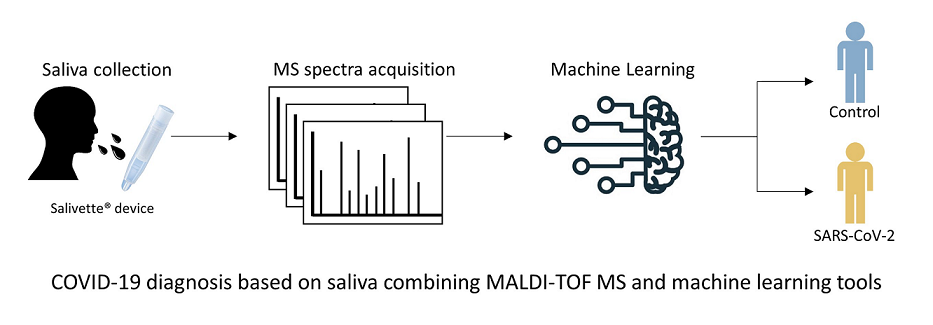
SARS-CoV-2 caused a large outbreak since its emergence in December 2019. The COVID-19 diagnosis became a priority to isolate and treat infected individuals in order to break the contamination chain. Currently, the reference test for COVID-19 diagnosis is the molecular detection (RT-qPCR) of the virus from nasopharyngeal swab (NPS) samples. Although this sensitive and specific test remains the gold standard, it has several limitations, such as the invasive collection method, the relative high cost and the duration of the test. Moreover, the material shortage to perform tests due to the discrepancy between the high demand for tests and the production capacities puts additional constraints on RT-qPCR. Here, we propose a PCR-free method for diagnosing SARS-CoV-2 based on Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) profiling and machine learning (ML) models from salivary samples. Kinetic saliva samples were collected at enrollment and ten and thirty days later (D0, D10 and D30), to assess the classification performance of the ML models compared to the molecular tests performed on NPS specimens. Spectra were generated using an optimized protocol of saliva collection and successive quality control steps were developed to ensure the reliability of spectra. A total of 360 averaged spectra were included in the study. At D0, the comparison of MS spectra from SARS-CoV-2 positive patients (n=105) with healthy healthcare controls (n=51) revealed nine peaks that significantly distinguished the two groups. Among the five ML models tested, Support Vector Machine with Linear Kernel (SVM-LK) provided the best performance on the training dataset (accuracy = 85.2 %, sensitivity = 85.1 %, specificity = 85.3 %, F1-Score = 85.1 %). The application of the SVM-LK model on independent datasets confirmed it performances with 88.9% and 80.8% of correct classification for samples collected at D0 and D30, respectively. Conversely, at D10, the proportion of correct classification fallen to 64.3%. The analysis of saliva samples by MALDI-TOF MS and ML appears as an interesting supplementary tool for COVID-19 diagnosis, despite the mitigated results obtained for convalescent patients (D10).