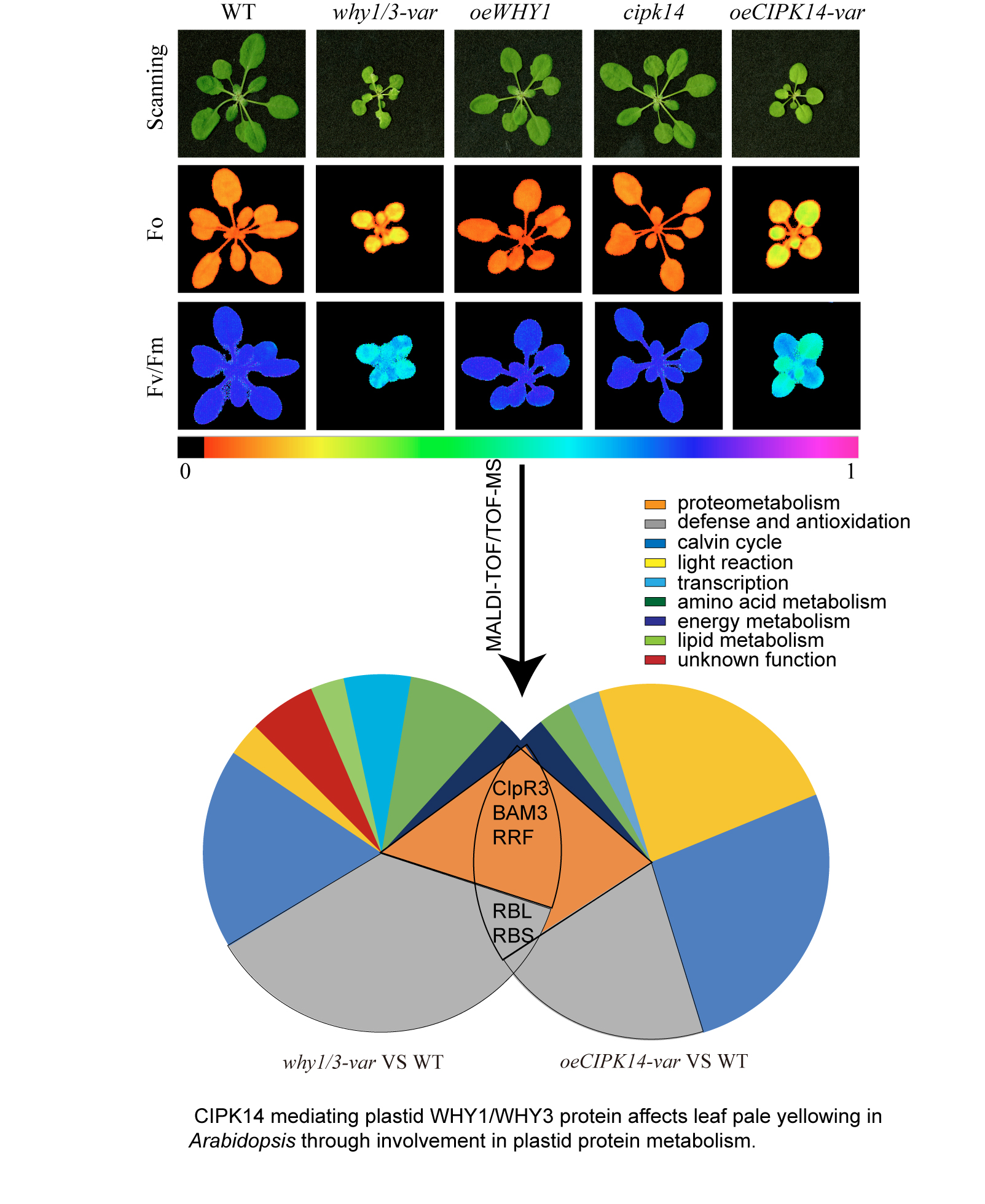
Leaf variegation pale yellowing is observed in the Calcineurin B-Like-Interacting Protein Kinase14 (CIPK14) overexpression line (oeCIPK14) and double knockout WHIRLY1/WHIRLY3 (why1/3) lines of Arabidopsis, the distribution of WHIRLY1 (WHY1) protein between plastids and the nucleus are affected by the phosphorylation of WHY1 by CIPK14. To elucidate the coregulation of CIPK14 and WHIRLY1/WHIRLY3 mediated leaf pale yellowing, a differential proteomic analysis is conducted between the oeCIPK14 variegated (oeCIPK14-var) line, why1/3 variegated (why1/3-var) line and wild type (WT). More than 800 protein spots are distinguished on each gel, 67 differential abundance proteins (DAPs) are identified by matrix-assisted laser desorption ionization-time of flight/time of flight mass spectrometry (MALDI-TOF/TOF-MS), of which, 34 DAPs are in the oeCIPK14-var, 33 DAPs are in the why1/3-var compared to WT. Five overlapping proteins differentially change both in the oeCIPK14-var and in the why1/3-var. They are ATP-dependent Clp protease proteolytic subunit-related protein 3 (ClpR3), Ribulose bisphosphate carboxylase large chain (RBL), Beta-amylase 3 (BAM3), Ribosome-recycling factor (RRF), Ribulose bisphosphate carboxylase small chain (RBS). Bioinformatics analysis show that most of DAPs are involved in photosynthesis, defense and antioxidation pathway, protein metabolism, amino acid metabolism, energy metabolism, malate biosynthesis, lipid metabolism and transcription. Thus, the photosystem parameters are measured that the content of chlorophyll, the photochemical efficiency of PSⅡ (Fv/Fm), and electron transport rates (ETR) decrease in the why1/3-var and oeCIPK14-var, but the non-photochemical quenching (NPQ) increases. Both mutants show high sensitivity to strong light. Based on the annotation of DAPs from both why1/3-var and oeCIPK14-var lines, we conclude that CIPK14 phosphorylation mediated WHY1 deficiency in plastids is related to impairment of protein metabolism leading to chloroplast dysfunction.