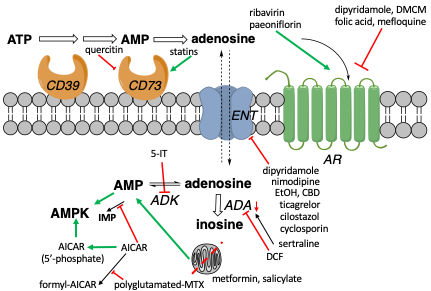
Many ligands directly target adenosine receptors (ARs). Here we review the effects of noncanonical AR drugs on adenosinergic signaling. Non-AR mechanisms include raising adenosine levels by inhibiting adenosine transport (e.g. ticagrelor, ethanol, cannabidiol), affecting intracellular metabolic pathways (e.g. methotrexate, nicotinamide riboside, salicylate, 5‐aminoimidazole‐4‐carboxamide riboside), or undetermined means (e.g. acupuncture). Yet other compounds bind ARs in addition to their canonical ‘on-target’ activity (e.g. mefloquine). The strength of experimental support for an adenosine-related role in a drug’s effects varies widely. AR knockout mice are the ‘gold standard’ method for investigating an AR role, but few drugs have been tested in these mice. Given the interest in AR modulation for treatment of cancer, CNS, immune, metabolic, cardiovascular, and musculoskeletal conditions, it is informative to consider AR and non-AR adenosinergic effects of approved drugs and conventional treatments.