Submitted:
27 January 2023
Posted:
29 January 2023
You are already at the latest version
Abstract

Keywords:
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Calculation of the Genomic Coverage by the Cyclooctaoxygen Sodium-Bridged Spermine Phosphate Epigenetic Shell of Interphase DNA in Bovine Lymphocytes
2.3. Calculation of the Spermine Coverage of Highly Condensed Mitotic Metaphase DNA in HeLa S3 Cells
2.4. Calculation of the Polyamine Coverage of Maximally Condensed Mitotic Late Anaphase/Early Telophase DNA in Murine Cryptal Enterocytes
2.5. Calculation of the Hydrogen Selenite (HSeO3−) Coverage of Human Euchromatin DNA Specifically at ATG Start Codon Sequences
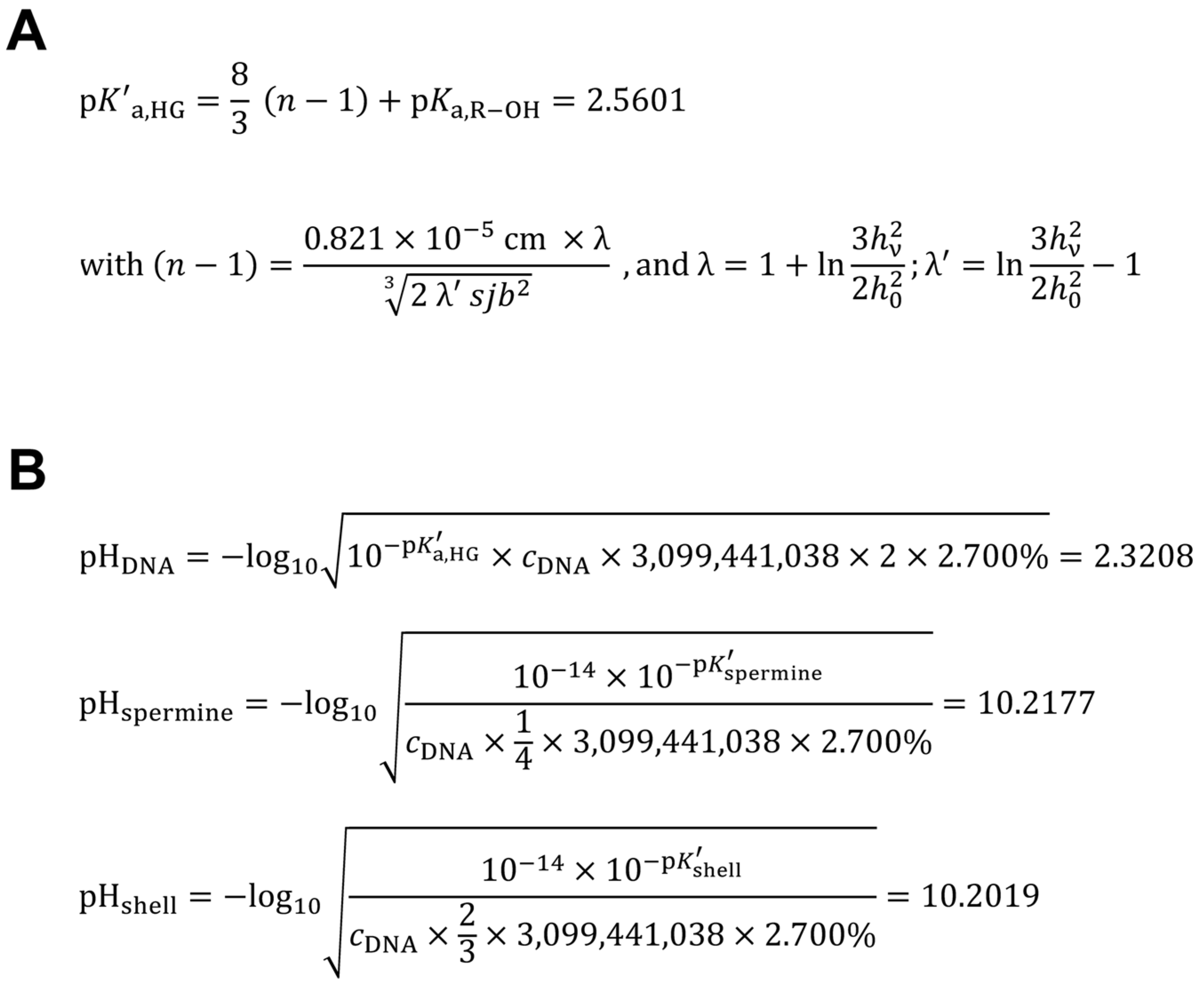
2.6. Calculation of the Apparent Acid Dissociation Constant of the Human Genome DNA
2.7. Calculation of the Hypothetical Intranuclear Micro-pH Mediated by Single Spermine Occupation of Human Interphase Euchromatin
2.8. Calculation of the Theoretical Intranuclear Micro-pH Mediated by Sperminium Hydrogen Phosphate/Cyclooctaoxygen Sodium Complex Occupation of Human Interphase Euchromatin
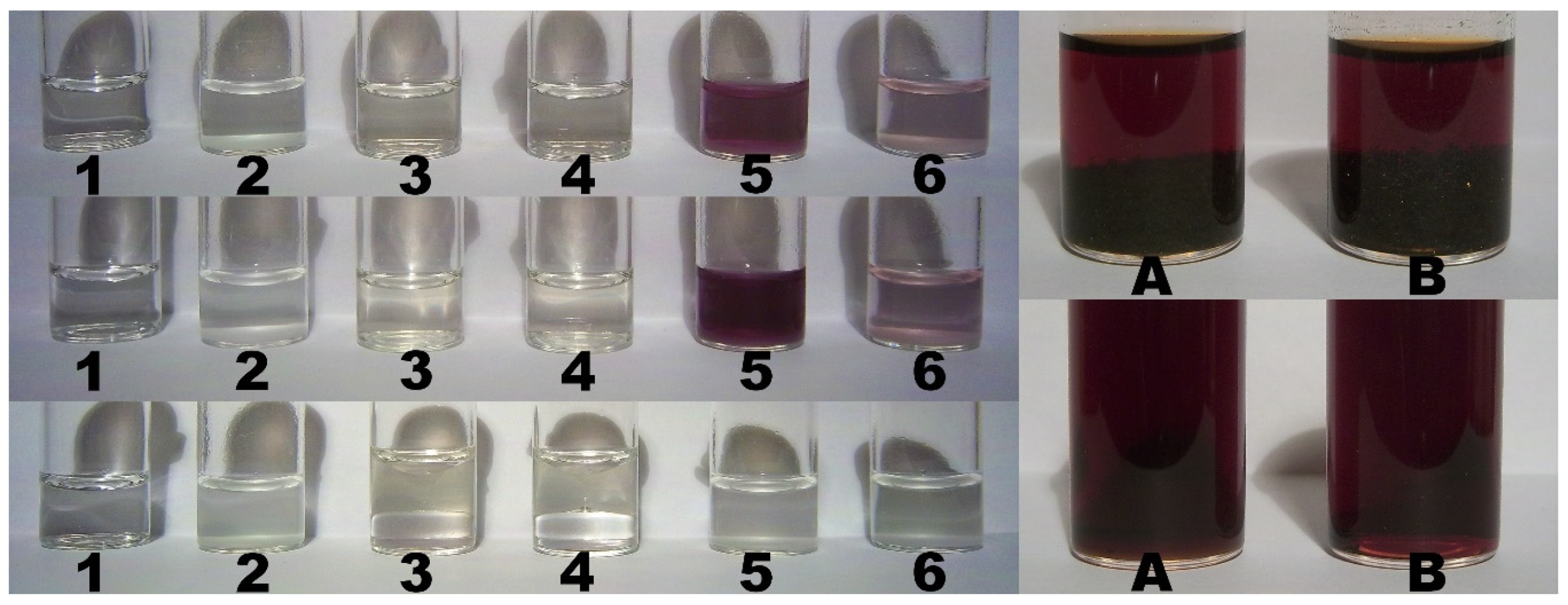
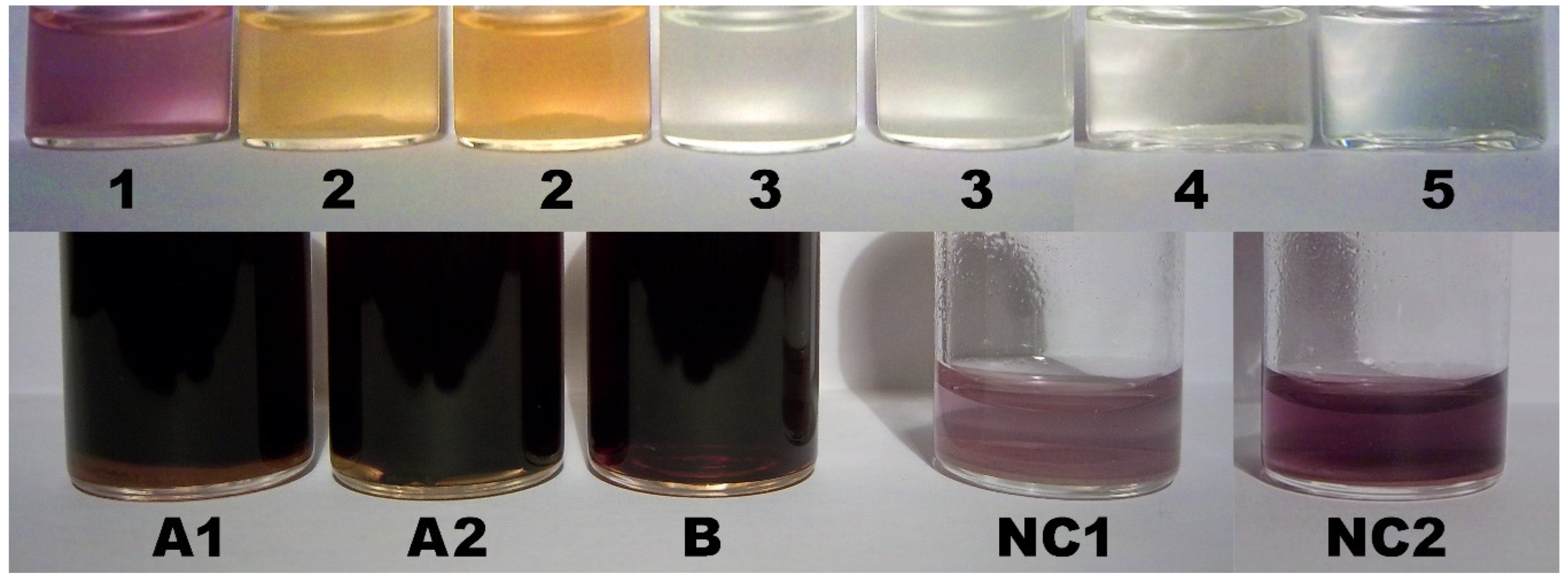
2.9. Color Assay for Cyclo-O8-Na+ Contained in RC – Destruction of Cyclo-O8-Na+ by the Glyphosate Metabolite (Aminomethyl)Phosphonic Acid
2.10. Control Color Assay for Potential Reduction of Elemental Iodine by the Glyphosate Metabolite (Aminomethyl)Phosphonic Acid
2.11. Color Assay for Cyclo-O8-Na+ Contained in RC – Destruction of Cyclo-O8-Na+ by Glyphosate and ROUNDUP®
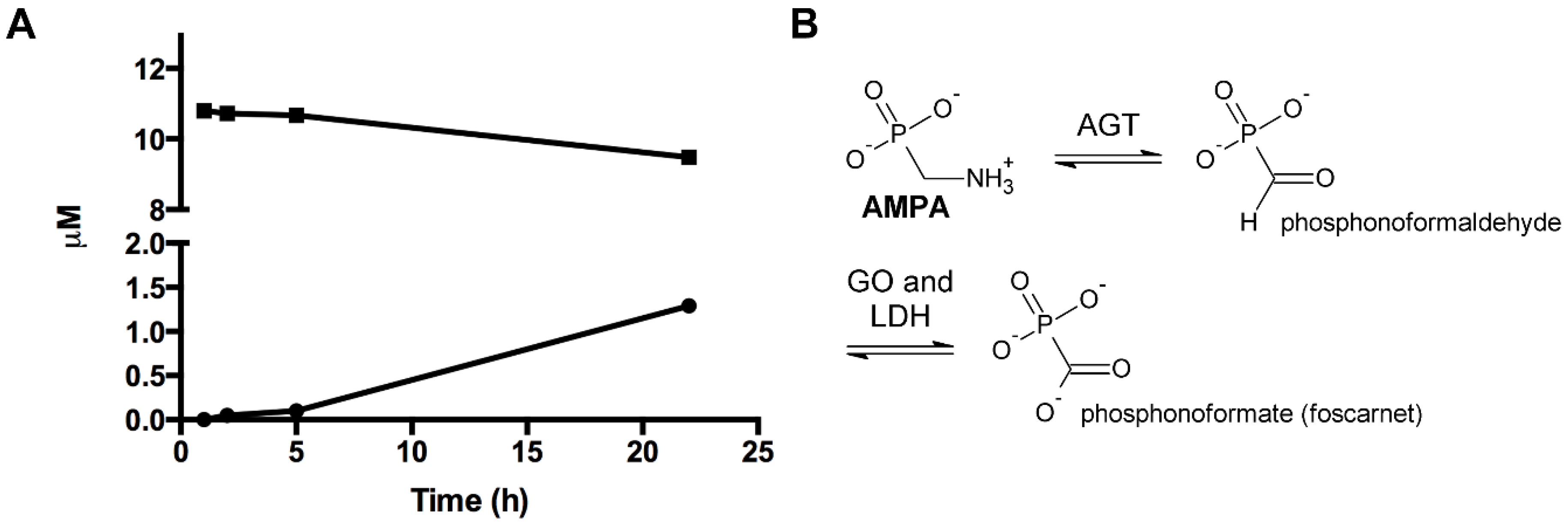
2.12. Enzyme Assay of the Glyphosate Metabolite (Aminomethyl)Phosphonic Acid with Human Mitochondrial γ-Aminobutyric Acid Transaminase
2.13. Enzyme Assay of the Glyphosate Metabolite (Aminomethyl)Phosphonic Acid with Human Wild-Type Alanine:Glyoxylate Aminotransferase
3. Results
3.1. Calculation of the Genomic Coverage by the Cyclooctaoxygen Sodium-Bridged Spermine Phosphate Epigenetic Shell of Interphase DNA in Bovine Lymphocytes
3.2. Calculation of the Spermine Coverage of Highly Condensed Mitotic Metaphase DNA in HeLa S3 Cells
3.3. Calculation of the Polyamine Coverage of Maximally Condensed Mitotic Late Anaphase/Early Telophase DNA in Murine Cryptal Enterocytes
3.4. Calculation of the Hydrogen Selenite (HSeO3−) Coverage of Human Euchromatin DNA Specifically at ATG Start Codon Sequences
3.5. Calculation of the Apparent Acid Dissociation Constant of the Human Genome DNA, and the Intranuclear Micro-pH Mediated by Single Spermine Occupation, in Comparison to Spermine Phosphate/Cyclooctaoxygen Sodium Complex Occupation, of Human Interphase Euchromatin
3.6. Color Assay for Cyclo-O8-Na+ Contained in RC – Destruction of Cyclo-O8-Na+ by the Glyphosate Metabolite (Aminomethyl)Phosphonic Acid
3.7. Color Assay for Cyclo-O8-Na+ Contained in RC – Destruction of Cyclo-O8-Na+ by Glyphosate and ROUNDUP®
3.8. Enzymatic Investigations with the Glyphosate Metabolite (Aminomethyl)Phosphonic Acid
4. Discussion
5. Conclusions
Acknowledgments
References
- van Leeuwenhoek, Observationes D. Anthonii Lewenhoeck, de natis è semine genitali animalculis, Philos. Trans. R. Soc. Lond. 12 (1677‒1678) 1040‒1046.
- O. Rosenheim, The isolation of spermine phosphate from semen and testis, Biochem. J. 18 (1924) 1253‒1262. [CrossRef]
- M.S. Cooke, M.D. Evans, M. Dizdaroglu, J. Lunec, Oxidative DNA damage: mechanisms, mutation, and disease, FASEB J. 17 (2003) 1195‒1214. [CrossRef]
- I. Kirmes, et al., A transient ischemic environment induces reversible compaction of chromatin, Genome Biol. 16 (2015), 246. [CrossRef]
- A.J. Kesel, C.W. Day, C.M. Montero, R.F. Schinazi, A new oxygen modification cyclooctaoxygen binds to nucleic acids as sodium crown complex, Biochim. Biophys. Acta 1860 (2016) 785‒794. [CrossRef]
- K. Igarashi, K. Kashiwagi, Polyamines: mysterious modulators of cellular functions, Biochem. Biophys. Res. Commun. 271 (2000) 559‒564. [CrossRef]
- H. Takata, et al., Chromatin compaction protects genomic DNA from radiation damage, PLoS One 8 (2013), e75622. [CrossRef]
- L.R. Ferguson, N. Karunasinghe, S. Zhu, A.H. Wang, Selenium and its’ role in the maintenance of genomic stability, Mutat. Res. 733 (2012) 100‒110. [CrossRef]
- G.B. Segel, G.R. Cokelet, M.A. Lichtman, The measurement of lymphocyte volume: importance of reference particle deformability and counting solution tonicity, Blood 57 (1981) 894‒899.
- R. Kuse, S. Schuster, H. Schübbe, S. Dix, K. Hausmann, Blood lymphocyte volumes and diameters in patients with chronic lymphocytic leukemia and normal controls, Blut (Berl.) 50 (1985) 243‒248. [CrossRef]
- C.R. Sipe, A.D. Chanana, E.P. Cronkite, G.L. Gulliani, D.D. Joel, Studies on lymphocytes XIII. Nuclear volume measurement as a rapid approach to estimate proliferative fraction, Scand. J. Haematol. 16 (1976) 196‒201.
- National Center for Biotechnology Information (NCBI), Bos_taurus_UMD_3.1.1, https://www.ncbi.nlm.nih.gov/assembly/GCF_000003055.6 (Bethesda, MD, 2016), retrieved 2016/11/12.
- S. Watanabe, K. Kusama-Eguchi, H. Kobayashi, K. Igarashi, Estimation of polyamine binding to macromolecules and ATP in bovine lymphocytes and rat liver, J. Biol. Chem. 266 (1991) 20803‒20809. [CrossRef]
- National Center for Biotechnology Information (NCBI), GRCh38.p14, https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.40 (Bethesda, MD, 2022), retrieved 2022/10/16.
- A. Adey, et al., The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line, Nature 500 (2013) 207‒211. [CrossRef]
- M. Macville, et al., Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping, Cancer Res. 59 (1999) 141‒150.
- M.H. Goyns, Polyamine content of a non-aqueously isolated chromosome preparation, Exp. Cell Res. 122 (1979) 377‒380. [CrossRef]
- S. Jain, G. Zon, M. Sundaralingam, Base only binding of spermine in the deep groove of the A-DNA octamer d(GTGTACAC), Biochemistry 28 (1989) 2360‒2364. [CrossRef]
- M. Egli, L.D. Williams, Q. Gao, A. Rich, Structure of the pure-spermine form of Z-DNA (magnesium free) at 1-Å resolution, Biochemistry 30 (1991) 11388‒11402. [CrossRef]
- H. Ohishi, et al., Interaction between the left-handed Z-DNA and polyamine-2. The crystal structure of the d(CG)3 and spermidine complex, FEBS Lett. 391 (1996) 153‒156.
- H. Ohishi, Y. Tozuka, Z. Da-Yang, T. Ishida, K. Nakatani, The rare crystallographic structure of d(CGCGCG)2: the natural spermidine molecule bound to the minor groove of left-handed Z-DNA d(CGCGCG)2 at 10 °C, Biochem. Biophys. Res. Commun. 358 (2007) 24‒28. [CrossRef]
- I.L. Cameron, N.K.R. Smith, T.B. Pool, Element concentration changes in mitotically active and postmitotic enterocytes. An X-ray microanalysis study, J. Cell Biol. 80 (1979) 444‒450. [CrossRef]
- National Center for Biotechnology Information (NCBI), C3H_HeJ_v1, https://www.ncbi.nlm.nih.gov/assembly/738461 (Bethesda, MD, 2016), retrieved 2016/11/12.
- S. Sarhan, N. Seiler, On the subcellular localization of the polyamines, Biol. Chem. Hoppe-Seyler 370 (1989) 1279‒1284. [CrossRef]
- M.P. Viola-Magni, P.B. Gahan, J. Pacy, Phospholipids in plant and animal chromatin, Cell Biochem. Funct. 3 (1985) 71‒78. [CrossRef]
- R. Hurst, et al., Establishing optimal selenium status: results of a randomized, double-blind, placebo-controlled trial, Am. J. Clin. Nutr. 91 (2010) 923‒931. [CrossRef]
- R. Muecke, et al., Whole blood selenium levels and selenium supplementation in patients treated in a family doctor practice in Golßen (state of Brandenburg, Germany): a laboratory study, Integr. Cancer Ther. 17 (2018) 1132‒1136.
- Y.-C. Park, P.D. Whanger, Toxicity, metabolism and absorption of selenite by isolated rat hepatocytes, Toxicology 100 (1995) 151‒162. [CrossRef]
- S.E. Calvo, D.J. Pagliarini, V.K. Mootha, Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans, Proc. Natl. Acad. Sci. U. S. A. 106 (2009) 7507‒7512. [CrossRef]
- M.L. Crowe, X.-Q. Wang, J.A. Rothnagel, Evidence for conservation and selection of upstream open reading frames suggests probable encoding of bioactive peptides, BMC Genomics 7 (2006), 16. [CrossRef]
- M. Kozak, Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs, Nucleic Acids Res. 12 (1984) 857-872. [CrossRef]
- W.D. Kumler, J.J. Eiler, The acid strength of mono and diesters of phosphoric acid. The n-alkyl esters from methyl to butyl, the esters of biological importance, and the natural guanidine phosphoric acids, J. Am. Chem. Soc. 65 (1943) 2355‒2361. [CrossRef]
- A. Katchalsky, J. Gillis, Theory of the potentiometric titration of polymeric acids, Recl. Trav. Chim. Pays-Bas 68 (1949) 879‒897. [CrossRef]
- W. Kuhn, H. Kuhn, Die Frage nach der Aufrollung von Fadenmolekülen in strömenden Lösungen, Helv. Chim. Acta 26 (1943) 1394‒1465.
- M. Mandelkern, J.G. Elias, D. Eden, D.M. Crothers, The dimensions of DNA in solution, J. Mol. Biol. 152 (1981) 153‒161. [CrossRef]
- R. Langridge, et al., The molecular configuration of deoxyribonucleic acid. II. Molecular models and their Fourier transforms, J. Mol. Biol. 2 (1960) 38‒64.
- A. Mermer, P. Starynowicz, Charge-density distribution in potassium dihydrogen phosphoglycolate – a comparison of phosphate and phosphonate groups, Acta Crystallogr. B 68 (2012) 625‒635. [CrossRef]
- D. Aikens, et al., The interactions between nucleic acids and polyamines. II. Protonation constants and 13C-NMR chemical shift assignments of spermidine, spermine, and homologs, Biophys. Chem. 17 (1983) 67‒74. [CrossRef]
- L.W. Tari, A.S. Secco, Base-pair opening and spermine binding—B-DNA features displayed in the crystal structure of a gal operon fragment: implications for protein–DNA recognition, Nucleic Acids Res. 23 (1995) 2065‒2073. [CrossRef]
- N. Wiberg (Ed.), Holleman–Wiberg, Lehrbuch der Anorganischen Chemie, 101. ed., Walter de Gruyter, Berlin, New York, 1995, p. 771.
- D.S.M. Shor, E.A. Struys, B.M. Hogema, K.M. Gibson, C. Jakobs, Development of a stable-isotope dilution assay for γ-aminobutyric acid (GABA) transaminase in isolated leukocytes and evidence that GABA and β-alanine transaminases are identical, Clin. Chem. 47 (2001) 525‒531. [CrossRef]
- Cellini, R. Montioli, S. Bianconi, J.P. López-Alonso, C. Borri Voltattorni, Construction, purification and characterization of untagged human liver alanine-glyoxylate aminotransferase expressed in Escherichia coli, Protein Pept. Lett. 15 849 (2008) 153‒159. [CrossRef]
- E. Oppici, et al., Crystal structure of the S187F variant of human liver alanine: glyoxylate aminotransferase associated with primary hyperoxaluria type I and its functional implications, Proteins 81 (2013) 1457‒1465.
- A.G. Matera, R.M. Terns, M.P. Terns, Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs, Nat. Rev. Mol. Cell. Biol. 8 (2007) 209‒220. [CrossRef]
- Llères, J. James, S. Swift, D.G. Norman, A.I. Lamond, Quantitative analysis of chromatin compaction in living cells using FLIM–FRET, J. Cell Biol. 187 (2009) 481‒496. [CrossRef]
- C.T. Rhodes, Determination of micro-pH in solid drug delivery systems, Drug Dev. Ind. Pharm. 25 (1999) 1221‒1222.
- F.H. Herbstein, M. Kapon, W. Schwotzer, Crystal structure of tetrakis(phenacetin) dihydrogentetraiodide dihydrate {[H5C2OC6H4N(H)C(CH3)=O]4 • H2I4 • 2 H2O}, Helv. Chim. Acta 66 (1983) 35‒43.
- P.B. Hitchcock, et al., Preparation of new vanadium(II) iodides and crystal structure of hexakis(acetonitrile)vanadium(II) (tetraiodide), J. Chem. Soc., Dalton Trans. (1994) 3683‒3687. [CrossRef]
- E.E. Genser, R.E. Connick, Exchange of iodide ion with triiodide ion studied by nuclear magnetic resonance, J. Chem. Phys. 58 (1973) 990‒996. [CrossRef]
- H.-U. Schenck, P. Simak, E. Haedicke, Structure of polyvinylpyrrolidone-iodine (povidone-iodine), J. Pharm. Sci. 68 (1979) 1505‒1509. [CrossRef]
- F.H. Herbstein, M. Kapon, Zigzag chains of alternating iodine molecules and triiodide ions in crystalline (phenacetin)2 • HI5, Nature Phys. Sci. 239 (1972) 153‒154.
- W. Saenger, The structure of the blue starch–iodine complex, Naturwissenschaften 71 (1984) 31‒36. [CrossRef]
- P.H. Svensson, L. Kloo, Synthesis, structure, and bonding in polyiodide and metal iodide–iodine systems, Chem. Rev. 103 (2003) 1649‒1684.
- C.D.S. Tomlin (Ed.), The Pesticide Manual: A World Compendium, Incorporating the Agrochemicals Handbook, eleventh ed., British Crop Protection Council, Farnham, Surrey, UK, 1997.
- K.R. Terpstra, A.J.J. Woortman, J.C.P. Hopman, Yellow dextrins: evaluating changes in structure and colour during processing, Starch/Stärke 62 (2010) 449‒457. [CrossRef]
- B. Cellini, M. Bertoldi, R. Montioli, A. Paiardini, C. Borri Voltattorni, Human wild-type alanine:glyoxylate aminotransferase and its naturally occurring G82E variant: functional properties and physiological implications, Biochem. J. 408 (2007) 39‒50. [CrossRef]
- H. Schuster, G. Schramm, W. Zillig, Die Struktur der Ribonucleinsäure aus Tabakmosaikvirus, Z. Naturforsch. B 11 (1956) 339‒345. [CrossRef]
- M.A. Lever, et al., A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types, Front. Microbiol. 6 (2015), 476. [CrossRef]
- E.H. Binns, The dissociation constant of phenol in water between 25°C and 60°C, Trans. Faraday Soc. 55 (1959) 1900‒1903. [CrossRef]
- Y. Takeda, et al., Determination of protonation sites in thermospermine and in some other polyamines by 15N and 13C nuclear magnetic resonance spectroscopy, Eur. J. Biochem. 130 (1983) 383‒389. [CrossRef]
- C.H. Waddington, The epigenotype, Endeavour (Engl. Ed. Lond.) 1 (1942) 18‒20.
- F.H.C. Crick, J.D. Watson, The complementary structure of deoxyribonucleic acid, Proc. R. Soc. Lond. A Math. Phys. Sci. 223 (1954) 80‒96. [CrossRef]
- M.K. Schlegel, L.-O. Essen, E. Meggers, Duplex structure of a minimal nucleic acid, J. Am. Chem. Soc. 130 (2008) 8158‒8159. [CrossRef]
- U. Heinemann, H. Lauble, R. Frank, H. Blöcker, Crystal structure analysis of an A-DNA fragment at 1.8 Å resolution: d(GCCCGGGC), Nucleic Acids Res. 15 (1987) 9531‒9550. [CrossRef]
- P.K. Mandal, S. Venkadesh, N. Gautham, Structure of the tetradecanucleotide d(CCCCGGTACCGGGG)2 as an A-DNA duplex, Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68 (2012) 393‒399. [CrossRef]
- W. Saenger, W.N. Hunter, O. Kennard, DNA conformation is determined by economics in the hydration of phosphate groups, Nature 324 (1986) 385‒388. [CrossRef]
- P. Leder, M. Nirenberg, RNA codewords and protein synthesis, II. Nucleotide sequence of a valine RNA codeword, Proc. Natl. Acad. Sci. U. S. A. 52 (1964) 420‒427. [CrossRef]
- P.S. Sunkara, S. Ramakrishna, K. Nishioka, P.N. Rao, The relationship between levels and rates of synthesis of polyamines during mammalian cell cycle, Life Sci. 28 (1981) 1497‒1506. [CrossRef]
- G. Li, G. Sudlow, A.S. Belmont, Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning, J. Cell Biol. 140 (1998) 975‒989. [CrossRef]
- T.J. Thomas, U.B. Gunnia, T. Thomas, Polyamine-induced B-DNA to Z-DNA conformational transition of a plasmid DNA with (dG-dC)n insert, J. Biol. Chem. 266 (1991) 6137‒6141.
- H. Deng, V.A. Bloomfield, J.M. Benevides, G.J. Thomas Jr., Structural basis of polyamine–DNA recognition: spermidine and spermine interactions with genomic B-DNAs of different GC content probed by Raman spectroscopy, Nucleic Acids Res. 28 (2000) 3379‒3385. [CrossRef]
- A. Rich, S. Zhang, Z-DNA: the long road to biological function, Nat. Rev. Genet. 4 (2003) 566‒572. [CrossRef]
- S.C. Ha, K. Lowenhaupt, A. Rich, Y.-G. Kim, K.K. Kim, Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases, Nature 437 (2005) 1183‒1186. [CrossRef]
- L. Flohé, J.R. Andreesen, R. Brigelius-Flohé, M. Maiorino, F. Ursini, Selenium, the element of the moon, in life on earth, IUBMB Life 49 (2000) 411‒420.
- J.J. Berzelius, Lettre de M. Berzelius à M. Berthollet sur deux Métaux nouveaux, Ann. Chim. Phys. (Paris) 7 (1817) 199‒206.
- S. Borah, P.P. Kumar, Ab initio molecular dynamics study of Se(IV) species in aqueous environment, Phys. Chem. Chem. Phys. 18 (2016) 26755‒26763. [CrossRef]
- S. Nafisi, M. Montazeri, F. Manouchehri, The effect of Se salts on DNA structure, J. Photochem. Photobiol. B 113 (2012) 36‒41. [CrossRef]
- S. Nafisi, F. Manouchehri, M. Montazeri, RNA adducts with Na2SeO4 and Na2SeO3 – Stability and structural features, J. Mol. Struct. 1006 (2011) 547‒552. [CrossRef]
- J. Wu, G.H. Lyons, R.D. Graham, M.F. Fenech, The effect of selenium, as selenomethionine, on genome stability and cytotoxicity in human lymphocytes measured using the cytokinesis-block micronucleus cytome assay, Mutagenesis 24 (2009) 225‒232. [CrossRef]
- A. Graupner, et al., Genotoxic effects of two-generational selenium deficiency in mouse somatic and testicular cells, Mutagenesis 30 (2015) 217‒225. [CrossRef]
- K.M. Abdo, National Toxicology Program (NTP). Technical report on toxicity studies of sodium selenate and sodium selenite (CAS Nos. 13410-01-0 and 10102-18-8) administered in drinking water to F344/N rats and B6C3F1 mice (NIH Publication 94-3387), Toxic. Rep. Ser. 38 (1994) 1‒127.
- M.L. Rueppel, B.B. Brightwell, J. Schaefer, J.T. Marvel, Metabolism and degradation of glyphosate in soil and water, J. Agric. Food Chem. 25 (1977) 517‒528. [CrossRef]
- C.M. Benbrook, Trends in glyphosate herbicide use in the United States and globally, Environ. Sci. Eur. 28 (2016), 3. [CrossRef]
- R.P. Holmes, D.G. Assimos, Glyoxylate synthesis, and its modulation and influence on oxalate synthesis, J. Urol. 160 (1998) 1617‒1624.
- Helgstrand, et al., Trisodium phosphonoformate, a new antiviral compound, Science 201 (1978) 819‒821. [CrossRef]
- C.L.K. Sabourin, J.M. Reno, J.A. Boezi, Inhibition of eucaryotic DNA polymerases by phosphonoacetate and phosphonoformate, Arch. Biochem. Biophys. 187 (1978) 96‒101. [CrossRef]
- S. Khan, S. Ahmed, Role of swi7H4 mutant allele of DNA polymerase α in the DNA damage checkpoint response, PLoS One 10 (2015), e0124063. [CrossRef]
- S. Rosa, P. Shaw, Insights into chromatin structure and dynamics in plants, Biology 2 (2013) 1378‒1410. [CrossRef]
- H. Zeng, Selenium as an essential micronutrient: roles in cell cycle and apoptosis, Molecules 14 (2009) 1263‒1278. [CrossRef]
- G.D. Frenkel, Effects of sodium selenite and selenate on DNA and RNA synthesis in vitro, Toxicol. Lett. 25 (1985) 219‒223. [CrossRef]
- L.A. Loeb, B.D. Preston, Mutagenesis by apurinic/apyrimidinic sites. Ann. Rev. Genet. 20 (1986) 201‒230.
- C.O. Miller, F. Skoog, F.S. Okumura, M.H. von Saltza, F.M. Strong, Isolation, structure and synthesis of kinetin, a substance promoting cell division, J. Am. Chem. Soc. 78 (1956) 1375‒1380. [CrossRef]
- J. Barciszewski, G.E. Siboska, B.O. Pedersen, B.F.C. Clark, S.I.S. Rattan, A mechanism for the in vivo formation of N6-furfuryladenine, kinetin, as a secondary oxidative damage product of DNA, FEBS Lett. 414 (1997) 457‒460.
- J. Barciszewski, F. Massino, B.F.C. Clark, Kinetin—a multiactive molecule, Int. J. Biol. Macromol. 40 (2007) 182‒192.
- W.D. Fuller, R.A. Sanchez, L.E. Orgel, Studies in prebiotic synthesis. VI. Synthesis of purine nucleosides, J. Mol. Biol. 67 (1972) 25‒33.
- M.-C. Maurel, O. Convert, Chemical structure of a prebiotic analog of adenosine, Orig. Life Evol. Biosph. 20 (1990) 43‒48. [CrossRef]
- T.J.L. Mustard, D.J. Mack, J.T. Njardarson, P.H.-Y. Cheong, Mechanism and the origins of stereospecificity in copper-catalyzed ring expansion of vinyl oxiranes: a traceless dual transition-metal-mediated process, J. Am. Chem. Soc. 135 (2013) 1471‒1475. [CrossRef]
- E.A. Ilardi, J.T. Njardarson, Ring expansions of vinyloxiranes, -thiiranes, and -aziridines: synthetic approaches, challenges, and catalytic success stories, J. Org. Chem. 78 (2013) 9533‒9540. [CrossRef]
- V.J. Koller, et al., Cytotoxic and DNA-damaging properties of glyphosate and Roundup in human-derived buccal epithelial cells, Arch. Toxicol. 86 (2012) 805‒813. [CrossRef]
- R.H. Coupe, S.J. Kalkhoff, P.D. Capel, C. Gregoire, Fate and transport of glyphosate and aminomethylphosphonic acid in surface waters of agricultural basins, Pest Manag. Sci. 68 (2012) 16‒30. [CrossRef]

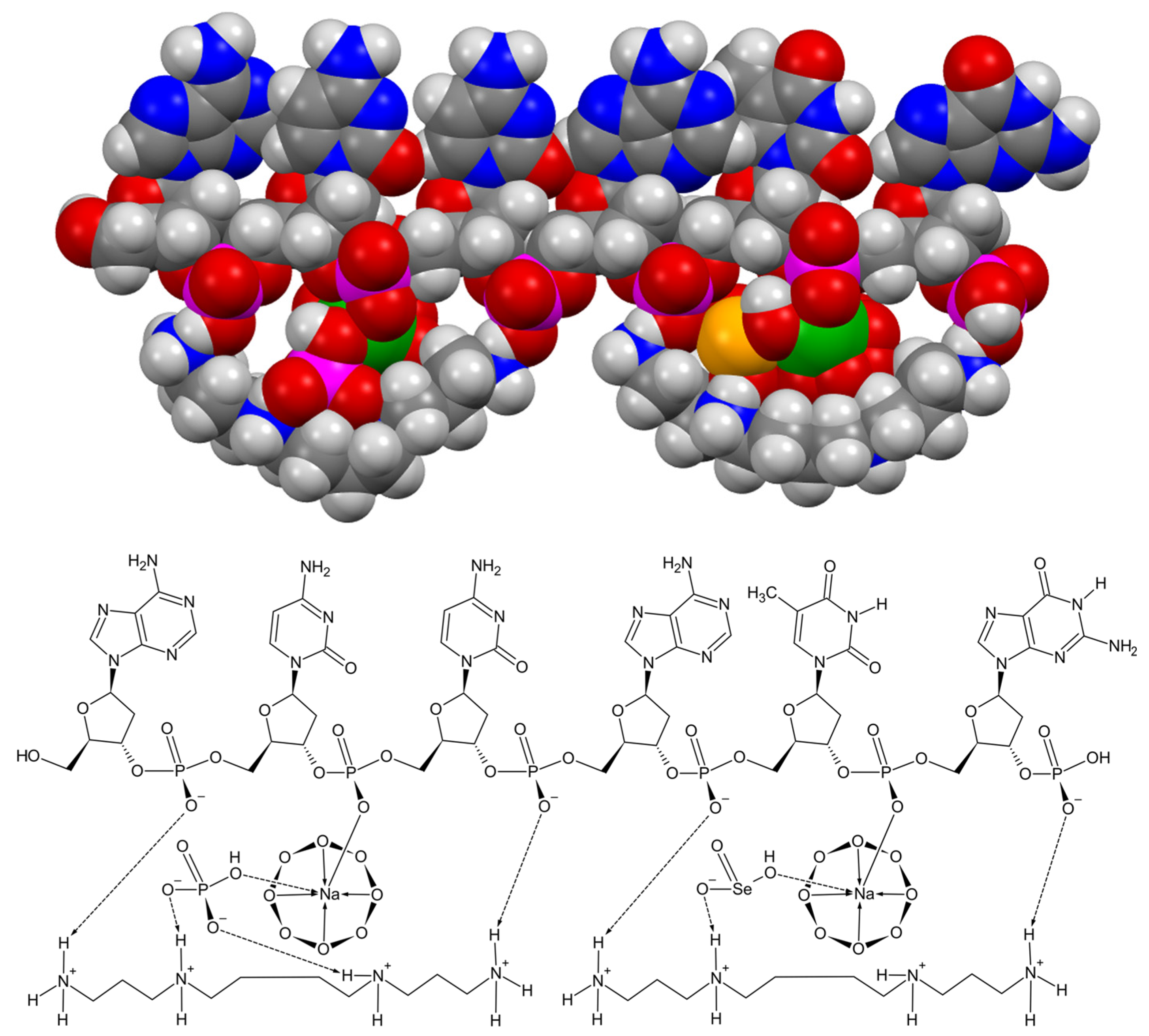
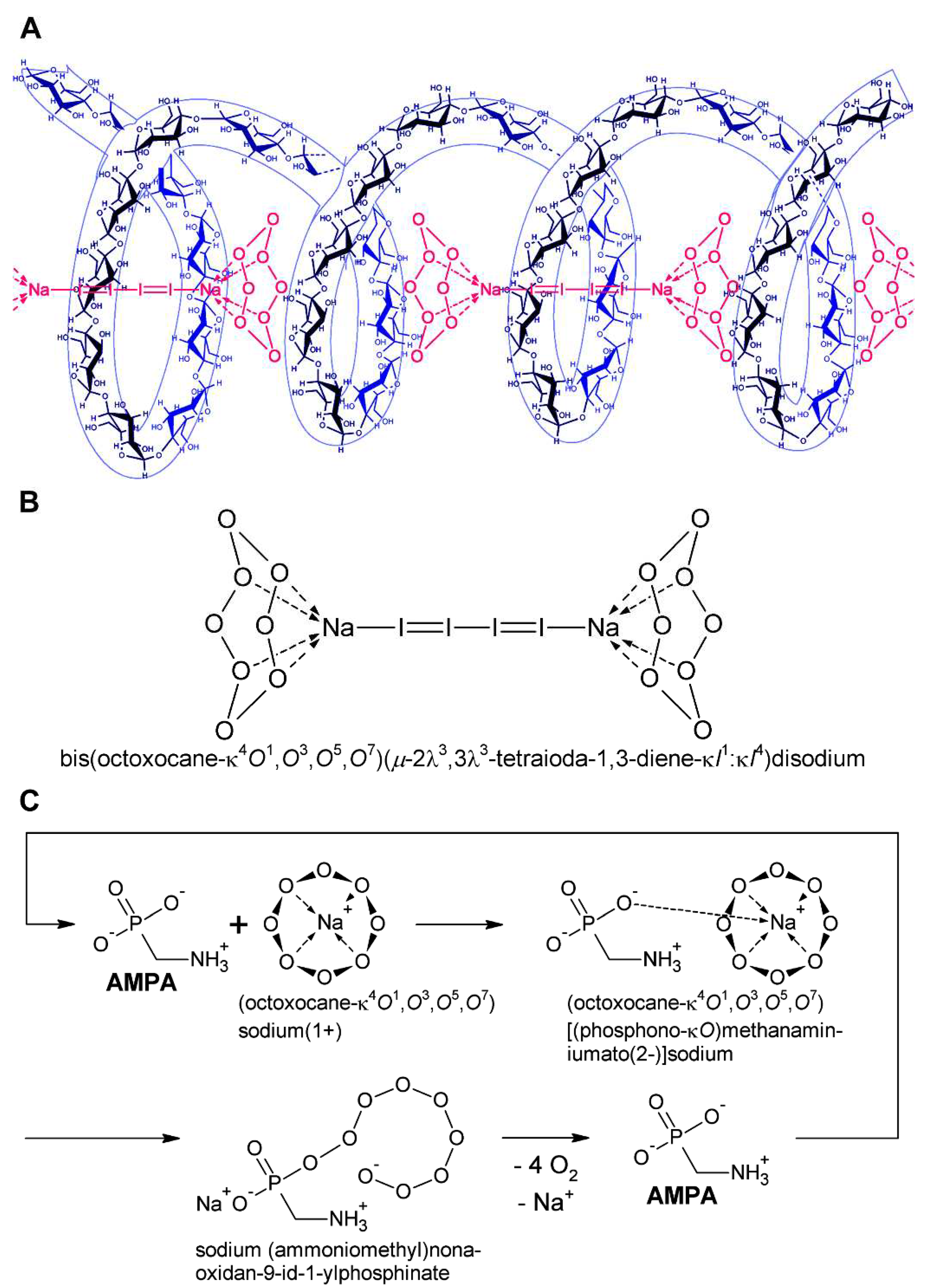
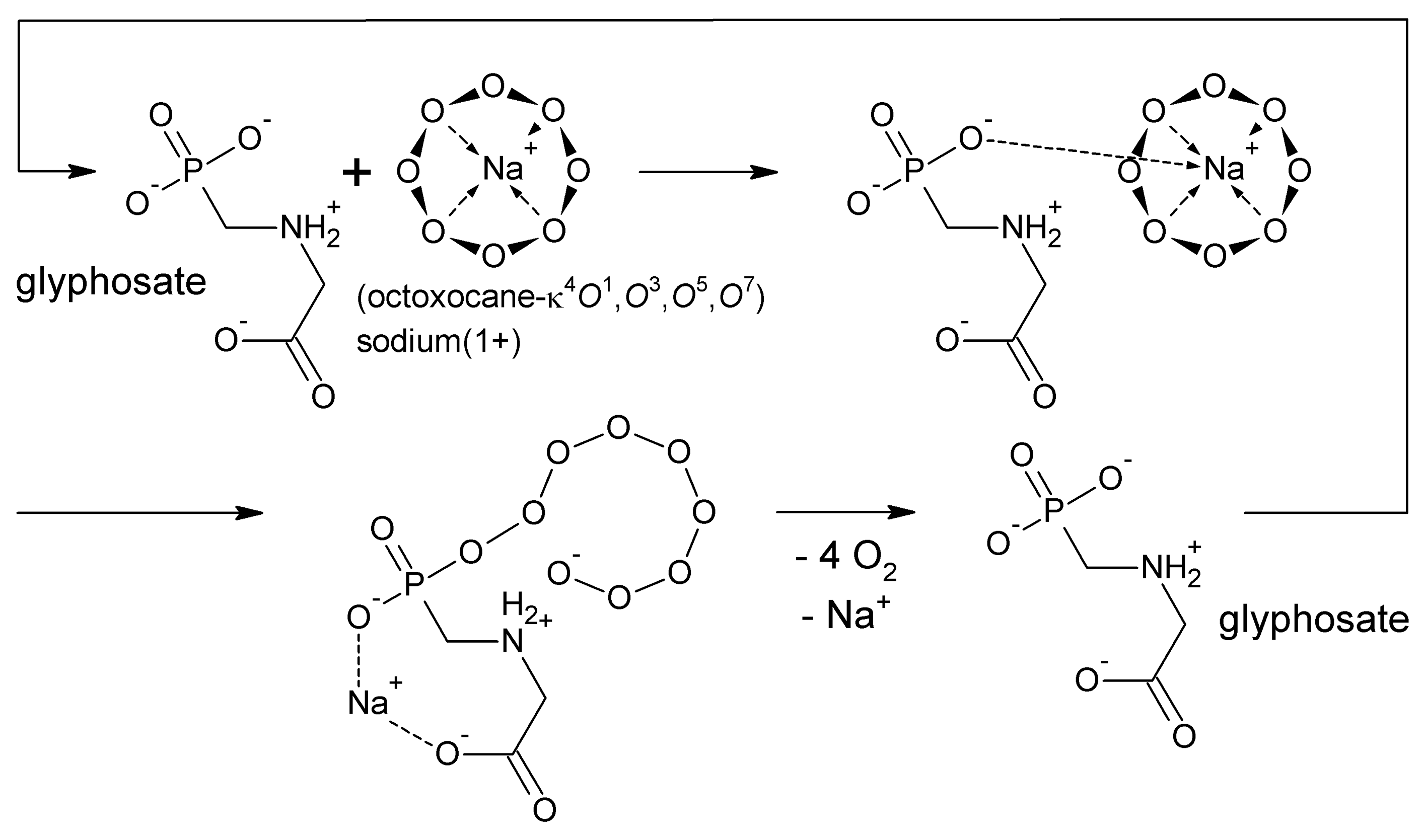
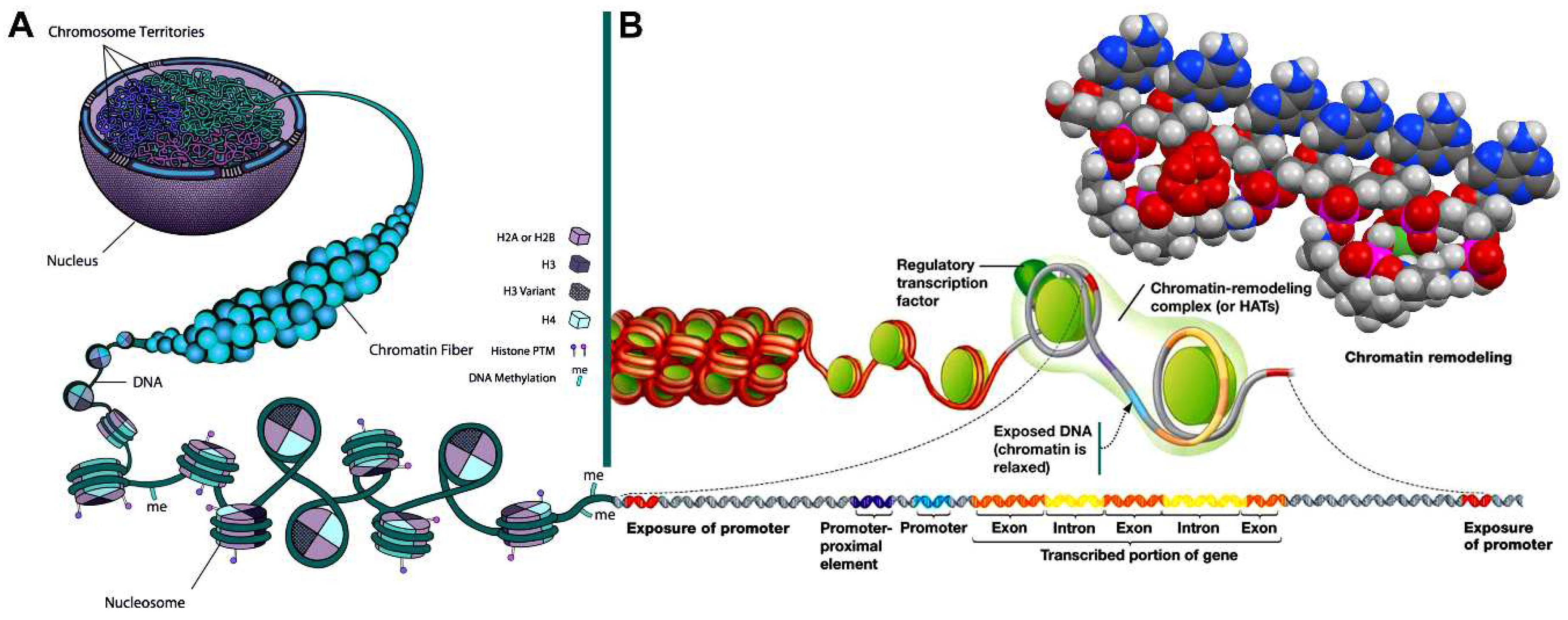
| Cell cycle phase | Heterochromatin | Euchromatin | |||
|---|---|---|---|---|---|
| Interphase | Function (concise) | Spermine-occupation? | Cyclo-O8-Na+-occupation? | Spermine-occupation? | Cyclo-O8-Na+-occupation? |
| G0 (Gap 0) | Resting and quiescence | No | No | Yes (2 ×) – With cyclo-O8-Na+ | Yes (2 ×) – On ‘open’ Chr |
| G1 (Gap 1) | Transcription and histone synthesis | No | No | Yes (2 ×) – With cyclo-O8-Na+ | Yes (2 ×) – On ‘open’ Chr |
| S (Synthesis) | DNA synthesis | No | No | Yes (2 ×) – With cyclo-O8-Na+ | Yes (2 ×) – On ‘open’ Chr |
| G2 (Gap 2) | Translation | No | No | Yes (2 ×) – With cyclo-O8-Na+ | Yes (2 ×) – On ‘open’ Chr |
| Mitosis | Function (concise) | Spermine-occupation? | Cyclo-O8-Na+-occupation? | Spermine-occupation? | Cyclo-O8-Na+-occupation? |
| Prophase | Chr condenses into chromosomes, nucleolus disappears | Yes (1 ×) – Condensing Chr | No | Yes (1 ×) – Condensing Chr | No |
| Prometaphase | Kinetochore and polar microtubules attach, mitotic spindle formed, nucleus disappears | Yes (1 ×) – Condensed Chr | No | Yes (1 ×) – Condensed Chr | No |
| Metaphase | Centrosomes pull chromosomes, chromosome centromeres line up at metaphase plate | Yes (1 ×) – Highly condensed Chr | No | Yes (1 ×) – Highly condensed Chr | No |
| Anaphase | Chromosomes break at centromeres, sister chromatids separated by microtubules | Yes (1 ×) – Maximally condensed Chr in late anaphase | No | Yes (1 ×) – Maximally condensed Chr in late anaphase | No |
| Telophase | Chr reformed from chromosomes, nucleus and nucleolus reappear | Yes (1 ×) – Maximally condensed Chr in early telophase | No | Yes (1 ×) – Maximally condensed Chr in early telophase | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




