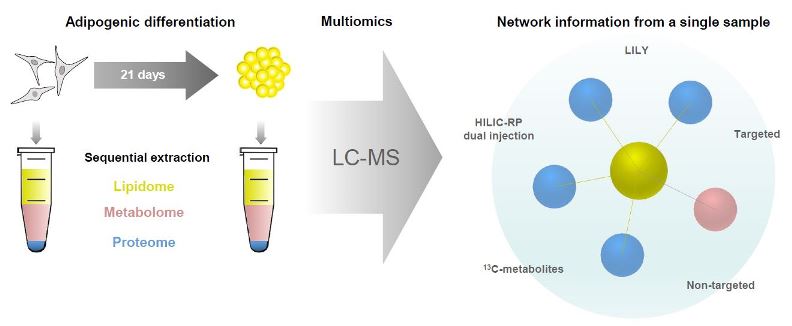
The molecular study of fat cell development in the human body is essential for our understanding of obesity and related diseases. Mesenchymal stem/stromal cells (MSC) are the ideal source to study fat formation as they are the progenitors of adipocytes. In this work, we used human MSCs, received from surgery waste, and differentiate them into fat adipocytes. The combination of several layers of information coming from lipidomics, metabolomics and proteomics enabled comprehensive analysis of the biochemical pathways in adipogenesis. Simultaneous analysis of metabolites, lipids and proteins in cell culture is challenging due to the compound’s chemical difference so that most studies involve separate analysis with unimolecular strategies. In this study, we employed a multimolecular approach using a two–phase extraction to monitor the crosstalk between lipid metabolism and protein-based signaling in a single sample (~105 cells). We developed an innovative analytical workflow including standardization with in-house produced 13C-isotopically labeled compounds, hyphenated high-end mass spectrometry (high-resolution Orbitrap MS) and chromatography (HILIC, RP) for simultaneous untargeted screening and targeted quantification. Metabolite and lipid concentrations ranged over 3-4 orders of magnitude and were detected down to the low fmol (absolute on column) level. Biological validation and data interpretation of the multiomics workflow was performed based on proteomics network reconstruction, metabolic modelling (MetaboAnalyst 4.0) and pathway analysis (OmicsNet). Comparing MSCs and adipocytes, we observed significant regulation of different metabolites and lipids such as triglycerides, gangliosides and carnitine with 113 fully reprogrammed pathways. The observed changes are in accordance with literature findings dealing with adipogenic differentiation of MSC. These results are a proof of principle for the power of multimolecular extraction combined with orthogonal LC-MS assays and network construction. Considering the analytical and biological validation performed in this study, we conclude that the proposed multiomics workflow is ideally suited for comprehensive follow-up studies on adipogenesis and is fit for purpose for different applications.