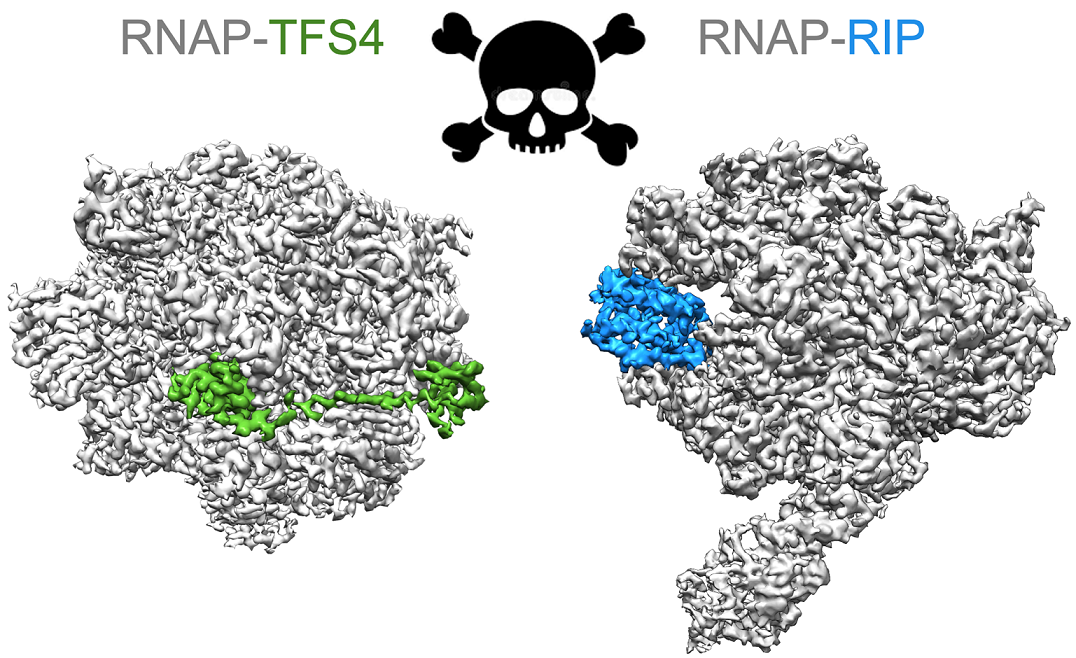
Multisubunit RNA polymerases (RNAP) carry out transcription in all domains of life; during vi-rus infection, RNAPs are targeted by transcription factors encoded by either the cell or the virus, resulting in the global repression of transcription with distinct outcomes for different host-virus combinations. These repressors serve as versatile molecular probes to study RNAP mechanisms, as well as they aid the exploration of druggable sites for the development of new antibiotics. Here, we review the mechanisms and structural basis of RNAP inhibition by the viral repressor RIP and the crenarchaeal negative regulator TFS4, which follow distinct strategies. RIP operates by occluding the DNA-binding channel and mimicking the initiation factor TFB/TFIIB. RIP binds tightly to the clamp and locks it into one fixed position, thereby preventing conformational oscil-lations that are critical for RNAP function as it progresses through the transcription cycle. TFS4 engages with RNAP in a similar manner to transcript cleavage factors such as TFS/TFIIS through the NTP-entry channel; TFS4 interferes with the trigger loop and bridge helix within the active site by occlusion and allosteric mechanisms, respectively. The conformational changes of RNAP described above are universally conserved and are also seen in inactive dimers of eukaryotic RNAPI and several inhibited RNAP complexes of both bacterial and eukaryotic RNA polymer-ases, including inactive states that precede transcription termination. A comparison of target sites and inhibitory mechanisms reveals that proteinaceous repressors and RNAP-specific antibiotics use surprisingly common ways to inhibit RNAP function.