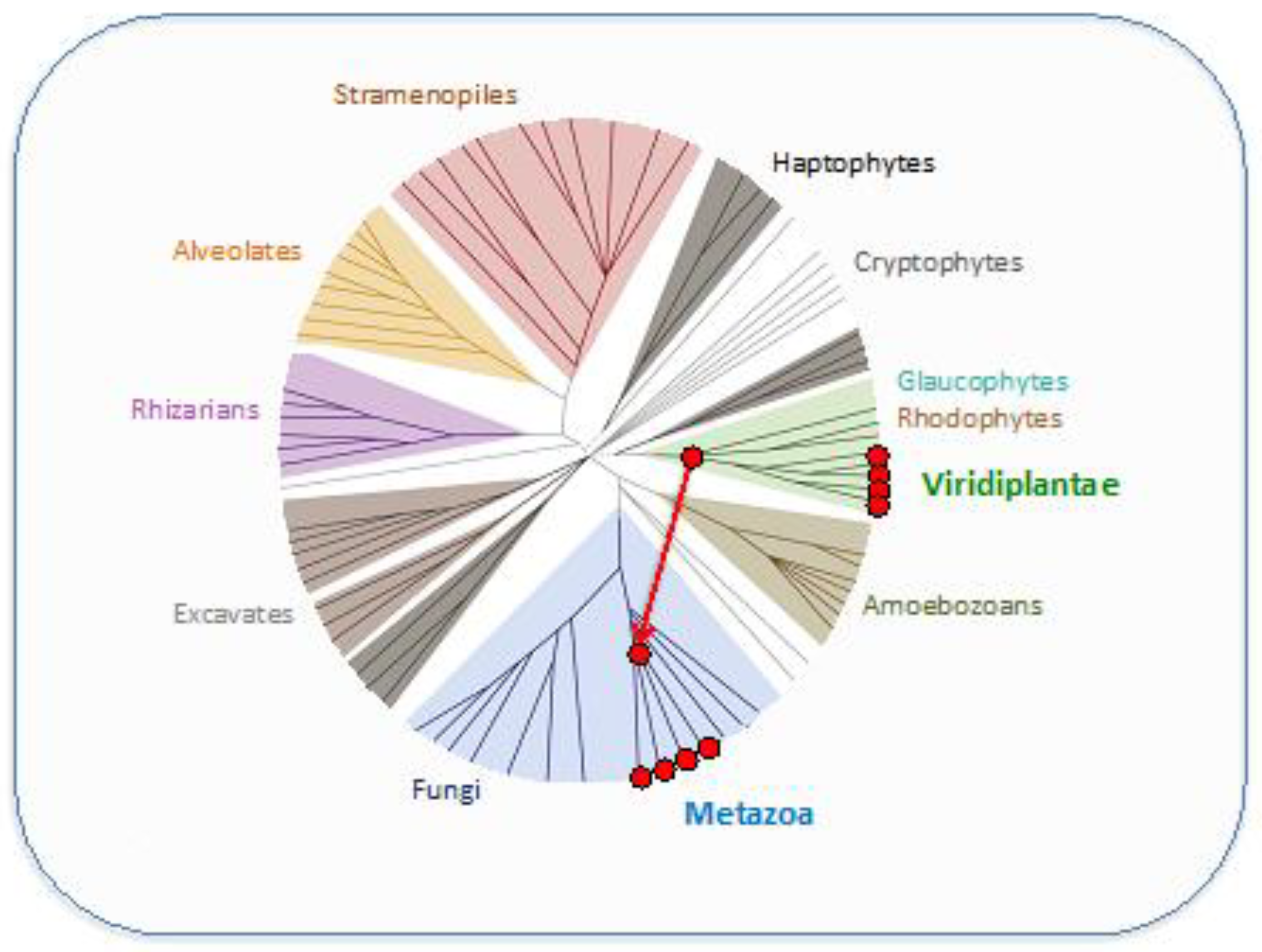
The VARL domain is postulated to be specific to the volvocine VARL gene family but contains a SAND domain similar to that seen in plants and animals. The evolutionary origins of the VARL family and its relationships with other SAND-domain containing proteins is unclear (Nedelcu, 2019). Interestingly, SAND domain-containing proteins appear to be restricted to Viridiplantae (green algae and land plants; aka green plants) and Metazoa. Specifically, SAND-containing sequences were detected in both lineages within Viridiplantae: Streptophyta (land plants and their closest green algal relatives, the Charophytes) and Chlorophyta (green algae in the Chlorophyceae, Trebouxiohyceae, Ulvophyceae, and the Prasinophytes – a paraphyletic group of early-diverged single-celled lineages) (
Figure 6). However, although SAND-containing proteins were identified in all metazoan groups (from early diverged lineages such as sponges, ctenophores and cnidarians, to mammals), SAND sequences could not be detected in their closest relatives (choanoflagellates, filasterians, ichthyosporeans) (Nedelcu, 2019).
Among SAND-containing proteins, the SAND domain is found either alone (as in the
regA gene family) or in various combinations with one or more domain types (
Figure 6). Notably, with the exception of single-SAND domain proteins, no domain architecture is shared between the animal and green plant lineages. Furthermore, the range and distribution of architectures are very different between the two lineages. For instance, a rich toolkit of SAND-containing proteins with various (and complex) architectures is present in green algae (including the volvocine species); but only two architecture types are found in land plants. On the other hand, only one or two architectures are found in early-diverged metazoans (sponges and cnidarians) – but a richer repertoire (six or more types) is present in vertebrates (
Figure 6). Moreover, phylogenetic analyses did not reveal any orthologous relationships between SAND domain-containing sequences from the animal and green plant lineages (Nedelcu, 2019). In addition, this limited distribution of SAND sequences is intriguing. Such a “patchy” distribution (
Figure 7) is considered indicative of lateral gene transfer (LGT) (Keeling & Palmer, 2008), and this was also suggested to be the case for SAND (Nedelcu, 2019). Interestingly, phylogenetic analyses and the presence of a specific sequence motif suggests that animal SAND-containing proteins evolved through an LGT event from a VARL-like sequence (Nedelcu, 2019).
Metazoans, streptophytes, and volvocine algae evolved multicellular development independently ca. 600 MYA, >700 MYA, and 200 MYA, respectively (Herron et al., 2009; Knoll, 2011). The fact that they all employ SAND-containing proteins in important developmental processes (involving the regulation of cell proliferation and differentiation) suggests that SAND-containing proteins have been co-opted and deployed into similar developmental processes in parallel. Thus, it is possible that the independent evolution of complex development in these lineages involved the parallel deployment of ancestral sequences containing the SAND domain. The presence of SAND-containing proteins in single-celled green algae (
Figure 6) indicates that the ancestral role of this domain was in the regulation of gene expression outside a multicellular context. Below, we review the role of such a SAND domain-containing sequence in a single-celled volvocine species and its co-option into regulation of cell proliferation and somatic cell differentiation during the evolution of the volvocine
regA family.
Functional Evolution of regA Through Co-option of a Life History Trade-off Gene
regA has been known to act as a master regulator of somatic cell differentiation in
V. carteri for over two decades (M. M. Kirk et al., 1999). However, it is still not known how
regA is differentially regulated and how exactly it acts to suppress division in cells that fall under the 8 µm threshold size at the end of embryogenesis. Evolutionary approaches can provide alternative or additional means to gain insight into the function of a gene. Based on its role in suppressing reproduction in somatic cells, it has been hypothesized that
regA evolved from a gene that was involved in trading off reproduction for survival (i.e., a life history trade-off gene) in the single-celled ancestors of
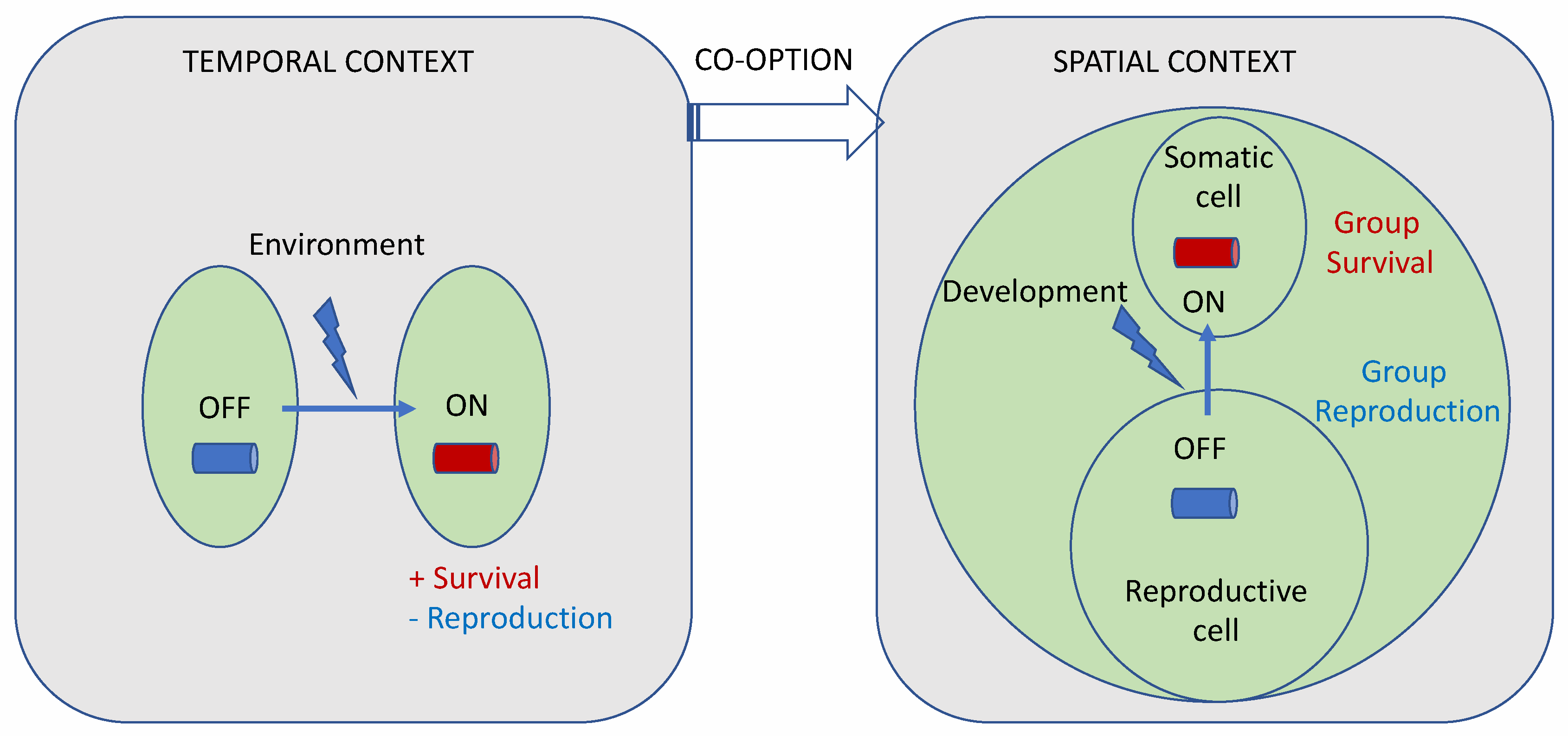
V. carteri. Specifically, such a gene could have been co-opted through changing its expression from a temporal context (in response to an environmental cue) into a spatial context (in response to a developmental cue) (Nedelcu & Michod, 2006) (
Figure 8).
Life history trade-offs in single celled organisms
Although single-cell organisms, by definition, do not possess specialized cell types, they are nevertheless capable of differentiating themselves into various cell states in response to environmental changes. These capabilities include both simply switching between activities enhancing survival, growth, or reproduction during their lifetime, as well as adopting more stable and functionally distinct states (e.g., gametes or spores). The need to switch between survival and reproduction to increase overall fitness reflects one of the main trade-offs that characterize the life-history of all organisms (Bochdanovits & De Jong, 2004; Flatt, 2020; Stearns, 1989). The mechanistic basis of life history trade-offs can be resource availability, structural constraints, cellular processes or genes that affect two sets of activities in opposite ways, or a combination of such factors (Bochdanovits & De Jong, 2004; Flatt, 2020; Flatt & Heyland, 2011; Hood et al., 2018; Hughes & Leips, 2017; Koch et al., 2021; Leroi, 2001; Monaghan et al., 2009; Roff & Fairbairn, 2007; Villellas & García, 2018; Zera & Harshman, 2001). Life history trade-offs can be amplified during certain environmental conditions – such as nutrient limitation, when survival is prioritized over reproduction (Villellas & García, 2018; Zera & Harshman, 2001).
In photosynthetic organisms, including volvocine algae, nutrient limitation induces a series of physiological changes – known as acclimation – characterized by the downregulation of photosynthesis to avoid oxidative damage associated with imbalances between light excitation and available reducing power (Wykoff et al., 1998). In single-celled volvocine algae, such acclimation processes that increase survival also result in a temporary cessation of growth (dependent on photosynthesis) and reproduction (dependent on growth) (Grossman, 2000).
Chlamydomonas RLS1 is a life history trade-off gene induced by environmental cues
As noted earlier, the closest homolog of regA in C. reinhardtii is RLS1 (Duncan et al., 2007; Nedelcu & Michod, 2006). Consistent with the proposed hypothesis that regA evolved from a life-history trade-off gene, initial studies showed that RLS1 expression is upregulated under nutrient (phosphorous or sulfur) and light deprivation, which require suppression of reproduction to increase survival (Nedelcu, 2009; Nedelcu & Michod, 2006). Later, to confirm that RLS1 acts as a bona fide life history trade-off gene, the reproduction and survival of an RLS1 mutant during phosphate deprivation was investigated (Saggere et al., 2022). As expected, the RLS1 mutant was not able to suppress its reproduction when phosphate was limited. In fact, the population size of the mutant exceeded that of the wild-type. However, this short-term immediate reproductive advantage was counteracted by a loss in long-term viability, arguing that RLS1 is a genuine life history trade-off gene (Saggere et al., 2022).
Theoretically, the suppression of reproduction in nutrient-limiting conditions can involve three distinct scenarios (Saggere et al., 2022). First, the suppression of reproduction and increased survival can be a direct response to the re-allocation of nutrients, possibly involving a trade-off between protein biosynthesis (growth) and energy metabolism (survival) (Bochdanovits & De Jong, 2004). Alternatively, reproduction could be suppressed in response to nutrient stress-induced production of reactive oxygen species (ROS); such oxidative stress could trigger a temporary cell cycle arrest (suppression of reproduction) to repair the ROS-induced DNA damage. Lastly, the suppression of reproduction can be induced by redox signals associated with imbalances between excitation energy and reducing power under nutrient deprivation (Pfannschmidt et al., 2009). Such signals trigger the downregulation of photosynthesis to avoid potential photo-oxidative damage, which will increase survival but also limit cell growth and thus reproduction. Available data are consistent with the latter scenario (Saggere et al., 2022). Thus, mechanistically, RLS1 appears to be induced by a redox imbalance/signal, and once expressed, RLS1 could act through the downregulation of photosynthesis. Consistent with this model, the induction of RLS1 coincides with the down-regulation of a light-harvesting chloroplast protein-coding gene, and the experimental inhibition of the photosynthetic electron transport can induce the expression of RLS1 (Nedelcu, 2009).
V. carteri regA retained the ancestral environmental regulation
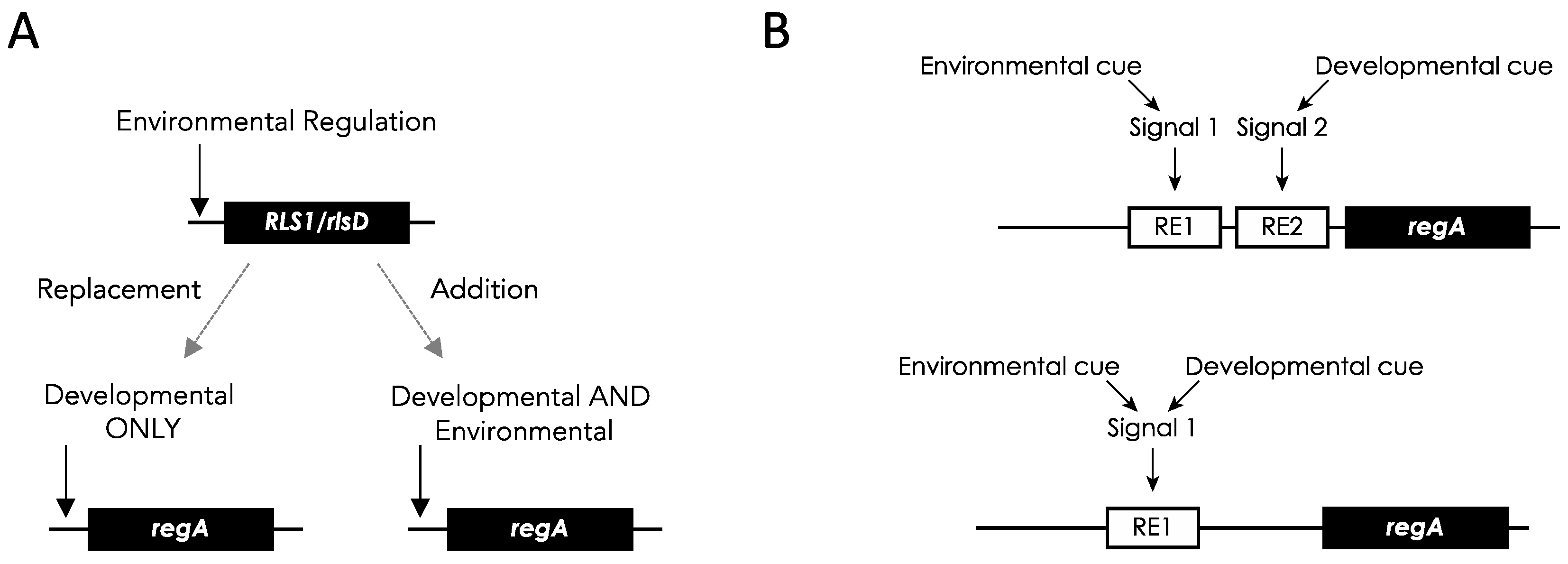
But how did the regulation of
regA in somatic cells evolve from the environmentally induced regulation of its
RLS1-like progenitor? To address this question, König & Nedelcu (2020a) proposed two scenarios (
Figure 9): (i) a new developmental regulation replaced the ancestral environmental regulation of
RLS1/rlsD in the
regA paralog, or (ii) a new developmental regulation was added to the ancestral regulation. The second scenario predicted that in addition to its developmental expression,
regA can also be induced in response to environmental cues. Recently, it was shown that
regA can, indeed, be expressed in both developmental and environmental contexts (scenario ii in
Figure 9A). Specifically,
regA was induced in response to light exposure following a period of extended dark in a
V. carteri mutant that lacks cell differentiation and a functional RegA protein, but still expresses
regA developmentally (König & Nedelcu, 2020). Furthermore, because the duration of both dark and light exposure affected
regA expression levels, it is likely that the environmental induction is triggered by a metabolic imbalance (König & Nedelcu, 2020).
Co-option of an environmentally regulated gene into a developmental master regulator
To address how the new developmental regulation evolved, two models have been proposed: a completely new signaling pathway evolved, or the ancestral signaling pathway was co-opted in a developmental context (König & Nedelcu, 2020). The two models make different predictions as to the regulation of
regA in
V. carteri (
Figure 9B).
The first model predicts that new
cis-regulatory and/or
trans-acting elements were added to the ancestral
RLS1/rlsD gene regulation. Although the developmental regulation of
regA in
V. carteri is known to involve intronic
cis-regulatory elements (Stark et al., 2001), their sequence as well as the
trans-acting factors binding to them are still unknown. Also, nothing is yet known about the regulatory sequences of
RLS1. Notably,
RLS1 does not share
regA’s exon-intron structure and/or similar intronic sequences; thus, there is no direct correspondence between the intronic silencers and enhancers in
regA and potential regulatory elements in
RLS1 (
Figure 3).
Changes in the deployment of trans-acting factors, cis-regulatory elements (de novo, or via modification of pre-existing elements), or a combination of both have been proposed to have taken place during the evolution of morphological innovations in animals (Koshikawa et al., 2015). Similarly, the acquisition of new, distal promoters was invoked in the co-option of ancestral cAMP signaling genes for new developmental roles in the social amoebae, Dictyostelium discoideum (Alvarez-Curto et al., 2005). The presence of a similar mechanism in volvocine algae would argue for a general role for changes in gene regulation during evolution of major morphological innovations. Genes with dual regulation are often seen as intermediate steps during the sub-functionalization process that results in two specialized genes (e.g., (Zhang, 2003)). It will be of interest to know if regA’s paralogs (rlsA, rlsB, rlsO, and rlsC) have retained the ancestral regulation or whether they specialized into specific developmental roles. A preliminary investigation found that the genes rlsB and rlsC are co-expressed with regA during development but the functional significance of this is not yet known (Harryman, 2012). Likewise, a preliminary investigation using RNAi to knockdown rlsA, rlsB and rlsC expression in V. carteri did not result in any discernable vegetative phenotype. The mechanisms underlying these results and their implications are not yet clear and require further investigation (Ortega Escalante, 2018).
The second model predicts that the same intracellular signal that triggers the environmental expression of
RLS1 in
C. reinhardtii is also triggered in
V. carteri’s small cells at the end of embryogenesis.
RLS1 is likely part of the general acclimation response (Nedelcu, 2009). Consequently,
RLS1 might be induced by an energetic imbalance mediated by a redox signal (e.g., NADPH/NADP+, reactive oxygen species – ROS) (e.g., (Strand et al., 2017)). A similar signal could be responsible for the observed induction of
regA in
V. carteri cells exposed to light after long periods of dark (König & Nedelcu, 2020). If the same signal is also produced in the small cells at the end of embryogenesis (induced by a different imbalance caused by small cell size),
regA can be expressed in a developmental context using the same environmental regulatory elements. The postulated developmental signal can be triggered by an imbalance between membrane-bound proteins (e.g., electron transport carriers, ion transporters) and soluble factors (e.g., NADP+) as the surface to volume ratio in these small cells is in favor of membrane proteins (see
Figure 2 in (Nedelcu, 2009)). Interestingly, the environmental induction of
regA is also affected by cell size (König & Nedelcu, 2020). If this model is correct, deciphering the exact signal involved in the environmental induction of
regA could provide insights into elucidating the long-standing question as to how small cell size determines somatic cell fate in
V. carteri.
Notably, the conditions that induced regA in regA mutant cells lacking a functional RegA protein ultimately resulted in the activation of programmed cell death (PCD), while wild-type somatic cells (harboring a functional RegA protein) show little response in terms of environmental regA induction or loss in viability (König & Nedelcu, 2020). Also, wild-type somatic cells are unaffected by heat-stress while regA mutant cells as well as wild-type gonidia (which do not express regA) undergo PCD (Cameron-Pack et al., 2022; Nedelcu, 2006). Altogether, these findings suggest that in addition to its role in suppressing the reproduction of somatic cells, the presence of a functional RegA protein in somatic cells confers (directly or indirectly) resistance to environmental stress.
But if somatic cells are already protected (see (König & Nedelcu, 2020) for discussion) by the presence of the developmentally expressed RegA protein, why is regA’s environmental regulation in V. carteri still maintained? It has been proposed that, similar to RLS1 in C. reinhardtii, regA plays a direct role in the response to stress in gonidia (König & Nedelcu, 2020). For instance, under nutrient deprivation, gonidia stop growing and undergo a temporary cessation of reproduction. At the cell level, the inhibition of gonidia growth and reproduction can be an acclimation response that prevents the accumulation of ROS-inducing damage and thus ensures survival. At the multicellular level, this is an adaptive response that is costly in the terms of immediate reproduction but beneficial in terms of offspring quality since it avoids ROS-induced DNA damage and mutations in the gonidia. Nevertheless, when damage is extensive (such as during heat stress), PCD is the best adaptive response as it eliminates potentially damaged gonidia and/or prevents the transmission of deleterious mutations to offspring (Nedelcu, 2006).
Preliminary investigations into the effects of knocking down or overexpressing rlsD during V. carteri development are also compelling (Jimenez Marin, 2023; Ortega Escalante, 2018). Knocking down rlsD expression appears to result in colonies with reduced size or with germ-cells that divide but fail to complete development. In contrast, overexpression of rlsD results in the development of “somagonidia” where germ cells do not grow to their normally large size but are still larger than soma, and exhibit soma-like characteristics such as the presence of eyespots and flagella (Ortega Escalante, 2018). The gene expression profiles of wild-type V. carteri compared to those with rlsD overexpression show down-regulation of ribosome and photosynthesis-related genes (Jimenez Marin, 2023). Together, these results suggest that rlsD plays an important complementary role to regA in regulating cell growth, supporting the idea that the ancestral functions of RLS1/rlsD were sub-functionalized and/or neo-functionalized as regA evolved following its duplication from RLS1/rlsD (Jimenez Marin, 2023).
The general role of stress and life history trade-off genes in the re-organization of fitness during the evolution of multicellularity
Stress responses take on special significance during evolutionary transitions in individuality more generally. As discussed in the Introduction, during an evolutionary transition, in which individuals form groups that evolve into a new kind of individual, fitness must be reorganized so that it becomes a property of the group. The fitness opportunities of the previous individual must be significantly reduced or eliminated. The readjustment of fitness components during stressful periods provides a useful substrate for the reorganization of fitness (König & Nedelcu, 2020; Michod & Nedelcu, 2003; Nedelcu, 2009; Nedelcu & Michod, 2006).
The finding that regA is a master developmental regulator that (i) evolved from a stress-induced gene, (ii) still manifests its ancestral environmental regulation and (iii) confers stress protection offers a direct link between stress responses and the early evolution of somatic cell differentiation. In stressful environments, fitness is compromised, and fitness components must be adjusted to meet the challenge if the individual is to survive. In particular, survival will often be prioritized over reproduction in the short term. We suggest that the ability to re-organize fitness components under stress can be co-opted during the evolution of multicellularity and can contribute to the evolutionary potential and stability of multicellular lineages.
In V. carteri, the co-option of a life history trade-off gene that can suppress reproduction at the cell level increased the survival of the multicellular individual, since non-dividing somatic cells maintain flagellar motility throughout the life cycle. However, as regA evolved from a life history trade-off gene expressed under stress, the permanent suppression of reproduction also provides somatic cells with survival cell-level benefits in terms of enhanced resistance to environmental stress. Indeed, the lack of a functional RegA protein (such as in regA mutants) makes cells more sensitive to environmental stress (Cameron-Pack et al., 2022). This increased sensitivity to stress can also contribute to the stability of the multicellular individual as such regA mutants that regain reproductive capabilities (and negatively affect the fitness of the multicellular individual) will incur a cost in survival (Cameron-Pack et al., 2022). On the other hand, the developmental repression of regA expression in gonidia is required to allow the reproduction of the individual. However, since regA has maintained its ancestral environmental regulation, gonidia can adaptively respond to environmental stress by inducing either a temporary cell cycle arrest (to prevent or repair damage) or PCD (to prevent the transmission of deleterious mutations to offspring).
As life history trade-offs are common in single-celled organisms (e.g., (Ferenci, 2016; Lang et al., 2009; Wenger et al., 2011; Wolf et al., 2015)), it has been suggested that similar co-option of life history trade-off genes with antagonistic effects on survival and reproduction have taken place during the evolution of multicellularity in other lineages (Cameron-Pack et al., 2022). Notably, the de-differentiation and increased proliferation of cancer cells was also shown to result in increased sensitivity to nutrient stress due to their failure to trade-off cell proliferation for maintenance in stressful environments (Lee et al., 2012; Raffaghello et al., 2008). This trade-off, likely inherited from the unicellular ancestors of animals, might have contributed to the stability of the multicellular individuals during the early animal evolution and might be involved in the purging of most pre-cancerous cells (Cameron-Pack et al., 2022). Recently, the evolution of several developmental processes – from aggregative multicellularity in D. discoideum to the differentiation of decidual stromal cells in placental mammals, has been linked to pre-existing ancestral stress-responses (Nedelcu & Michod, 2020; Schaap, 2016; Wagner et al., 2019).
Future directions
Despite the unprecedented suitability of the volvocine algae and the regA family to understand the mechanistic and genetic basis for the transition to a new higher-level individual through the reorganization of fitness components, there are many unknowns yet to be addressed. For instance, while the role regA plays in cellular differentiation is clear in V. carteri f. nagariensis, the specific details of how it carries out this function are not well understood. Expression of regA is regulated by cell size at the end of cleavage in V. carteri (Kirk et al., 1993), but how cell size induces the expression of regA is unknown. Similarly, how regA regulates its target genes and what those target genes are is unresolved.
The function of regA is even less clear in other volvocine algae species. Cellular differentiation arose three to five times in the Volvocaceae (Grochau-Wright et al., 2017; Herron & Michod, 2008; Lindsey et al., 2021) leaving open the question of whether regA was co-opted to control differentiation multiple times or if different genes control differentiation in other volvocine species with somatic cells. Intriguingly, regA is found in species lacking somatic cells, but its function in undifferentiated species is not known (Grochau-Wright et al., 2017). Some volvocine algae thought to lack differentiation, like Eudorina, can develop somatic-like cells under environmental stress (Davison & Michod, 2021). Whether or not regA is involved in this phenomenon is currently unknown as well. A better understanding of how RLS1 acts and is regulated in Chlamydomonas and volvocine species that lack cellular differentiation will help fill in the gaps of how regA was co-opted to control somatic cell differentiation as well. For instance, the cis-regulatory elements of RLS1 are undescribed. Identifying these regulatory elements and comparing them to the cis-regulatory elements of V. carteri regA would illuminate how the environmentally induced regulation of RLS1 was co-opted during the evolution of somatic cell differentiation.
The function of the regA cluster genes other than regA is also a mystery. Preliminary studies have shown that at least rlsB and rlsC have a similar expression pattern as regA during V. carteri development (Harryman, 2012) which suggests that the regA cluster as a whole may be involved in development and differentiation, but more work needs to be done to fully assess this idea (Ortega Escalante, 2018). Interestingly, Z. I. Grochau-Wright et al. (2021) found that a morphological mutant of V. powersii that has fewer cells and a lower soma to germ ratio than the WT strain has a mutation in its rlsB gene. But transformation of the WT-rlsB gene into this mutant did not lead to morphological rescue. Thus, the cause of the mutant’s altered phenotype and functional significance of the rlsB mutation (if any) remains unclear.
Very little is known about the structure and evolution of VARL genes outside of the reg cluster due to less attention and less effort put into cloning and sequencing these genes. However whole genome sequence data is available for several species that requires further analysis and annotation to determine the total number of VARL genes present, specifically Eudorina elegans, Yamagishiella unicocca, Volvox reticuliferus, and Volvox africanus (Hamaji et al., 2018; Yamamoto et al., 2021). In addition, Lindsey et al. (2021) generated a large transcriptomic dataset for 47 volvocine algae species that could potentially be mined for VARL gene family members. Searching this already available data for VARL genes could expand our knowledge of the structure and evolution of VARL genes outside of the reg sub-family substantially.
Also of interest is understanding how species in the genus
Astrephomene control their cellular differentiation.
Astrephomene species are the only volvocine algae outside of the Volvocaceae that have cellular differentiation, but unlike
Volvox and
Pleodorina species the somatic cells of
Astrephomene are located in the posterior of the colony due to the lack of an inversion process during development (Yamashita et al., 2016). In addition,
Astrephomene does not possess the
regA gene and evolved somatic cells independently of other volvocine algae (Grochau-Wright et al., 2017; Lindsey et al., 2021; Yamashita et al., 2021)(
Figure 2).
A key lesson from studying RLS1 and regA in the volvocine algae is that life-history genes may be co-opted for cell differentiation of soma, in particular, environmentally controlled growth suppression genes can be co-opted to provide the genetic basis for developmental control of cell differentiation. The need for single-celled organisms to respond to times of stress by decreasing their growth to support their survival provides the foundation for the evolution of somatic cells in a multicellular organism (Nedelcu, 2009; Nedelcu & Michod, 2006; Olson & Nedelcu, 2016). RegA is the best-known example of such a life history gene, however, as regA is specific to the volvocine algae clade, it would be useful to know if genes with similar life-history functions were co-opted to produce soma in other clades that made the transition to multicellularity. Similarly, the evolution of specialized reproductive and worker castes during the transition to eusociality is analogous to the evolution of germ and soma during the transition to multicellularity. Thus, it may be fruitful to investigate the co-option of stress response life history genes during the transition to eusociality as well.
Summary
The evolution of cellular differentiation is a key event during the transition from single-celled to multicellular life. Specialized germ cells and somatic cells reorganize the two essential components of fitness between different cell types, thereby transferring fitness from the cell-level to the multicellular level. Understanding the genetic basis for the evolution of cellular differentiation during the unicellular-to-multicellular transitions is a major challenge of evolutionary biology. Co-option of life history trade-off genes present in unicellular organisms that differentially affect survival and reproductive functions in response to the environment is one route for the evolution of genes controlling cellular differentiation. The regA-like gene family of the volvocine green algae is an unrivaled model system to study this co-option due to the recent origin of multicellularity in this clade and presence of extant relatives at different levels of multicellular complexity.
The
regA gene from
V. carteri f.
nagariensis is the type-gene for the VARL gene family specific to the volvocine algae lineage. The common ancestor of
V. carteri and
C. reinhardtii likely had several VARL gene family members, one of which was
RLS1. The
RLS1 gene duplicated several times to give rise to the
regA gene cluster in the common ancestor of the Volvocaceae, setting the stage for the functional co-option of
regA during the evolution of cellular differentiation as well as other lineage-specific changes to
regA cluster genes (
Figure 2). The co-option of
RLS1’s functions into a
regA-like gene responsible for somatic cell differentiation likely involved the simulation of the ancestral environmentally induced signal into a developmental context (
Figure 9).
The defining feature of all VARL genes is the presence of the VARL domain which contains a conserved SAND domain. The SAND domain is found in other green algae, land plants, and animals but appears to be missing from other eukaryotic lineages. This indicates the possibility that the SAND domain was horizontally transferred between green algae and animals early in eukaryotic evolution where it appears to have been co-opted multiple times independently in a variety of developmentally important functions.
Overall, we argue that the co-option of life history trade-off genes during the transition to multicellularity underlies the re-organization of fitness between soma and germ to optimize various aspects of fitness at the multicellular level and contribute to the stability of the multicellular individual. Although all organisms must have such stress response trade-off genes, it remains to be determined whether or how fitness reorganization and co-option of life history genes during the evolution of specialized cell types applies to other lineages that evolved multicellularity independently.
Author Contributions
Conceptualization, AN; Writing – original draft, ZGW, AN, RM; Writing – review & editing, ZGW, AN, RM. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Sciences and Engineering Research Council of Canada; and the National Science Foundation, grant NSF DEB 2029999.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Alvarez-Curto, E., Rozen, D. E., Ritchie, A. V., Fouquet, C., Baldauf, S. L., & Schaap, P. (2005). Evolutionary origin of cAMP-based chemoattraction in the social amoebae. Proceedings of the National Academy of Sciences of the United States of America, 102(18), 6385–6390. [CrossRef]
- Arakaki, Y., Kawai-Toyooka, H., Hamamura, Y., Higashiyama, T., Noga, A., Hirono, M., Olson, B. J. S. C., & Nozaki, H. (2013). The simplest integrated multicellular organism unveiled. PloS One, 8(12), e81641. [CrossRef]
- Babinger, K., Hallmann, A., & Schmitt, R. (2006). Translational control of regA, a key gene controlling cell differentiation in Volvox carteri. Development (Cambridge, England), 133, 4045–4051. [CrossRef]
- Baran, G. (1984). Analysis of somatic cell differentiation in Volvox carteri f. nagariensis. University of Virginia.
- Barker, H. E., Smyth, G. K., Wettenhall, J., Ward, T. A., Bath, M. L., Lindeman, G. J., & Visvader, J. E. (2008). Deaf-1 regulates epithelial cell proliferation and side-branching in the mammary gland. BMC Developmental Biology, 8(1), 94. [CrossRef]
- Bochdanovits, Z., & De Jong, G. (2004). Antagonistic pleiotropy for life-history traits at the gene expression level. Proceedings of the Royal Society B: Biological Sciences, 271(SUPPL. 3), S75–S78. [CrossRef]
- Bonner, J. T. (1998). The origins of multicellularity. Integrative Biology: Issues, News, and Reviews, 1(1), 27–36. [CrossRef]
- Boraas, M. E., Seale, D. B., & Boxhorn, J. E. (1998). Phagotrophy by flagellate selects for colonial prey: A possible origin of multicellularity. Evolutionary Ecology, 12(1973), 153–164. [CrossRef]
- Bottomley, M. J. J., Collard, M. W. W., Huggenvik, J. I. I., Liu, Z., Gibson, T. J. J., & Sattler, M. (2001). The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nature Structural Biology, 8(7), 626–633. [CrossRef]
- Brunet, T., & King, N. (2017). The Origin of Animal Multicellularity and Cell Differentiation. Developmental Cell, 43(2), 124–140. [CrossRef]
- Buss, LeoW. (1987). The Evolution of Individuality. Princeton University Press.
- Cameron-Pack, M. E., König, S. G., Reyes-Guevara, A., Reyes-Prieto, A., & Nedelcu, A. M. (2022). A personal cost of cheating can stabilize reproductive altruism during the early evolution of clonal multicellularity. Biology Letters, 18(6), 20220059. [CrossRef]
- Carles, C. C. (2005). ULTRAPETALA1 encodes a SAND domain putative transcriptional regulator that controls shoot and floral meristem activity in Arabidopsis. Development, 132(5), 897–911. [CrossRef]
- Carles, C. C., & Fletcher, J. C. (2009). The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes and Development, 23(23), 2723–2728. [CrossRef]
- Chen, L.-Q., Luo, J.-H., Cui, Z.-H., Xue, M., Wang, L., Zhang, X.-Y., Pawlowski, W. P., & He, Y. (2017). ATX3, ATX4, and ATX5 Encode Putative H3K4 Methyltransferases and Are Critical for Plant Development. Plant Physiology, 174(3), 1795–1806. [CrossRef]
- Choi, G., Przybylska, M., & Straus, D. (1996). Three abundant germ line-specific transcripts in Volvox carteri encode photosynthetic proteins. Current Genetics, 30(4), 347–355. http://www.ncbi.nlm.nih.gov/pubmed/8781179.
- Coleman, A. W. (2012). A Comparative Analysis of the Volvocaceae (Chlorophyta). Journal of Phycology, 48(3), 491–513. [CrossRef]
- Davison, D. R., & Michod, R. E. (2021). Phenotypic Plasticity and Evolutionary Transitions in Individuality. In D. W. Pfennig (Ed.), Phenotypic Plasticity & Evolution: Causes, Consequences, Controversies (pp. 241–266). CRC Press.
- Duncan, L., Nishii, I., Harryman, A., Buckley, S., Howard, A., Friedman, N. R., & Miller, S. M. (2007). The VARL gene family and the evolutionary origins of the master cell-type regulatory gene, regA, in Volvox carteri. Journal of Molecular Evolution, 65(1), 1–11. [CrossRef]
- Duncan, L., Nishii, I., Howard, A., Kirk, D., & Miller, S. M. (2006). Orthologs and paralogs of regA, a master cell-type regulatory gene in Volvox carteri. Current Genetics, 50(1), 61–72. [CrossRef]
- Featherston, J., Arakaki, Y., Hanschen, E. R., Ferris, P. J., Michod, R. E., Olson, B. J. S. C., Nozaki, H., & Durand, P. M. (2018). The 4-celled tetrabaena socialis nuclear genome reveals the essential components for genetic control of cell number at the origin of multicellularity in the volvocine lineage. Molecular Biology and Evolution, 35(4), 855–870. [CrossRef]
- Ferenci, T. (2016). Trade-off Mechanisms Shaping the Diversity of Bacteria. Trends in Microbiology, 24(3), 209–223. [CrossRef]
- Flatt, T. (2020). Life-history evolution and the genetics of fitness components in drosophila melanogaster. Genetics, 214(1), 3–48. [CrossRef]
- Flatt, T., & Heyland, Andreas. (2011). Mechanisms of Life History Evolution: the Genetics and Physiology of Life History Traits and Trade-Offs. Oxford University Press.
- Fletcher, J. C. (2001). The ULTRAPETALA gene controls shoot and floral meristem size in Arabidopsis. Development (Cambridge, England), 128(8), 1323–1333.
- Grochau-Wright, Z. I. (2019). THE ORIGIN AND EVOLUTION OF THE REG CLUSTER IN THE VOLVOCINE GREEN ALGAE: A MODEL SYSTEM FOR THE EVOLUTION OF CELLULAR DIFFERENTIATION [PhD Dissertation]. University of Arizona.
- Grochau-Wright, Z. I., Ferris, P. J., Tumberger, J., Jiménez-Marin, B., Olson, B. J. S. C., & Michod, R. E. (2021). Characterization and Transformation of reg Cluster Genes in Volvox powersii Enable Investigation of Convergent Evolution of Cellular Differentiation in Volvox. Protist, 172, 125834. [CrossRef]
- Grochau-Wright, Z. I., Hanschen, E. R., Ferris, P. J., Hamaji, T., Nozaki, H., Olson, B. J. S. C., & Michod, R. E. (2017). Genetic Basis for Soma is Present in Undifferentiated Volvocine Green Algae. Journal of Evolutionary Biology, 30, 1205–1218.
- Grosberg, R. K., & Strathmann, R. R. (2007). The Evolution of Multicellularity: A Minor Major Transition? Annual Review of Ecology, Evolution, and Systematics, 38(1), 621–654. [CrossRef]
- Grossman, A. (2000). Acclimation of Chlamydomonas reinhardtii to its nutrient environment. In Protist (Vol. 151, Issue 3, pp. 201–224). Elsevier GmbH. [CrossRef]
- Hamaji, T., Kawai-Toyooka, H., Uchimura, H., Suzuki, M., Noguchi, H., Minakuchi, Y., Toyoda, A., Fujiyama, A., Miyagishima, S., Umen, J. G., & Nozaki, H. (2018). Anisogamy evolved with a reduced sex-determining region in volvocine green algae. Communications Biology, 1(1), 1–7. [CrossRef]
- Hanschen, E. R., Davison, D. R., Grochau-Wright, Z. I., & Michod, R. E. (2017). Evolution of individuality: a case study in the volvocine green algae. Philosophy, Theory, and Practice in Biology, 9(January), 3. [CrossRef]
- Hanschen, E. R., Ferris, P. J., & Michod, R. E. (2014). Early Evolution of the Genetic Basis for Soma in the Volvocaceae. Evolution, 68(7), 2014–2025.
- Hanschen, E. R., Marriage, T. N., Ferris, P. J., Hamaji, T., Toyoda, A., Fujiyama, A., Neme, R., Noguchi, H., Minakuchi, Y., Suzuki, M., Kawai-Toyooka, H., Smith, D. R., Luria, V., Karger, A., Kirschner, M. W., Sparks, H., Anderson, J., Bakaric, R., Durand, P. M., … Olson, B. J. S. C. (2016). The Gonium pectorale genome demonstrates co-option of cell cycle regulation during the evolution of multicellularity. Nature Communications, 7, 11370. [CrossRef]
- Harryman, A. (2012). Investigating the roles of regA and related genes in the evolution of multicellularity in the volvocine green algae [Doctor of Philosophy]. University of Maryland, Baltimore County.
- Herron, M. D., Borin, J. M., Boswell, J. C., Walker, J., Chen, I. K., Knox, C. A., Boyd, M., Rosenzweig, F., & Ratcliff, W. C. (2019). De novo origins of multicellularity in response to predation. Scientific Reports, 1–9. [CrossRef]
- Herron, M. D., Conlin, P. L., & Ratcliff, W. C. (2022). The Evolution of Multicellularity. CRC Press. [CrossRef]
- Herron, M. D., Hackett, J. D., Aylward, F. O., & Michod, R. E. (2009). Triassic origin and early radiation of multicellular volvocine algae. Proceedings of the National Academy of Sciences of the United States of America, 106(9), 3254–3258. [CrossRef]
- Herron, M. D., & Michod, R. E. (2008). Evolution of complexity in the volvocine algae: transitions in individuality through Darwin’s eye. Evolution; International Journal of Organic Evolution, 62(2), 436–451. [CrossRef]
- Hood, W. R., Zhang, Y., Mowry, A. V, Hyatt, H. W., & Kavazis, A. N. (2018). Life History Trade-offs within the Context of Mitochondrial Hormesis. Integrative and Comparative Biology, 58(3), 567–577. [CrossRef]
- Hughes, K. A., & Leips, J. (2017). Pleiotropy, constraint, and modularity in the evolution of life histories: insights from genomic analyses. Annals of the New York Academy of Sciences, 1389(1), 76–91. [CrossRef]
- Huskey, R. J., & Griffin, B. E. (1979). Genetic Control of Somatic Cell Differentiation in Volvox. Developmental Biology, 72, 226–235.
- Huskey, R. J., Griffin, B. E., Cecil, P. O., & Callahan, A. M. (1979). A Preliminary Genetic Investigation of Volvox carteri. Genetics, 91(2), 229–244. [CrossRef]
- Jimenez Marin, L. B. (2023). Gene loss, co-option and the evolution of developmental complexity in the volvocine algae [PhD Dissertation]. Kansas State University.
- Keeling, P. J., & Burki, F. (2019). Progress towards the Tree of Eukaryotes. Current Biology, 29(16), R808–R817. [CrossRef]
- Keeling, P. J., & Palmer, J. D. (2008). Horizontal gene transfer in eukaryotic evolution. Nature Reviews Genetics, 9(8), 605–618. [CrossRef]
- Kirk, D. L. (1998). Volvox: Molecular-Genetic Origins of Multicellularity and Cellular Differentiation. Cambridge University Press.
- Kirk, D. L. (2001). Germ-soma differentiation in Volvox. Developmental Biology, 238(2), 213–223. [CrossRef]
- Kirk, D. L. (2005). A twelve-step program for evolving multicellularity and a division of labor. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 27(3), 299–310. [CrossRef]
- Kirk, M. M., Ransick, A., McRae, S. E., & Kirk, D. L. (1993). The Relationship between Cell Size and Cell Fate in Volvox carteri. The Journal of Cell Biology, 123(1), 191–208.
- Kirk, M. M., Stark, K., Miller, S. M., Müller, W., Taillon, B. E., Gruber, H., Schmitt, R., & Kirk, D. L. (1999). regA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development, 126(4), 639–647. http://www.ncbi.nlm.nih.gov/pubmed/9895312.
- Klein, B., Wibberg, D., & Hallmann, A. (2017). Whole transcriptome RNA-Seq analysis reveals extensive cell type-specific compartmentalization in Volvox carteri. BMC Biology, 15(1), 1–22. [CrossRef]
- Knoll, A. H. (2011). The Multiple Origins of Complex Multicellularity. Http://Dx.Doi.Org/10.1146/Annurev.Earth.031208.100209, 39, 217–239. [CrossRef]
- Koch, R. E., Buchanan, K. L., Casagrande, S., Crino, O., Dowling, D. K., Hill, G. E., Hood, W. R., McKenzie, M., Mariette, M. M., Noble, D. W. A., Pavlova, A., Seebacher, F., Sunnucks, P., Udino, E., White, C. R., Salin, K., & Stier, A. (2021). Integrating Mitochondrial Aerobic Metabolism into Ecology and Evolution. Trends in Ecology and Evolution, 36(4), 321–332. [CrossRef]
- König, S. G., & Nedelcu, A. M. (2020). The genetic basis for the evolution of soma: Mechanistic evidence for the co-option of a stress-induced gene into a developmental master regulator. Proceedings of the Royal Society B: Biological Sciences, 287(1940). [CrossRef]
- Koshikawa, S., Giorgianni, M. W., Vaccaro, K., Kassner, V. A., Yoder, J. H., Werner, T., & Carroll, S. B. (2015). Gain of cis-regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proceedings of the National Academy of Sciences of the United States of America, 112(24), 7524–7529. [CrossRef]
- Koufopanou, V. (1994). The Evolution of Soma in the Volvocales. The American Naturalist, 143(5), 907–931.
- Kulkarni, M., Shakes, D. C., Guevel, K., & Smith, H. E. (2012). SPE-44 Implements Sperm Cell Fate. PLoS Genetics, 8(4), e1002678. [CrossRef]
- Lang, G. I., Murray, A. W., & Botstein, D. (2009). The cost of gene expression underlies a fitness trade-off in yeast. Proceedings of the National Academy of Sciences, 106(14), 5755–5760. [CrossRef]
- Lee, C., Raffaghello, L., Brandhorst, S., Safdie, F. M., Bianchi, G., Martin-Montalvo, A., Pistoia, V., Wei, M., Hwang, S., Merlino, A., Emionite, L., de Cabo, R., & Longo, V. D. (2012). Fasting Cycles Retard Growth of Tumors and Sensitize a Range of Cancer Cell Types to Chemotherapy. Science Translational Medicine, 4(124), 124ra27-124ra27. [CrossRef]
- Leroi, A. M. (2001). Molecular signals versus the Loi de Balancement. In Trends in Ecology and Evolution (Vol. 16, Issue 1, pp. 24–29). Elsevier Ltd. [CrossRef]
- Lindsey, C. R., Rosenzweig, F., & Herron, M. D. (2021). Phylotranscriptomics points to multiple independent origins of multicellularity and cellular differentiation in the volvocine algae. BMC Biology, 19(1). [CrossRef]
- Matt, G. Y., & Umen, J. G. (2018). Cell-Type Transcriptomes of the Multicellular Green Alga Volvox carteri Yield Insights into the Evolutionary Origins of Germ and Somatic Differentiation Programs. G3: Genes|Genomes|Genetics, 8, 531–550. [CrossRef]
- Maynard Smith, J., & Szathmáry, E. (1995). The Major Transitions in Evolution. Oxford University Press.
- Meissner, M., Stark, K., Cresnar, B., Kirk, D. L., & Schmitt, R. (1999). Volvox germline-specific genes that are putative targets of RegA repression encode chloroplast proteins. Current Genetics, 36(6), 363–370. http://www.ncbi.nlm.nih.gov/pubmed/10654090.
- Michod, R. E. (1999). Darwinian Dynamics. Princeton University Press.
- Michod, R. E. (2006a). The group covariance effect and fitness trade-offs during evolutionary transitions in individuality. PNAS, 103(24), 9113–9117.
- Michod, R. E. (2006b). On the transfer of fitness from the cell to the multicellular organism. Biology & Philosophy, 20(5), 967–987. [CrossRef]
- Michod, R. E., & Nedelcu, A. M. (2003). On the reorganization of fitness during evolutionary transitions in individuality. Integrative and Comparative Biology, 43(1), 64–73. [CrossRef]
- Monaghan, P., Metcalfe, N. B., & Torres, R. (2009). Oxidative stress as a mediator of life history trade-offs: Mechanisms, measurements and interpretation. In Ecology Letters (Vol. 12, Issue 1, pp. 75–92). Ecol Lett. [CrossRef]
- Nakagawa, T. T. K. U. T. N. Y. (2008). GMEB1, a novel endogenous caspase inhibitor, prevents hypoxia- and oxidative stress-induced neuronal apoptosis. Neuroscience Letters, 438, 34–37. [CrossRef]
- Nedelcu, A. M. (2006). Evidence for p53-like-mediated stress responses in green algae. FEBS Letters, 580(13), 3013–3017. [CrossRef]
- Nedelcu, A. M. (2009). Environmentally induced responses co-opted for reproductive altruism. Biology Letters, 5(6), 805–808. [CrossRef]
- Nedelcu, A. M. (2019). Independent evolution of complex development in animals and plants: deep homology and lateral gene transfer. Development Genes and Evolution, 25–34. [CrossRef]
- Nedelcu, A. M., & Michod, R. E. (2003). Evolvability, modularity, and individuality during the transition to multicellularity in volvocalean green algae. In G. Schlosser & G. P. Wagner (Eds.), Modularity in development and evolution. University of Chicago Press.
- Nedelcu, A. M., & Michod, R. E. (2006). The evolutionary origin of an altruistic gene. Molecular Biology and Evolution, 23(8), 1460–1464. [CrossRef]
- Nedelcu, A. M., & Michod, R. E. (2020). Stress Responses Co-Opted for Specialized Cell Types During the Early Evolution of Multicellularity. BioEssays, 42(5), 2000029. [CrossRef]
- Nozaki, H., Itoh, M., Watanabe, M. M., & Kuroiwa, T. (1996). Ultrastructure of the vegetative colonies and systematic position of basichlamys (Volvocales, Chlorophyta). European Journal of Phycology, 31(1), 67–72. [CrossRef]
- Nozaki, H., Mahakham, W., Athibai, S., Yamamoto, K., Takusagawa, M., Misumi, O., Herron, M. D., Rosenzweig, F., & Kawachi, M. (2017). Rediscovery of the species of ‘ancestral Volvox’’: morphology and phylogenetic position of Pleodorina sphaerica (Volvocales, Chlorophyceae) from Thailand.’ Phycologia, 56(4), 469–475. [CrossRef]
- Nozaki, H., Misawa, K., Kajita, T., Kato, M., Nohara, S., & Watanabe, M. M. (2000). Origin and evolution of the colonial volvocales (Chlorophyceae) as inferred from multiple, chloroplast gene sequences. Molecular Phylogenetics and Evolution, 17(2), 256–268. [CrossRef]
- Olson, B. J. S. C., & Nedelcu, A. M. (2016). Co-option during the evolution of multicellularity and developmental complexity in the volvocine green algae. Current Opinion in Genetics and Development, 39, 107–115. [CrossRef]
- Ortega Escalante, J. A. (2018). Investigation of Volvox carteri Cell Differentiation and Its Evolution Through Functional Analysis of regA and regA Homologues [PhD Dissertation]. University of Maryland, Baltimore County.
- Pfannschmidt, T., Bräutigam, K., Wagner, R., Dietzel, L., Schröter, Y., Steiner, S., & Nykytenko, A. (2009). Potential regulation of gene expression in photosynthetic cells by redox and energy state: approaches towards better understanding. Annals of Botany, 103(4), 599. [CrossRef]
- Radhakrishnan, K., Bhagya, K. P., Kumar, A. T., Devi, A. N., Sengottaiyan, J., & Kumar, P. G. (2016). Autoimmune Regulator ( AIRE ) Is Expressed in Spermatogenic Cells, and It Altered the Expression of Several Nucleic-Acid-Binding and Cytoskeletal Proteins in Germ Cell 1 Spermatogonial (GC1-spg) Cells. Molecular & Cellular Proteomics, 15(8), 2686–2698. [CrossRef]
- Raffaghello, L., Lee, C., Safdie, F. M., Wei, M., Madia, F., Bianchi, G., & Longo, V. D. (2008). Starvation-dependent differential stress resistance protects normal but not cancer cells against high-dose chemotherapy. Proceedings of the National Academy of Sciences, 105(24), 8215–8220. [CrossRef]
- Ransick, A. (1991). Reproductive cell specification during Volvox obversus development. Developmental Biology, 143(1), 185–198. http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WDG-4DKTPB0-J&_user=441497&_coverDate=01/31/1991&_rdoc=1&_fmt=high&_orig=search&_sort=d&_docanchor=&view=c&_acct=C000020998&_version=1&_urlVersion=0.
- Ransick, A. (1993). Specification of reproductive cells in Volvox. In A. Spradling (Ed.), Evolutionary Conservation of Developmental Mechanisms: 50th Symposium of the Society for Developmental Biology, Marquette University, June 20–23, 1991. (pp. 55–70). Wiley-Liss.
- Ratcliff, W. C., Denison, R. F., Borrello, M., & Travisano, M. (2012). Experimental evolution of multicellularity. Proceedings of the National Academy of Sciences, 109(5), 1595–1600. [CrossRef]
- Ratcliff, W. C., Herron, M. D., Howell, K., Pentz, J. T., Rosenzweig, F., & Travisano, M. (2013). Experimental evolution of an alternating uni- and multicellular life cycle in Chlamydomonas reinhardtii. Nature Communications, 4(May), 2742. [CrossRef]
- Roff, D. A., & Fairbairn, D. J. (2007). The evolution of trade-offs: Where are we? Journal of Evolutionary Biology, 20(2), 433–447. [CrossRef]
- Rose, C. J., Hammerschmidt, K., & Rainey, P. B. (2020). Experimental evolution of nascent multicellularity: Recognizing a Darwinian transition in individuality. BioRxiv. [CrossRef]
- Saggere, R. M. S., Lee, C. W. J., Chan, I. C. W., Durnford, D. G., & Nedelcu, A. M. (2022). A life-history trade-off gene with antagonistic pleiotropic effects on reproduction and survival in limiting environments. Proceedings of the Royal Society B, 289(1967). [CrossRef]
- Schaap, P. (2016). Evolution of developmental signalling in Dictyostelid social amoebas. In Current Opinion in Genetics and Development (Vol. 39, pp. 29–34). Elsevier Ltd. [CrossRef]
- Schaller, C. E., Wang, C. L., Beck-engeser, G., Goss, L., Scott, H. S., Anderson, M. S., & Wabl, M. (2008). Expression of Aire and the Early Wave of Apoptosis in Spermatogenesis. The Journal of Immunology, 180(3), 1338–1343. [CrossRef]
- Sessoms, A. H., & Huskey, R. J. (1973). Genetic Control of Development in Volvox: Isolation and Characterization of Morphogenetic Mutants. Proceedings of the National Academy of Sciences, 70(5), 1335–1338.
- Sharpe, S. C., Eme, L., Brown, M. W., & Roger, A. J. (2014). Timing the Origins of Multicellular Eukaryotes Through Phylogenomics and Relaxed Molecular Clock Analyses. In A. M. Nedelcu & I. Ruiz-Trillo (Eds.), Evolutionary Transitions to Multicellular Life (pp. 3–30). Springer.
- Stark, K., Kirk, D. L., & Schmitt, R. (2001). Two enhancers and one silencer located in the introns of regA control somatic cell differentiation in Volvox carteri. Genes & Development, 15, 1449–1460. [CrossRef]
- Starr, R. C. (1970). Control of differentiation in Volvox. Dev. Biol., 4, 59–100.
- Stearns, S. C. (1989). Trade-Offs in Life-History Evolution. Functional Ecology, 3(3), 259–268. [CrossRef]
- Strand, D. D., Livingston, A. K., Satoh-Cruz, M., Koepke, T., Enlow, H. M., Fisher, N., Froehlich, J. E., Cruz, J. A., Minhas, D., Hixson, K. K., Kohzuma, K., Lipton, M., Dhingra, A., & Kramer, D. M. (2017). Defects in the Expression of Chloroplast Proteins Leads to H2O2 Accumulation and Activation of Cyclic Electron Flow around Photosystem I. Frontiers in Plant Science, 7, 2073. [CrossRef]
- Tam, L., & Kirk, D. L. (1991). Identification of Cell-Type-Specific Characterization of their Expression Genes of Volvox carteri and during the Asexual Life Cycle. Developmental Biology, 145, 51–66.
- Umen, J. G. (2020). Volvox and volvocine green algae. EvoDevo, 11(1), 7–15. [CrossRef]
- Veraksa, A., Kennison, J., & McGinnis, W. (2002). DEAF-1 function is essential for the early embryonic development of Drosophila. Genesis, 33(2), 67–76. [CrossRef]
- Villellas, J., & García, M. B. (2018). Life-history trade-offs vary with resource availability across the geographic range of a widespread plant. Plant Biology, 20(3), 483–489. [CrossRef]
- Wagner, G. P., Erkenbrack, E. M., & Love, A. C. (2019). Stress-Induced Evolutionary Innovation: A Mechanism for the Origin of Cell Types. BioEssays, 41, 1800188. [CrossRef]
- Wenger, J. W., Piotrowski, J., Nagarajan, S., Chiotti, K., Sherlock, G., & Rosenzweig, F. (2011). Hunger Artists: Yeast Adapted to Carbon Limitation Show Trade-Offs under Carbon Sufficiency. PLOS Genetics, 7(8), e1002202. [CrossRef]
- Wolf, J. B., Howie, J. A., Parkinson, K., Gruenheit, N., Melo, D., Rozen, D., & Thompson, C. R. L. (2015). Fitness Trade-offs Result in the Illusion of Social Success. Current Biology: CB, 25(8), 1086–1090. [CrossRef]
- Wykoff, D. D., Davies, J. P., Melis, A., & Grossman, A. R. (1998). The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiology, 117(1), 129–139. [CrossRef]
- Yamamoto, K., Hamaji, T., Kawai-Toyooka, H., Matsuzaki, R., Takahashi, F., Nishimura, Y., Kawachi, M., Noguchi, H., Minakuchi, Y., Umen, J. G., Toyoda, A., & Nozaki, H. (2021). Three genomes in the algal genus Volvox reveal the fate of a haploid sex-determining region after a transition to homothallism. PNAS, 118(21), e2100712118. [CrossRef]
- Yamashita, S., Arakaki, Y., Kawai-Toyooka, H., Noga, A., Hirono, M., & Nozaki, H. (2016). Alternative evolution of a spheroidal colony in volvocine algae: developmental analysis of embryogenesis in Astrephomene (Volvocales, Chlorophyta). BMC Evolutionary Biology, 16(1), 243. [CrossRef]
- Yamashita, S., Yamamoto, K., Matsuzaki, R., Suzuki, S., Yamaguchi, H., Hirooka, S., Minakuchi, Y., Miyagishima, S. ya, Kawachi, M., Toyoda, A., & Nozaki, H. (2021). Genome sequencing of the multicellular alga Astrephomene provides insights into convergent evolution of germ-soma differentiation. Scientific Reports, 11(1). [CrossRef]
- Zera, A. J., & Harshman, L. G. (2001). The Physiology of Life History Trade-Offs in Animals. Annual Review of Ecology and Systematics, 32(1), 95–126. [CrossRef]
- Zhang, J. (2003). Evolution by gene duplication: An update. In Trends in Ecology and Evolution (Vol. 18, Issue 6, pp. 292–298). Elsevier Ltd. [CrossRef]
Figure 1.
Select species demonstrating volvocine green algae diversity. a) Chlamydomonas reinhardtii (scale bar 10 µm), b) Gonium pectorale (scale bar 10 µm), c) Eudorina elegans UTEX 1212 (scale bar 10 µm), d) Pleodorina californica (scale bar 25 µm), e) Volvox carteri f. nagariensis (scale bar 50 µm), f) V. carteri regenerator mutant showing dedifferentiated somatic cells (scale bar 50 µm). Images a – e reproduced from Grochau-Wright et al. (2017).
Figure 1.
Select species demonstrating volvocine green algae diversity. a) Chlamydomonas reinhardtii (scale bar 10 µm), b) Gonium pectorale (scale bar 10 µm), c) Eudorina elegans UTEX 1212 (scale bar 10 µm), d) Pleodorina californica (scale bar 25 µm), e) Volvox carteri f. nagariensis (scale bar 50 µm), f) V. carteri regenerator mutant showing dedifferentiated somatic cells (scale bar 50 µm). Images a – e reproduced from Grochau-Wright et al. (2017).
Figure 2.
Origin and evolution of the regA gene cluster in the volvocine algae. Phylogenetic tree shows relationships among selected volvocine algae species based on topology from Lindsey et al. (2021). Species in green have obligate somatic cells while species in black are undifferentiated; the number and position of origins of somatic cells are consistent with Z. Grochau-Wright et al., (2017) and Lindsey et al (2021). Currently available regA family gene sequences and assemblies are shown to the right. regA and rls genes are shown in green while nearby syntenic marker gene ACK2/ackB is shown in blue. Gene cluster diagrams show assembly status and completeness but are drawn to maintain alignment of homologs not to scale of actual genomic distances. Note that P. californica is assumed to possess and rlsC gene that has not yet been sequenced. Major events in the evolutionary history of the regA gene cluster marked on phylogeny: R = origin of regA gene cluster, 1 = loss of rlsO, 2 = Loss and reduplication of regA, rlsO, and/or rlsB in Y. unicocca, 3 = inversion of regA cluster relative to nearby syntenic genes in P. caudata, and 4 = transformation of rlsO into rlsN through domain duplication.
Figure 2.
Origin and evolution of the regA gene cluster in the volvocine algae. Phylogenetic tree shows relationships among selected volvocine algae species based on topology from Lindsey et al. (2021). Species in green have obligate somatic cells while species in black are undifferentiated; the number and position of origins of somatic cells are consistent with Z. Grochau-Wright et al., (2017) and Lindsey et al (2021). Currently available regA family gene sequences and assemblies are shown to the right. regA and rls genes are shown in green while nearby syntenic marker gene ACK2/ackB is shown in blue. Gene cluster diagrams show assembly status and completeness but are drawn to maintain alignment of homologs not to scale of actual genomic distances. Note that P. californica is assumed to possess and rlsC gene that has not yet been sequenced. Major events in the evolutionary history of the regA gene cluster marked on phylogeny: R = origin of regA gene cluster, 1 = loss of rlsO, 2 = Loss and reduplication of regA, rlsO, and/or rlsB in Y. unicocca, 3 = inversion of regA cluster relative to nearby syntenic genes in P. caudata, and 4 = transformation of rlsO into rlsN through domain duplication.
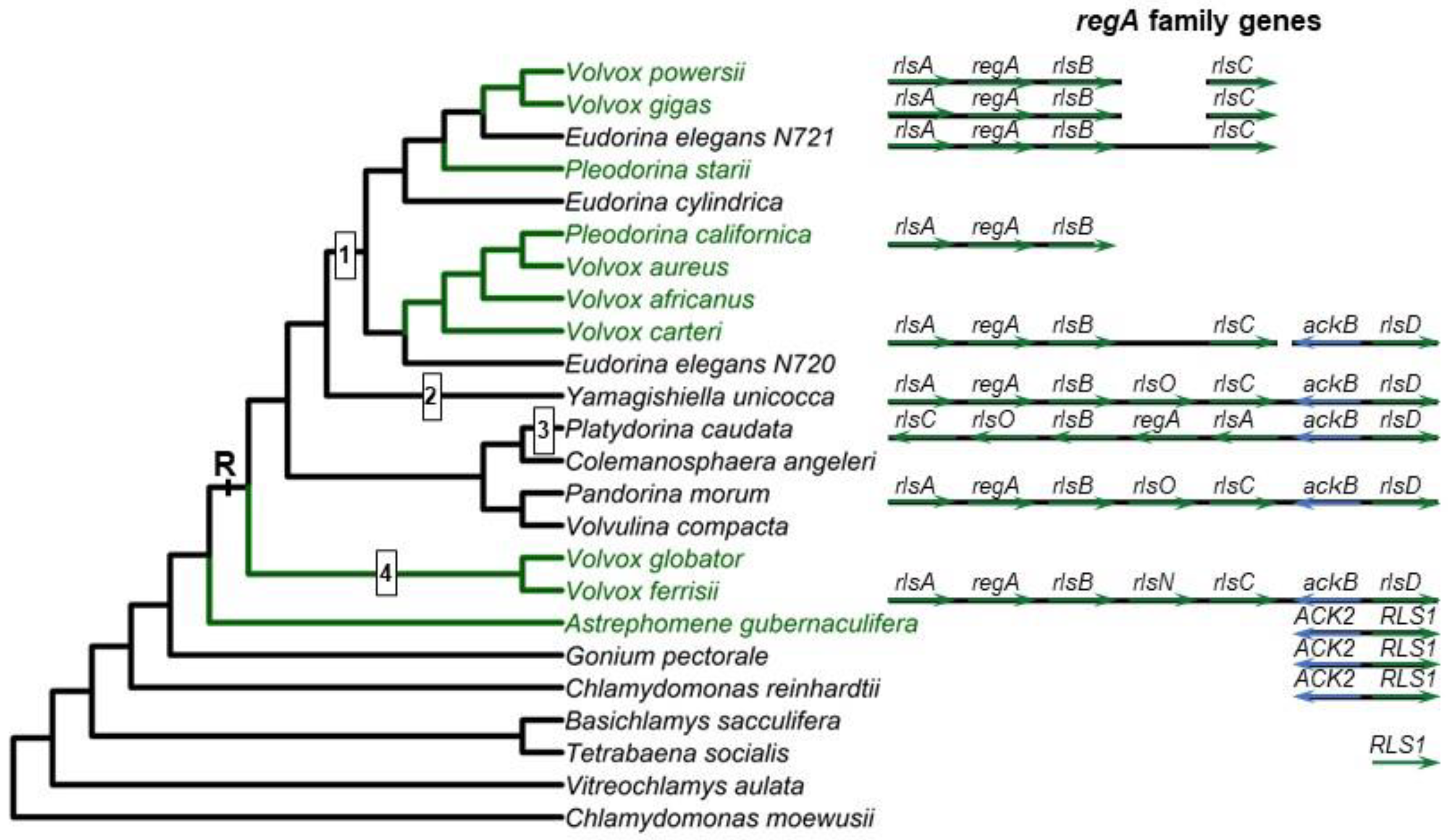
Figure 3.
Gene structure diagrams for regA and rlsD from V. carteri f. nagariensis and RLS1 from C. reinhardtii. Exons are shown as boxes with untranslated regions in white and coding regions in black. The portion of the coding region encoding the VARL domain is indicated with striped patterns. Introns are shown as lines between exons and the 42-bp minimal promoter of regA is shown before first exon and labeled with “P”. regA introns labeled with “E” and “S” denote locations of enhancer and silencer cis-regulatory elements respectively.
Figure 3.
Gene structure diagrams for regA and rlsD from V. carteri f. nagariensis and RLS1 from C. reinhardtii. Exons are shown as boxes with untranslated regions in white and coding regions in black. The portion of the coding region encoding the VARL domain is indicated with striped patterns. Introns are shown as lines between exons and the 42-bp minimal promoter of regA is shown before first exon and labeled with “P”. regA introns labeled with “E” and “S” denote locations of enhancer and silencer cis-regulatory elements respectively.
Figure 4.
Alignment of VARL domains from C. reinhardtii and V. carteri. Conserved regions annotated above alignment, variable linker region between N-terminal extension and core VARL SAND domain not shown. Vertical green lines show conserved intron positions. Figure adapted from Duncan et al. (2007a).
Figure 4.
Alignment of VARL domains from C. reinhardtii and V. carteri. Conserved regions annotated above alignment, variable linker region between N-terminal extension and core VARL SAND domain not shown. Vertical green lines show conserved intron positions. Figure adapted from Duncan et al. (2007a).
Figure 5.
Protein similarity plots for all regA cluster and RLS1/rlsD proteins based on syntenic position (rlsA, regA, rlsB, rlsO, rlsC and rlsD). Regions showing high similarity are highlighted with grey boxes. The two peaks in the shaded VARL region represent the N-terminal extension and core VARL domain separated by the less conserved linker region. Reproduced from Grochau-Wright et al. (2017).
Figure 5.
Protein similarity plots for all regA cluster and RLS1/rlsD proteins based on syntenic position (rlsA, regA, rlsB, rlsO, rlsC and rlsD). Regions showing high similarity are highlighted with grey boxes. The two peaks in the shaded VARL region represent the N-terminal extension and core VARL domain separated by the less conserved linker region. Reproduced from Grochau-Wright et al. (2017).
Figure 6.
Most common domain architectures in SAND domain-containing proteins present in the main Viridiplantae and Metazoa lineages (examples of proteins with known functions are indicated in brackets). Adapted from (Nedelcu, 2019).
Figure 6.
Most common domain architectures in SAND domain-containing proteins present in the main Viridiplantae and Metazoa lineages (examples of proteins with known functions are indicated in brackets). Adapted from (Nedelcu, 2019).
Figure 7.
Phylogenetic distribution of SAND sequences (red dots indicate occurrence; eukaryotic tree adapted from Keeling & Burki (2019)) and the hypothesized lateral gene transfer (red arrow) from a green algal ancestor to an early metazoan (adapted from (Nedelcu, 2019).
Figure 7.
Phylogenetic distribution of SAND sequences (red dots indicate occurrence; eukaryotic tree adapted from Keeling & Burki (2019)) and the hypothesized lateral gene transfer (red arrow) from a green algal ancestor to an early metazoan (adapted from (Nedelcu, 2019).
Figure 8.
Co-option of a life history gene that trades off reproduction for survival in a single-celled individual into a regulator of soma/germ cell differentiation in a multicellular individual through changing its expression from a temporal context (in response to an environmental cue) into a spatial context (in response to a developmental cue).
Figure 8.
Co-option of a life history gene that trades off reproduction for survival in a single-celled individual into a regulator of soma/germ cell differentiation in a multicellular individual through changing its expression from a temporal context (in response to an environmental cue) into a spatial context (in response to a developmental cue).
Figure 9.
A. Two potential scenarios envisioning the co-option of an ancestral environmentally regulated RLS1/rlsD-like gene into a developmentally regulated regA, involving either (i) the replacement of the ancestral environmental regulation or (ii) the addition of a new layer of regulation. B. Two models to account for the observed dual regulation (environmental and developmental) of regA in V. carteri involving (i) the acquisition of a new signal transduction pathway and regulatory element (RE2) or (ii) the co-option of the environmentally induced intracellular signal and regulatory element (RE1). Adapted from (König & Nedelcu, 2020).
Figure 9.
A. Two potential scenarios envisioning the co-option of an ancestral environmentally regulated RLS1/rlsD-like gene into a developmentally regulated regA, involving either (i) the replacement of the ancestral environmental regulation or (ii) the addition of a new layer of regulation. B. Two models to account for the observed dual regulation (environmental and developmental) of regA in V. carteri involving (i) the acquisition of a new signal transduction pathway and regulatory element (RE2) or (ii) the co-option of the environmentally induced intracellular signal and regulatory element (RE1). Adapted from (König & Nedelcu, 2020).
| 1 |
Huskey & Griffin, (1979) originally described a second regB locus based on linkage group analysis of regenerator mutants, but reexamination of regB mutants from members of the same research lab determined that regB mutants are not regenerator mutants and have a different mutant phenotype (Baran, 1984). Thus, in retrospect, all regenerator mutants can be mapped to the regA locus (D. L. Kirk, 1998). |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).