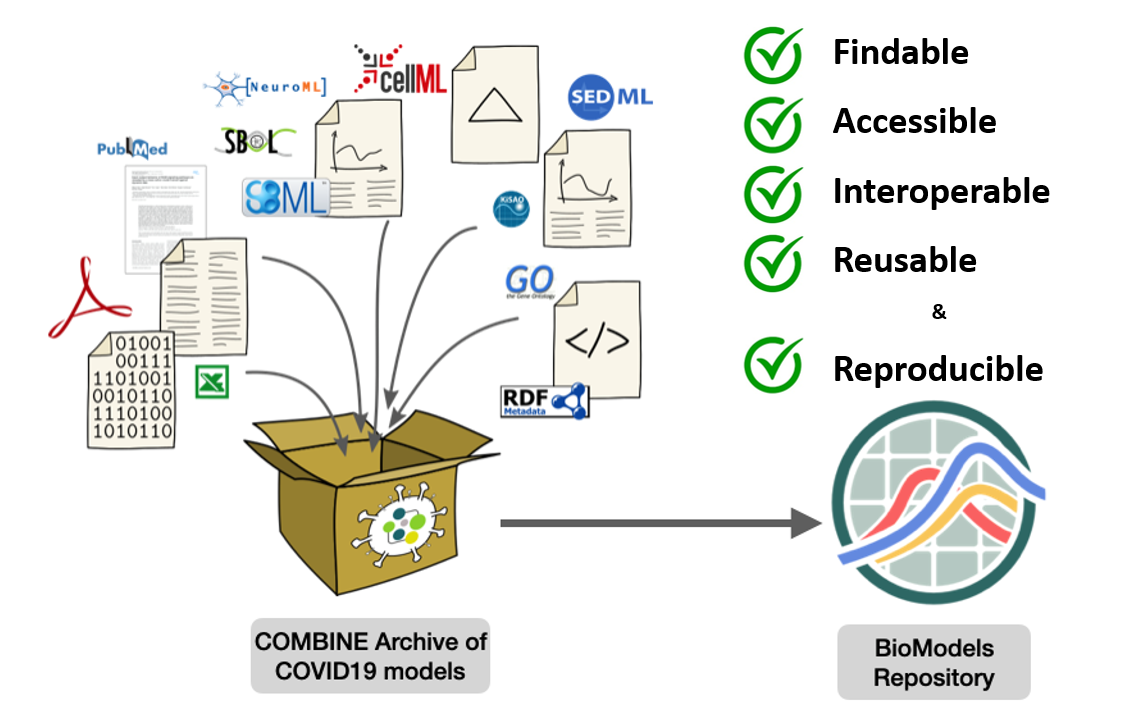
A major challenge for the dissemination, replication, and reuse of epidemiological forecasting studies during COVID-19 pandemics is the lack of clear guidelines and platforms to exchange models in a Findable, Accessible, Interoperable, and Reusable (FAIR) manner, facilitating reproducibility of research outcomes. During the beginning of pandemics, models were developed in diverse tools that were not interoperable, opaque without traceability and semantics, and scattered across various platforms - making them hard to locate, infer and reuse. In this work, we demonstrate that implementing the standards developed by the systems biology community to encode and share COVID-19 epidemiological models can serve as a roadmap to implement models as a tool in medical informatics, in general. As a proof-of-concept, we encoded and shared 24 epidemiological models using the standard format for model exchange in systems biology, annotated them with cross-references to data resources, packed up all associated files in COMBINE archives for easy sharing, and finally, disseminated the models through BioModels repository to significantly enhance their reproducibility and repurposing potential. We recommend the use of systems biology standards to encode and share models of epidemic and pandemic forecasts to improve their findability, accessibility, interoperability, reusability, and reproducibility.