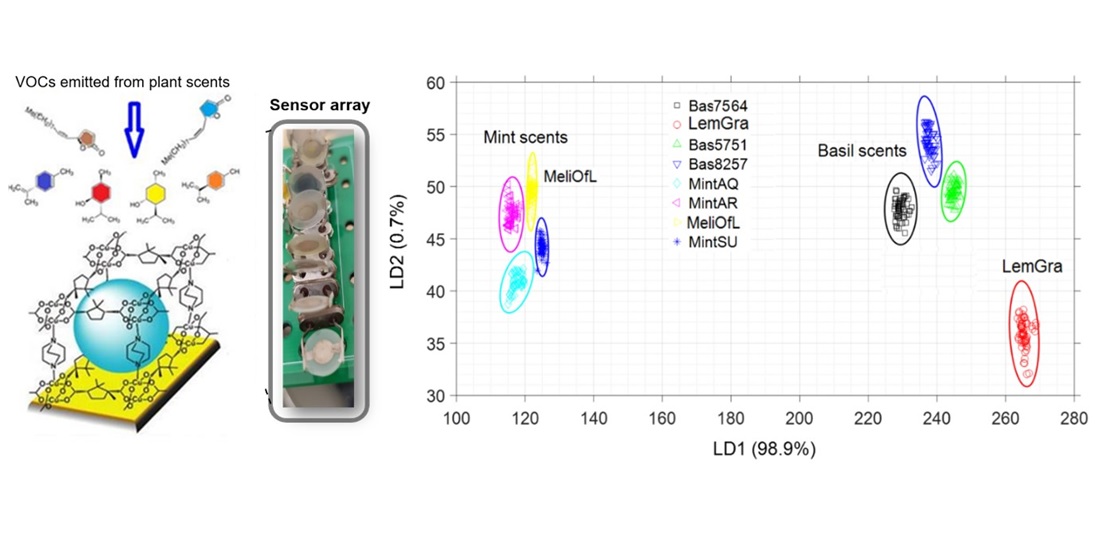
The Lamiaceae belong to the species-richest families of flowering plants and harbor many species used as herbs or for medicinal applications, such as Basils or Mints. Evolution of this group has been driven by chemical speciation, mainly of Volatile Organic Compounds (VOCs). The commercial use of these plants is characterized by a large extent of adulteration and surrogation. To authenticate and discern the species, is, thus, relevant for consumer safety, but usually requires cumbersome analytics, such as Gas Chromatography, often to be coupled with Mass Spectroscopy. We demon-strate here that quartz-crystal microbalance (QCM)-based electronic noses provide a very cost-efficient alternative, allowing for a fast, automated discrimination of scents emitted from leaves of different plants. To explore the range of this strategy, we used leaf material from four genera of Lamiaceae along with Lemongrass as similarly scented, but non-related outgroup. In order to unambiguously differentiate the scents from the different plants, the output of the 6 different SURMOF/QCM sensors was analyzed using machine learning (ML) methods, together with a thorough statistical analysis. The exposure and purging datasets (4 cycles) obtained from a QCM-based, low-cost homemade portable e-Nose were analyzed with Linear Discriminant Analysis (LDA) classification model. Prediction accuracies with repeating test measurements reached values of up to 90%. We show that it is not only possible to discern and identify plants on the genus level, but even to discriminate closely related sister clades within a genus (Basil), demonstrating that e-Noses are a powerful technology to safeguard consumer safety against the challenges of globalized trade.