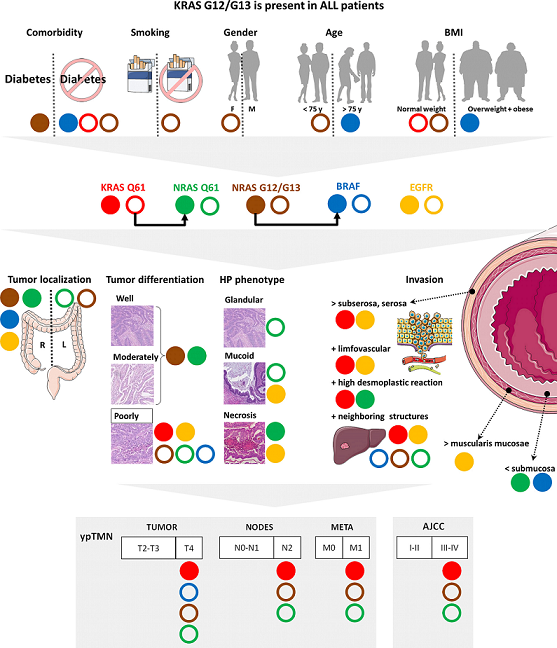
Background: Biomarker profiles should represent a coherent description of the colorectal cancer (CRC) stage and its evolution. Methods: Using droplet digital PCR, we detected the allelic frequencies (AF) of KRAS, NRAS, BRAF and EGFR mutations from 60 tumors. We employed a pair-wise association approach to estimate the risk involving AF mutations as outcome variables for clinical data and as predicting variables for tumor-staging. We evaluated correlations between mutations AFs and also between the mutations and histopathology features (tumor staging, inflammation, differentiation and invasiveness). Results: KRAS G12/G13 mutations were present in all patients. KRAS Q61 was significantly associated with poor differentiation, high desmoplastic reaction, invasiveness (ypT4) and metastasis (ypM1). NRAS and BRAF were associated with the right-side localization of tumors. Diabetic patients had a higher risk to exhibit NRAS G12/G13 mutations. BRAF’s presence limited the invasiveness in the submucosa, co-existing with NRAS G12/G13 mutations. Conclusions: The associations we found and the mutational AF we reported may help to understand disease processes and may be considered as potential CCR biomarker candidates. In addition, we propose representative mutation panels associated with specific clinical and histopathological features of CRC, as a unique opportunity to refine the degree of personalization of CRC treatment.