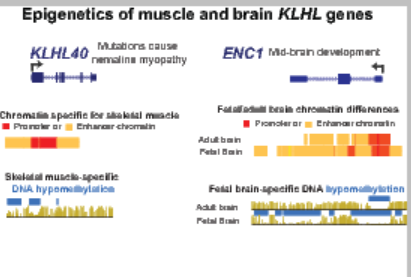
KLHL genes and the related KBTBD genes encode components of the Cullin-E3 ubiquitin ligase complex and typically target tissue-specific proteins for degradation, thereby affecting differentiation, homeostasis, metabolism, cell signaling, and the oxidative stress response. Despite their importance in normal cell function and in disease (KLHL40, KLHL41, KBTBD13, KEAP1, and ENC1), previous studies that examined epigenetic factors affecting transcription were predominantly limited to promoter DNA methylation. Using diverse tissue and cell culture whole-genome profiles, we examined 17 KLHL or KBTBD genes preferentially expressed in skeletal muscle or brain to identify tissue-specific enhancer and promoter chromatin, open chromatin (DNaseI hypersensitivity), and DNA hypomethylation. Sixteen of the 17 genes displayed muscle- or brain-specific enhancer chromatin in their gene bodies, and most exhibited specific intergenic enhancer chromatin as well. Seven genes were embedded in a super-enhancer. The enhancer chromatin regions typically displayed foci of DNA hypomethylation at peaks of open chromatin. In addition, we found evidence that gene neighbors (HHATL and FBXO32) of KLHL40 and KLHL38 harbor enhancer chromatin that likely upregulates the adjacent KLHL gene. Many KLHL/KBTBD genes had tissue-specific promoter chromatin at their 5’ ends, but surprisingly, two (KBTBD11 and KLHL31) had constitutively unmethylated promoter chromatin in their 3’ exons that overlaps a retrotransposed KLHL gene. Our findings demonstrate the importance of expanding epigenetic analyses beyond the 5’ ends of genes in studies of normal and abnormal gene regulation.