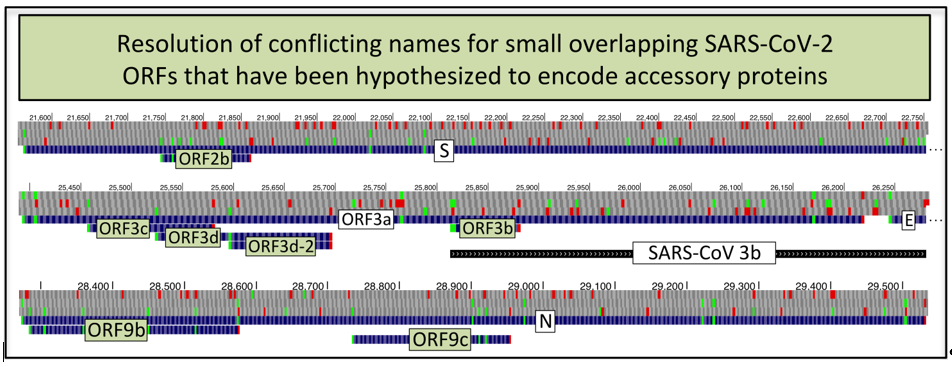
At least six small alternate-frame open reading frames (ORFs) overlapping well-characterized SARS-CoV-2 genes have been hypothesized to encode accessory proteins. Researchers have used different names for the same ORF or the same name for different ORFs, resulting in erroneous homological and functional inferences. We propose standard names for these ORFs and their shorter isoforms, developed in consultation with the Coronaviridae Study Group of the ICTV. We recommend calling the 39 codon Spike-overlapping ORF ORF2b; the 41, 57, and 22 codon ORF3a-overlapping ORFs ORF3c, ORF3d, and ORF3b; the 33 codon ORF3d isoform ORF3d-2; and the 97 and 73 codon Nucleocapsid-overlapping ORFs ORF9b and ORF9c. Finally, we document conflicting usage of the name ORF3b in 32 studies, and consequent erroneous inferences, stressing the importance of reserving identical names for homologs. We recommend that authors referring to these ORFs provide lengths and coordinates to minimize ambiguity due to prior usage of alternative names.