Submitted:
20 December 2025
Posted:
23 December 2025
You are already at the latest version
Abstract
Callus induction is frequently employed in rice breeding to either regenerate entire plants from callus cultures or incorporate desired features through genetic alteration. Plant growth regulators, especially auxin, are known for its role in the embryogenic callus proliferation and plant growth. In the present study, the effect of 2, 4-D in callus induction and plant regeneration in 11 rice varieties of Manipur was evaluated. Experimental results revealed that the optimum level of 2,4-D for callus induction varies for different rice varieties. In most of the rice varieties, the optimum dose of callus induction was found to be 2 mg/L. However, 3mg/L concentration of 2,4-D was found to be the most efficient dose in Mani-10, Mani-12, and Mani-13, while a 4 mg/L concentration of 2,4-D was most efficient for callus induction in Mani-14. Using 23 indel markers and 13 SSR markers, genetic diversity and marker trait association were also examined in the 11 rice genotypes. The SSR profile clearly indicates significant heterogeneity among rice accessions and also revealed four sub-populations or groups. Among all the rice genotypes examined, Mani-4 and Ma-ni-5 exhibited the greatest similarity, potentially attributable to their shared lineage. Marker trait association study reveals that the markers RM21, RM411, RM569, RCu4, and RCu5, with R2 values of 0.517, 0.451, 0.451, 0.604, and 0.604, respectively, were found to have genetic correlations with 2,4-D growth hormone concentrations of 0.5 mg/L. This study’s findings will help to conserve rice germplasm and build high-yielding, sustainable rice breeding programs to assure global food security.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Surface Sterilization of Seeds
2.3. Callus Induction in Media
2.4. Identification of Indica-Japonica Specific InDel Markers of Candidate Genes
2.5. Genomic DNA Isolation
2.6. PCR Amplification and Gel Electrophoresis
2.7. Allele Scoring and Diversity Analysis
2.8. Candidate-Based Association Analysis
2.9. Statistical Analysis
3. Results and Discussion
3.1. Callus Induction Frequency Analysis in 11 High-Yielding Indica Rice Varieties
3.2. Identification of Indica-Japonica Specific InDel Markers of Candidate Genes
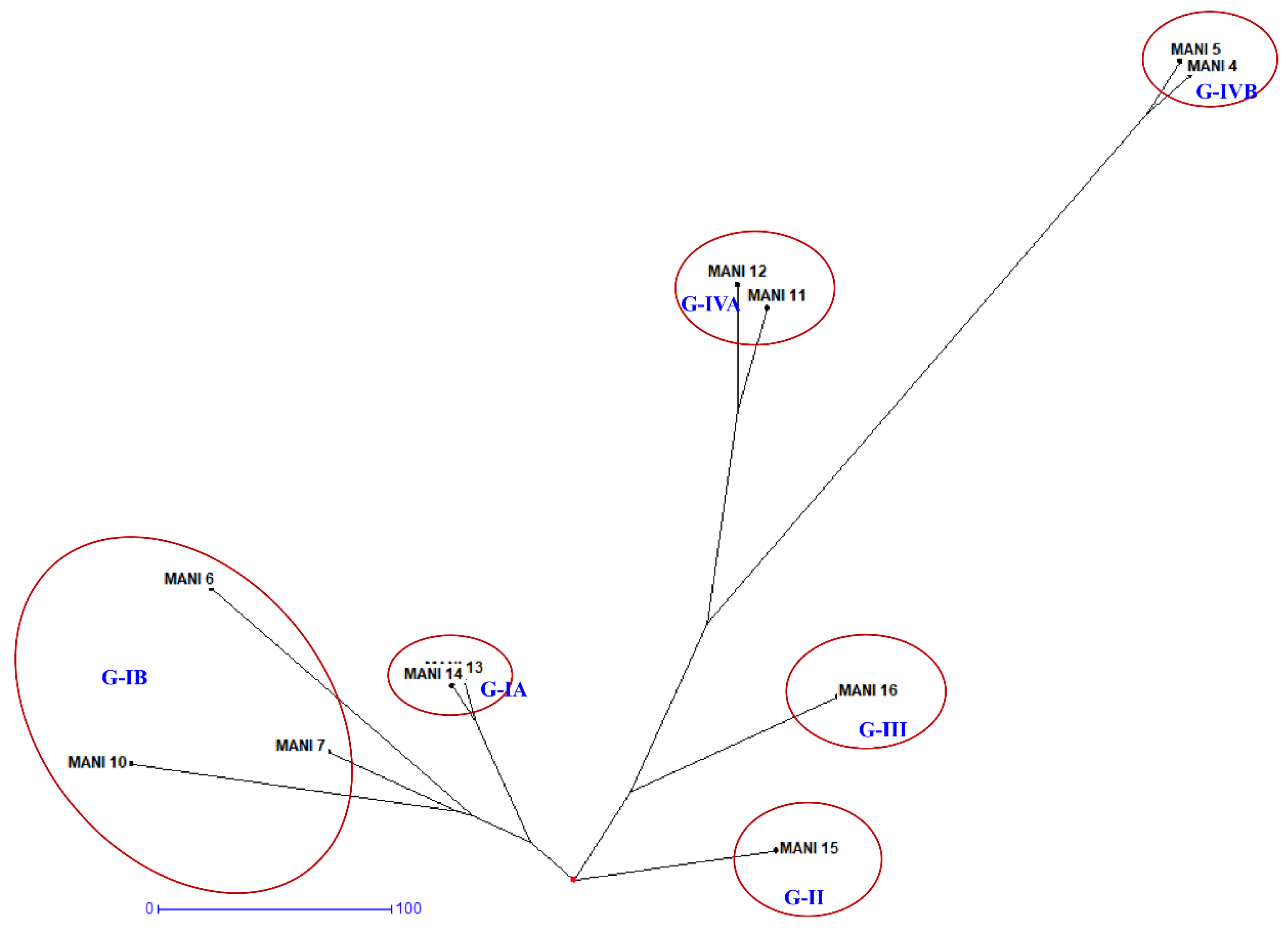
3.3. Genetic Diversity and Relationship Analysis
3.4. Candidate-Based Association Analysis for Mature Rice Seed Culturability
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| QTL | quantitative trait loci |
| 2,4-D | 2,4-dichlorophenoxyacetic acid |
| PVE | Percentage Variance Explained |
| SSR | Simple sequence repeats |
| CTAB | Cetyltrimethyl ammonium bromide |
| Hobs | Observed frequency |
| (HExp) | Expected frequency |
| PIC | Polymorphic information content |
| SEM | Standard error of the mean |
| MS Media | Murashige and Skoog media |
References
- Ngangkham, U.; Nath, M.; Dokku, P.; AmithaMithra, S.V.; Ramamurthy, S.; Singh, N.K.; Sharma, R.P.; Mohapatra, T. An EMS induced new sequence variant, TEMS5032 in the coding region of SRS3 gene leads to shorter grain length in rice (Oryza sativa L.). J. Appl. Genet. 2018, 59(4):377-389. [CrossRef]
- Redoña, E.D.; Mula, F.L. Some imperatives and challenges for rice biotechnology in Asian national agricultural research and extension systems. Asian Biotechnol. Dev. Rev. http://www. ris. org. in/abdr_ nov2005. htm. 2004.
- Tripathy, S.K. Optimization of culture variables for efficient callus induction and rapid plant regeneration in zinc rich rice (Oryza sativa L.) cv. “Chittimuthyalu”. J. Appl. Biol. Biotechnol. 2021, 9, 1-9.
- Wu, J.; Chang, X.; Li, C.; Zhang, Z.; Zhang, J.; Yin, C.; Ma, W.; Chen, H.; Zhou, F.; Lin, Y. QTLs related to rice callus regeneration ability: localization and effect verification of qPRR3. Cells 2022, 11, 4125. [CrossRef]
- Ali, J.; Nicolas, K.L.C.; Akther, S.; Torabi, A.; Ebadi, A.A.; Marfori-Nazarea, C.M.; Mahender, A. Improved anther culture media for enhanced callus formation and plant regeneration in rice (Oryza sativa L.). Plants 2021, 10, 839. [CrossRef]
- Yang, Y.S.; Zheng, Y.D.; Chen, Y.L.; Jian, Y.Y.; Improvement of plant regeneration from long-term cultured calluses of Taipei 309, a model rice variety in in vitro studies. Plant Cell Tissue Organ Cult. 1999, 57,199-206. [CrossRef]
- Lee, K.; Jeon, H.; Kim, M. Optimization of a mature embryo-based in vitro culture system for high-frequency somatic embryogenic callus induction and plant regeneration from japonica rice cultivars. Plant Cell Tissue Organ Cult. 2002, 71, 237-244. [CrossRef]
- Dabul, A.N.G.; Belefant-Miller, H.; RoyChowdhury, M.; Hubstenberger, J.F.; Lorence, A.; Phillips, G.C. Screening of a broad range of rice (Oryza sativa L.) germplasm for in vitro rapid plant regeneration and development of an early prediction system. In Vitro Cell. Dev. Biol.-Plant 2009, 45, 414-420. [CrossRef]
- Zaidi, M.A.; Narayanan, M.; Sardana, R.; Taga, I.; Postel, S.; Johns, R.; McNulty, M.; Mottiar, Y.; Mao, J.; Loit, E.; Altosaar, I. Optimizing tissue culture media for efficient transformation of different indica rice genotypes. Agron. Res. 2006, 4, 563-575.
- Zhang, K.; Su, J.; Xu, M.; Zhou, Z.; Zhu, X.; Ma, X.; Hou, J.; Tan, L.; Zhu, Z.; Cai, H.; Liu, F. A common wild rice-derived BOC1 allele reduces callus browning in indica rice transformation. Nat. Commun. 2020, 11, p.443.
- Taguchi-Shiobara, F.; Lin, S.Y.; Tanno, K.; Komatsuda, T.; Yano, M.; Sasaki, T.; Oka, S. Mapping quantitative trait loci associated with regeneration ability of seed callus in rice, Oryza sativa L. Theor. Appl. Genet. 1997, 95, 828-833. [CrossRef]
- Taguchi-Shiobara, F.; Yamamoto, T.; Yano, M.; Oka, S. Mapping QTLs that control the performance of rice tissue culture and evaluation of derived near-isogenic lines. Theor. Appl. Genet. 2006, 112, 968-976. [CrossRef]
- Nishimura, A.; Ashikari, M.; Lin, S.; Takashi, T.; Angeles, E.R.; Yamamoto, T.; Matsuoka, M. Isolation of a rice regeneration quantitative trait loci gene and its application to transformation systems. Proc. Natl. Acad. Sci. 2005, 102, 11940-11944. [CrossRef]
- Ozawa, K.; Kawahigashi, H. Positional cloning of the nitrite reductase gene associated with good growth and regeneration ability of calli and establishment of a new selection system for Agrobacterium-mediated transformation in rice (Oryza sativa L.). Plant Sci. 2006, 170, 384-393. [CrossRef]
- Zhao, L.; Zhou, H.; Lu, L.; Liu, L.; Li, X.; Lin, Y.; Yu, S.; Identification of quantitative trait loci controlling rice mature seed culturability using chromosomal segment substitution lines. Plant Cell Rep. 2009, 28, 247-256. [CrossRef]
- Li, S.; Yan, S.; Wang, A.H.; Zou, G.; Huang, X.; Han, B.; Qian, Q.; Tao, Y. Identification of QTLs associated with tissue culture response through sequencing-based genotyping of RILs derived from 93-11× Nipponbare in rice (Oryza sativa). Plant Cell Rep. 2013, 32, 103-116. [CrossRef]
- Zhang, K.; Yin, Z.; Xu, X.; Pu, C.; Li, Q.; Wu, D.; Sun, C.; Fu, Y. Quantitative trait loci for mature embryo culturability traits from Yuanjiang common wild rice (Oryza rufipogon Griff.). Indian J. Genet. Plant Breed. 2016, 76,167-172. [CrossRef]
- Doyle, J.; Doyle, J. Isolation of plant DNA from fresh tissue. Focus 1990,12, 13–15.
- Ngangkham, U.; Dash, S.; Parida, M.; Samantaray, S.; Nongthombam, D.; Yadav, M.K.; Kumar, A.; Chidambaranathan, P.; Katara, J.L.; Patra, B.C.; Bose, L.K. The potentiality of rice microsatellite markers in assessment of cross species transferability and genetic diversity of rice and its wild relatives. 3 Biotech. 2019. [CrossRef]
- Ngangkham, U.; Katara, J.L.; Shanmugavadivel, P.S.; Yadav, M.K.; Yadav, S.; Devachandra, N.; Samantaray, S.; Bose, L.K. Identification and characterization of polymorphic genic SSR markers between cultivated (Oryza sativa) and Indian wild rice (Oryza nivara). Indian J. Biotechnol. 2020, 19, 299–310.
- Stipoljev, S.; Safner, T.; Gančević, P.; Galov, A.; Stuhne, T.; Svetličić, I.; Grignolio, S.; Cassinello, J.; Šprem, N. Population structure and genetic diversity of non-native aoudad populations. Sci. Rep. 2021, 11, 12300. [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292.
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin software. https:// darwin. cirad. fr/ darwin. 2006.
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633-2635. [CrossRef]
- Hoque, M.E; Mansfield, J.W. Effect of genotype and explant age on callus induction and subsequent plant regeneration from root-derived callus of Indica rice genotypes. Plant Cell Tissue Organ Cult. 2004, 78, 217-223.
- Upadhyaya, G.; Sen, M.; Roy, A. In vitro callus induction and plant regeneration of rice (Oryza sativa L.) var. Sita, Rupali and Swarna Masuri. Asian J. Plant Sci. Res. 2015, 5, 24-27.
- Din, A.R.J.M.; Fauziah, I.A.; Sarmidi, M.R. Improvement of efficient in vitro regeneration potential of mature callus induced from Malayasian upland rice seed (Oryza sativa L. cv. Panderas). Saudi J. Biol. Sci. 2016, 23, S69-S77. [CrossRef]
- Mostafiz, S.B.; Wagiran, A. Efficient callus induction and regeneration in selected indica rice. Agronomy 2018, 8, 1-18. [CrossRef]
- Carsono, N.; Juwendah, E.; Sari, S.; Damayanti, F.; Rachmadi, M. Optimize 2,4-D concentration and callus induction time enhance callus proliferation and plant regeneration of three rice genotypes. Biodiversitas 2021, 22, 2555-2560. [CrossRef]
- Khan, M.N.M. In vitro callus induction of aromatic rice depends on the concentration of 2, 4-D. Malays. J. Halal Res. 2019, 2, 9-13.
- Abd Rahman, Z.; Seman, Z.A.; Othman, A.N.; Ab Ghaffar, M.B.; Ab Razak, S.; Yusof, M.F.M.; Nasir K.H.; Ahmad, K.; Chow, Y.L.; Subramaniam, S. Efficient callus induction and plant regeneration of Malaysian indica rice MR219 using anther culture. Biocatal. Agric. Biotechnol 2021, 31, 101865. [CrossRef]
- Kamolsukyeunyong, W.; Dabbhadatta, Y.; Jaiprasert, A.; Thunnom, B.; Poncheewin, W.; Wanchana, S.; Ruanjaichon, V.; Toojinda, T.; Burns P. Genome-wide association analysis identifies candidate loci for callus induction in rice (Oryza sativa L.). Plants 2024, 13, 2112. [CrossRef]
- Serrote, C.M.L.; Reiniger, L.R.S.; Silva, K.B.; dos Santos Rabaiolli, S.M.; Stefanel, C.M. Determining the polymorphism information content of a molecular marker. Gene 2020, 726, 144175. [CrossRef]
- Singh, I.M.; Ngangkham, U.; Sarika, K.; Devi, Y.S.; Singh, T.S.; Singh, T.B.; Singh, K.R.; Devi, E.L.; Kumar, M.; Nameirakpam, M.; Arambam, U.; Diviya, T.; Laha, R. Genetic diversity and DNA fingerprinting of rice varieties of Manipur using microsatellite markers. Electron. J. Plant Breed. 2024, 15, 504-514. [CrossRef]
- Devi, Y.S.; Ngangkham, U.; Singh, T.S.; Singh, T.B.; Singh, K.R.; Devi, E.L.; Roy, S.; Devanna, P.; Konsam, S.; Yengkhom, B.K.; Kumar, A.; Philanim W.S.; Laha, R. Genetic diversity and marker-trait association analysis in Manipur rice germplasm using microsatellite markers. Genet. Resour. Crop Evol. 2025, 72, 5311-5327. [CrossRef]
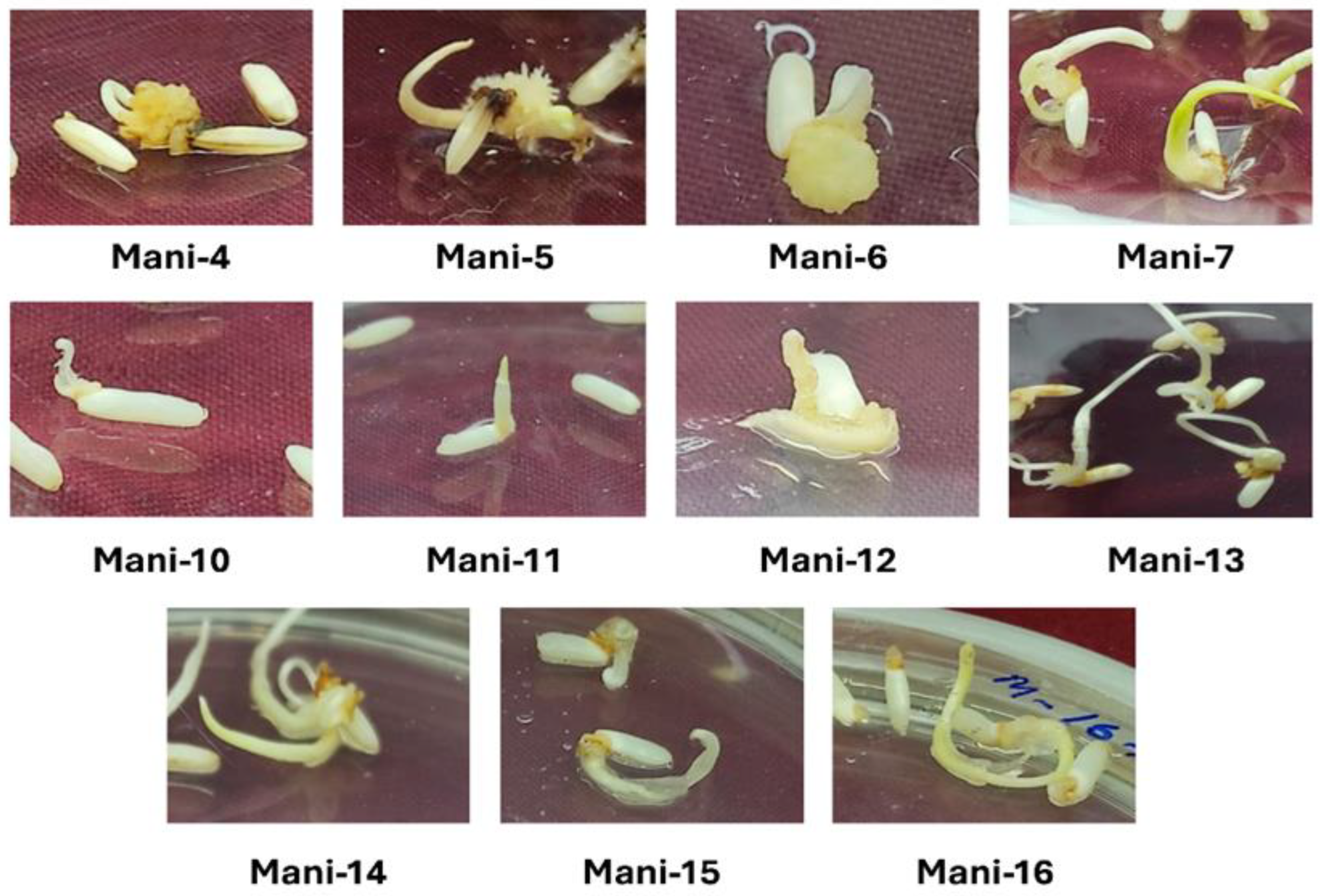
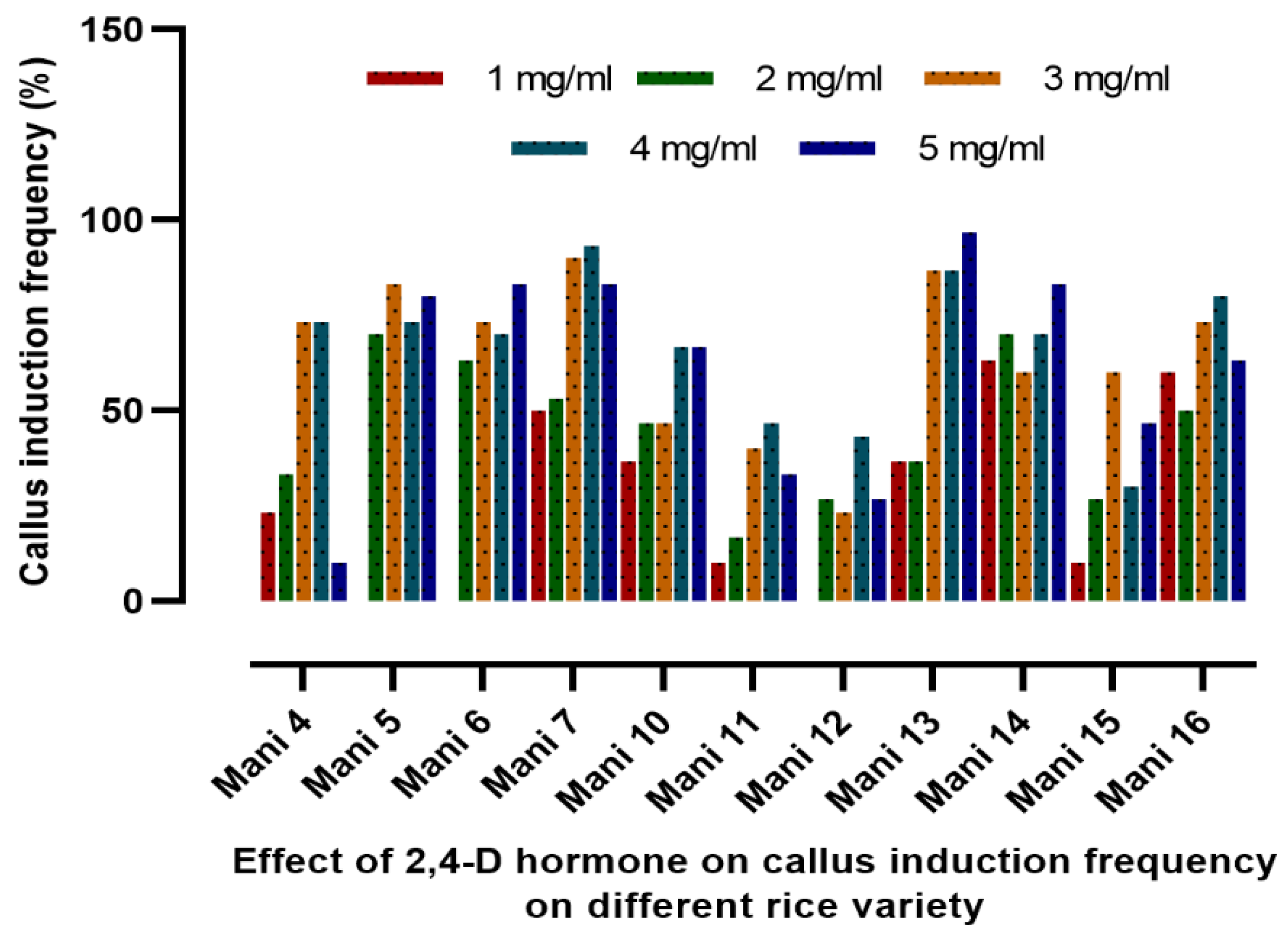
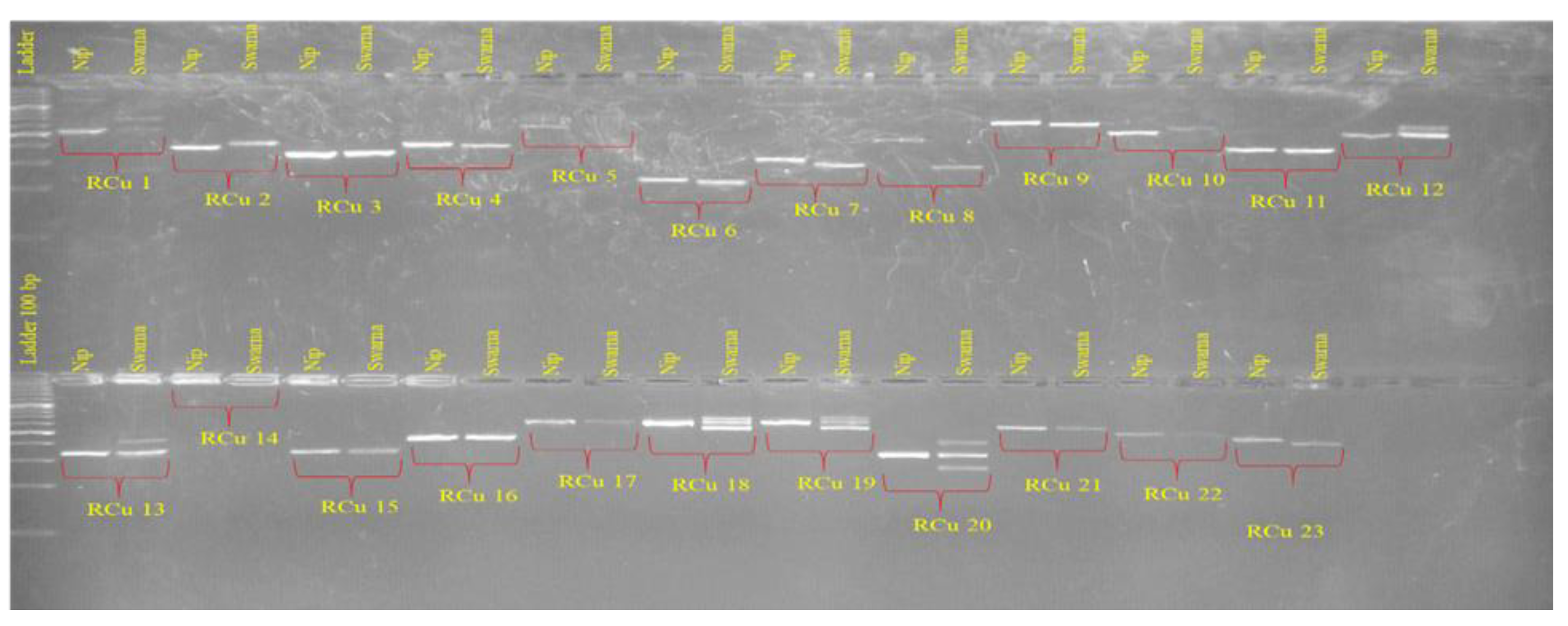
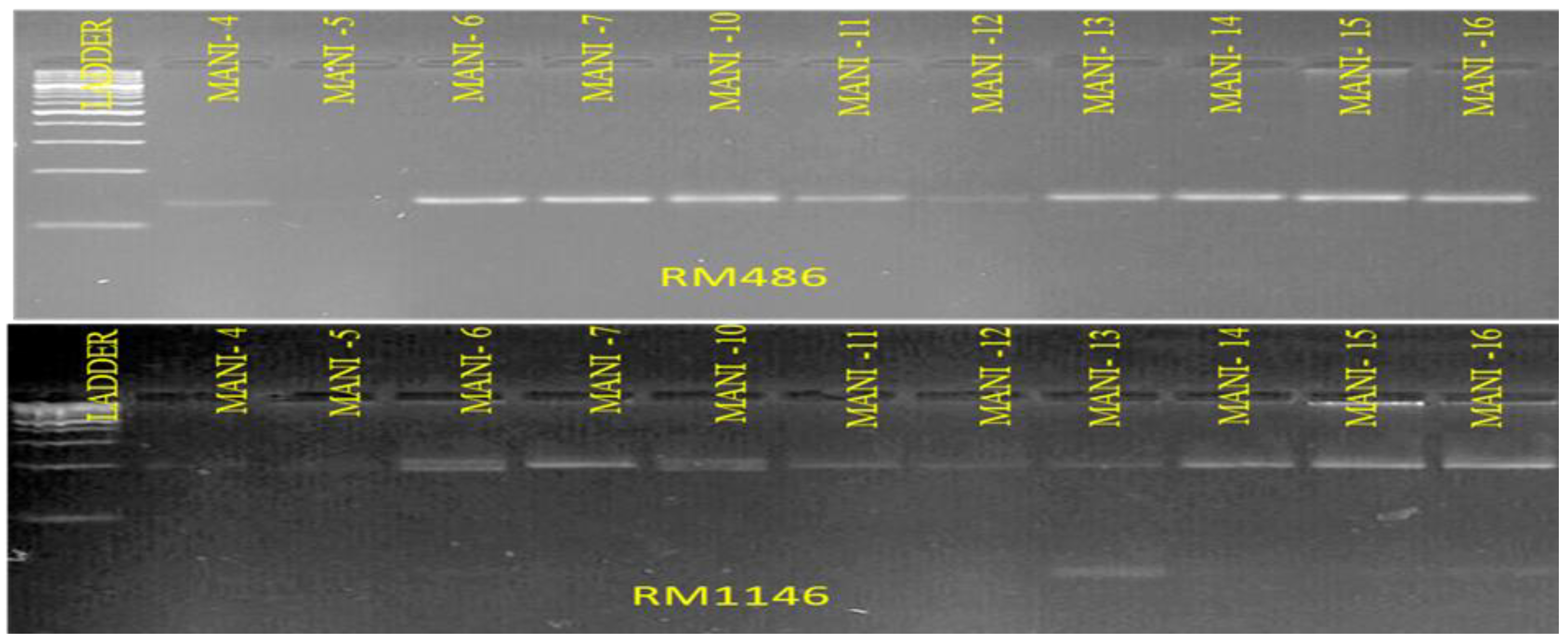
| Sl No. | Markers | Primer Sequences | PVE (%) |
| 1 | RM1146 | Forward-5′-TCTCCCTATTCCCGTGTAAATCG-3′ Reverse-5′-CCCGATGATCGATTGTACCTAGC-3′ |
33.93 |
| 2 | RM21 | Forward-5′-ACAGTATTCCGTAGGCACGG-3′ Reverse-5′-GCTCCATGAGGGTGGTAGAG-3′ |
4.18 |
| 3 | RM224 | Forward-5′-ATCGATCGATCTTCACGAGG-3′ Reverse-5′-TGCTATAAAAGGCATTCGGG-3′ |
4.64 |
| 4 | RM322 | Forward-5′-CAAGCGAAAATCCCAGCAG-3′ Reverse-5′-GATGAAACTGGCATTGCCTG-3′ |
9.02 |
| 5 | RM348 | Forward-5′-CATGAAGCTGTGTTGCTGTTGC-3′ Reverse-5′-CGCTACTAATAGCAGAGAGACCATCG-3′ |
15.17 |
| 6 | RM411 | Forward-5′-GTAGGAAATTCTTCGCCAGATGC-3′ Reverse-5′-CCGAGACTTGGAACAATCTTAGGC-3′ |
9.35 |
| 7 | RM414 | Forward-5′-CAAGGAAGATCTTGTGGACCATGC-3′ Reverse-5′-CTGCAGATGCAGAGGCAGAGG-3′ |
10.23 |
| 8 | RM42 | Forward-5′-ATCCTACCGCTGACCATGAG-3′ Reverse-5′-TTTGGTCTACGTGGCGTACA-3′ |
5.04 |
| 9 | RM467 | Forward-5′-TGTTGTCACATGAGATGGCTATGC-3′ Reverse-5′-GCTGACCTTGTGAGACGTTTAGACC-3′ |
19.20 |
| 10 | RM486 | Forward-5′-GCTTGCATTATGCGATTGTACTCC-3′ Reverse-5′-TGAGCTTTCTCAACAACGACTGC-3′ |
7.46 |
| 11 | RM569 | Forward-5′-CTGCGTCAGATTTCTCCTCTTCG-3′ Reverse-5′-ACATTCTCGCTTGCTCCTCTCG-3′ |
4.73 |
| 12 | RM570 | Forward-5′-AGAAATGGTGAAAGATGGTGCTACCG-3′ Reverse-5′-CTGAATGTTCTTCAACTCCCAGTGC-3′ |
10.35 |
| 13 | RM5746 | Forward-5′-CAGCTTCGGCAAAGCAAAGC-3′ Reverse-5′-CTCGCTACGTCGACTGATTTGG-3′ |
12.12 |
| Genotypes | 2,4-D (mg/L) |
No. of seeds inoculated (A) |
No. of callus-induced seeds (B) |
Callus induction frequency (%) (C) |
||
| 15 Days | 30 Days | C= B/A*100 (15 Days) | C= B/A*100 (30 Days) | |||
| Mani-4 | 0 | 30 | - | - | - | - |
| 0.5 | 30 | 6 | 7 | 20.0 | 23.33 | |
| 1 | 30 | 6 | 10 | 20.0 | 33.33 | |
| 2 | 30 | 22 | 22 | 73.33 | 73.33 | |
| 3 | 30 | 15 | 22 | 50.0 | 73.33 | |
| 4 | 30 | 2 | 3 | 6.67 | 10.0 | |
| Mani-5 | 0 | 30 | - | - | - | - |
| 0.5 | 30 | - | - | - | - | |
| 1 | 30 | 19 | 21 | 63.33 | 70.0 | |
| 2 | 30 | 24 | 25 | 80.0 | 83.33 | |
| 3 | 30 | 21 | 22 | 70.0 | 73.33 | |
| 4 | 30 | 23 | 24 | 76.67 | 80.0 | |
| Mani-6 | 0 | 30 | - | - | - | - |
| 0.5 | 30 | - | - | - | - | |
| 1 | 30 | 11 | 19 | 36.67 | 63.33 | |
| 2 | 30 | 17 | 22 | 56.67 | 73.33 | |
| 3 | 30 | 15 | 21 | 50.0 | 70.0 | |
| 4 | 30 | 12 | 25 | 40.0 | 83.33 | |
| Mani-7 | 0 | 30 | - | - | - | - |
| 0.5 | 30 | 7 | 15 | 23.33 | 50.0 | |
| 1 | 30 | 9 | 16 | 30.0 | 53.33 | |
| 2 | 30 | 24 | 27 | 80.0 | 90.0 | |
| 3 | 30 | 15 | 28 | 50.0 | 93.33 | |
| 4 | 30 | 15 | 25 | 50.0 | 83.33 | |
| Mani-10 | 0 | 30 | - | - | - | - |
| 0.5 | 30 | 3 | 11 | 10.0 | 36.67 | |
| 1 | 30 | 5 | 14 | 16.67 | 46.67 | |
| 2 | 30 | 5 | 14 | 16.67 | 46.67 | |
| 3 | 30 | 11 | 20 | 36.67 | 66.67 | |
| 4 | 30 | 8 | 20 | 26.67 | 66.67 | |
| Mani-11 | 0 | 30 | - | - | - | - |
| 0.5 | 30 | 1 | 3 | 3.33 | 10.0 | |
| 1 | 30 | 3 | 5 | 10.0 | 16.67 | |
| 2 | 30 | 8 | 12 | 26.67 | 40.0 | |
| 3 | 30 | 13 | 14 | 43.33 | 46.67 | |
| 4 | 30 | 6 | 10 | 20.0 | 33.33 | |
| Mani-12 | 0 | 30 | - | - | - | - |
| 0.5 | 30 | - | - | - | - | |
| 1 | 30 | 3 | 8 | 10.0 | 26.67 | |
| 2 | 30 | 5 | 7 | 16.67 | 23.33 | |
| 3 | 30 | 9 | 13 | 30.0 | 43.33 | |
| 4 | 30 | 6 | 8 | 20.0 | 26.67 | |
| Mani-13 | 0 | 30 | - | - | - | - |
| 0.5 | 30 | 3 | 11 | 10.0 | 36.67 | |
| 1 | 30 | 5 | 11 | 16.67 | 36.67 | |
| 2 | 30 | 13 | 26 | 46.67 | 86.67 | |
| 3 | 30 | 17 | 26 | 56.67 | 86.67 | |
| 4 | 30 | 11 | 29 | 36.67 | 96.67 | |
| Mani-14 | 0 | 30 | - | - | - | - |
| 0.5 | 30 | 15 | 19 | 50.0 | 63.33 | |
| 1 | 30 | 18 | 21 | 60.0 | 70.0 | |
| 2 | 30 | 14 | 18 | 46.67 | 60.0 | |
| 3 | 30 | 18 | 21 | 60.0 | 70.0 | |
| 4 | 30 | 15 | 25 | 50.0 | 83.33 | |
| Mani-15 | 0 | 30 | - | - | - | - |
| 0.5 | 30 | - | 3 | - | 10.0 | |
| 1 | 30 | 3 | 8 | 10.0 | 26.67 | |
| 2 | 30 | 13 | 18 | 43.33 | 60.0 | |
| 3 | 30 | 9 | 9 | 30.0 | 30.0 | |
| 4 | 30 | 8 | 14 | 26.67 | 46.67 | |
| Mani-16 | 0 | 30 | - | - | - | - |
| 0.5 | 30 | 17 | 18 | 56.67 | 60.0 | |
| 1 | 30 | 13 | 15 | 43.33 | 50.0 | |
| 2 | 30 | 19 | 22 | 63.33 | 73.33 | |
| 3 | 30 | 17 | 24 | 56.67 | 80.0 | |
| 4 | 30 | 15 | 19 | 50.0 | 63.33 | |
| Sl No. | Gene ID | Gene product | References |
| 1 | LOC_Os01g19470 | nodal modulator 1 precursor, putative, expressed | Zhang et al., 2019 |
| 2 | LOC_Os01g19480 | kelch motif family protein, putative, expressed | Zhang et al., 2019 |
| 3 | LOC_Os02g30900 | protein kinase domain containing protein, expressed | Zhang et al., 2019 |
| 4 | LOC_Os02g57240 | oxidoreductase, aldo/keto reductase family protein, putative, expressed | Zhang et al., 2019 |
| 5 | LOC_Os02g57250 | OsIAA10—Auxin-responsive Aux/IAA gene family member, expressed | Zhang et al., 2019 |
| 6 | LOC_Os02g57380 | thioredoxin, putative, expressed | Zhang et al., 2019 |
| 7 | LOC_Os03g60120 | AP2 domain containing protein, expressed | Zhang et al., 2019 |
| 8 | LOC_Os03g60130 | transcription elongation factor protein, putative, expressed | Zhang et al., 2019 |
| 9 | LOC_Os05g33890 | microtubule associated protein, putative, expressed | Zhang et al., 2019 |
| 10 | LOC_Os06g09290 | 26S protease regulatory subunit 7, putative, expressed | Zhang et al., 2019 |
| 11 | LOC_Os06g09320 | expressed protein | Zhang et al., 2019 |
| 12 | LOC_Os06g09330 | ubiquitin-conjugating enzyme, putative, expressed | Zhang et al., 2019 |
| 13 | LOC_Os06g09390 | AP2 domain containing protein, expressed | Zhang et al., 2019 |
| 14 | LOC_Os06g09450 | sucrose synthase, putative, expressed | Zhang et al., 2019 |
| 15 | LOC_Os03g05550 | expressed protein | Jiemin et al., 2022 |
| 16 | LOC_Os03g05570 | RING-H2 finger protein ATL3F, putative, expressed | Wu Jiemin et al., 2022 |
| 17 | LOC_Os03g05680 | histone demethylase JARID1C, putative, expressed | Wu Jiemin et al., 2022 |
| 18 | LOC_Os03g05690 | ZOS3-03—C2H2 zinc finger protein, expressed | Wu Jiemin et al., 2022 |
| 19 | LOC_Os03g05700 | expressed protein | Wu Jiemin et al., 2022 |
| 20 | LOC_Os03g05710 | acetyltransferase, GNAT family, putative, expressed | Wu Jiemin et al., 2022 |
| 21 | LOC_Os03g05750 | heavy-metal-associated domain-containing protein, putative, expressed | Wu Jiemin et al., 2022 |
| 22 | LOC_Os03g05760 | transcription factor Dp, putative, expressed | Wu Jiemin et al., 2022 |
| 23 | LOC_Os03g05780 | 4-coumarate--CoA ligase-like 7, putative, expressed | Wu Jiemin et al., 2022 |
| 24 | LOC_Os03g12820 | ATP8, putative, expressed | Zhang et al., 2020 |
| 25 | LOC_Os02g49370 | histone-like transcription factor and archaeal histone, putative, expressed | Guo Fu et al., 2023 |
| Sl No. | Gene Locus ID | InDels | InDel ID | Primer name | Primer Sequence (5′ to 3′) | amplicon (bp) |
| 1 | LOC_Os01g19480 | 1 | vg0111057118 | RCu1 | F-AGCAATCCCATCCACCTTGA R-GCGAGACGAATCTTTTAAGCCT |
350 |
| 2 | vg0111057704 | RCu2 | F-ACCCAACTTAGCCCTAGTGG R-CGTATTATCGGATGGAGAGGGT |
372 | ||
| 2 | LOC_Os02g30900 | 1 | vg0218445904 | RCu3 | F-AAGGAAAGACCAGGACGGAC R-TGCTCGGGATCGGATCTTC |
317 |
| 2 | vg0218446199 | RCu4 | F-AAGACGCGTATTAGTTGGGC R-AAGCAGCCCATCTCTCTCTC |
366 | ||
| 3 | LOC_Os02g57250 | 1 | vg0235077064 | RCu5 | F-GGCTGTATGATTGACGTGTTCA R-TTTCCAAACCATCCCACACG |
599 |
| 4 | LOC_Os02g57380 | 1 | vg0235157418 | RCu6 | F-ACCAGGTGGCCTCCTTAATC R-ACGAAAAGCGGCAAAAGACT |
212 |
| 5 | LOC_Os05g33890 | 1 | vg0519994856 | RCu7 | F-CTCATTTGTGCCGTGCTGTA R-ACCCTCATCATTTAGCTCGGT |
295 |
| 6 | LOC_Os06g09290 | 1 | vg0604671911 | RCu8 | F-CTGACCCACATGTCATTGACTC R-TAGTAGCAGATGTGGCACCC |
417 |
| 7 | LOC_Os06g09320 | 1 | vg0604685498 | RCu9 | F-TATATTCGCTGGGTTCGGCA R-GAGCACACAATGGCTACCTT |
600 |
| 8 | LOC_Os06g09390 | 1 | vg0604730040 | RCu10 | F-CCGGTCAAAAGCTGAGACAG R-TGTCCTGACGACACTTCTTTAC |
476 |
| 9 | LOC_Os06g09450 | 1 | vg0604802924 | RCu11 | F-TCCCCCCAAATTCCCCATTT R-TCCAAAGCTGACAATGGTGC |
352 |
| 10 | LOC_Os03g05550 | 1 | vg0302762832 | RCu12 | F-AGGTGGGACCATCGACAATT R-TACCTTCTGTCTGCCAGCAA |
466 |
| 2 | vg0302765163 | RCu13 | F-TCTAGTGCCCTTGTTCTGCA R-CTTCGTTGGTGTTGTTGGGT |
344 | ||
| 3 | vg0302765827 | RCu14 | F-CTGTTCTCACAGGCCAACAC R-AGCATGCTTAACCCTGGAGT |
339 | ||
| 11 | LOC_Os03g05570 | 1 | vg0302776484 | RCu15 | F-GCTCCACCCTCTATCTCGTC R-TTGTGTGCGTGCAATGTGTA |
335 |
| 12 | LOC_Os03g05680 | 1 | vg0302831140 | RCu16 | F-AGCGGATTTATGGCGTGTTG R-CAACGGGATTTTCAACGGGA |
418 |
| 2 | vg0302831393 | RCu17 | F-CCCGTTGAAAATCCCGTTGA R-TCACATACCGCGACTGGTC |
569 | ||
| 3 | vg0302832709 | RCu18 | F-TACAAAAGGAGGTGCGAGGT R-AGAGGAAGGGAAGGGAGGAT |
590 | ||
| 4 | vg0302832713 | RCu19 | F-TACAAAAGGAGGTGCGAGGT R-AGAGGAAGGGAAGGGAGGAT |
590 | ||
| 5 | vg0302832755 | RCu20 | F-TACAAAAGGAGGTGCGAGGT R-GCAAAACTAACCGCCCTTCT |
331 | ||
| 13 | LOC_Os03g12820 | 1 | vg0306898719 | RCu21 | F-TCGGGAGTTTACGGAGCTTT R-TATCATTGATTCACGCGGCC |
534 |
| 2 | vg0306899285 | RCu22 | F-GGCCGCGTGAATCAATGATA R-AAATTGAGCGTGGGCACTAC |
487 | ||
| 14 | LOC_Os02g49370 | 1 | vg0230166558 | RCu23 | F-TGCGAAGAAACGGATGGAAC R-TCATGCAGGCACAACGAAAA |
455 |
| Locus | k | HObs | HExp | PIC | F(Null) |
| RM1146 | 6 | 0.455 | 0.537 | 0.491 | 0.1121 |
| RM21 | 2 | 0.000 | 0.312 | 0.253 | 0.9818 |
| RM224 | 4 | 0.000 | 0.675 | 0.579 | 1.0000 |
| RM322 | 2 | 0.182 | 0.312 | 0.253 | 0.2414 |
| RM348 | 1 | 0.000 | 0.000 | 0.000 | ND |
| RM411 | 4 | 0.000 | 0.745 | 0.656 | 1.0000 |
| RM414 | 2 | 0.000 | 0.485 | 0.356 | 0.9989 |
| RM42 | 2 | 0.000 | 0.485 | 0.356 | 0.9989 |
| RM467 | 3 | 0.000 | 0.537 | 0.444 | 0.9996 |
| RM486 | 4 | 0.182 | 0.398 | 0.353 | 0.4071 |
| RM569 | 4 | 0.000 | 0.710 | 0.623 | 1.0000 |
| RM570 | 3 | 0.182 | 0.255 | 0.228 | 0.2547 |
| RM5746 | 1 | 0.000 | 0.000 | 0.000 | ND |
| RCu1 | 1 | 0.000 | 0.000 | 0.000 | ND |
| RCu2 | 3 | 0.000 | 0.658 | 0.551 | 1.0000 |
| RCu3 | 5 | 0.000 | 0.797 | 0.720 | 1.0000 |
| RCu4 | 2 | 0.000 | 0.519 | 0.373 | 0.9995 |
| RCu5 | 2 | 0.000 | 0.519 | 0.373 | 0.9995 |
| RCu6 | 1 | 0.000 | 0.000 | 0.000 | ND |
| RCu7 | 4 | 0.000 | 0.762 | 0.678 | 1.0000 |
| RCu8 | 3 | 0.000 | 0.554 | 0.473 | 0.9997 |
| RCu9 | 2 | 0.000 | 0.485 | 0.356 | 0.9989 |
| RCu10 | 2 | 0.000 | 0.485 | 0.356 | 0.9989 |
| RCu11 | 3 | 0.000 | 0.606 | 0.486 | 0.9999 |
| RCu12 | 1 | 0.000 | 0.000 | 0.000 | ND |
| RCu13 | 3 | 0.000 | 0.589 | 0.476 | 0.9999 |
| RCu14 | 2 | 0.273 | 0.247 | 0.208 | -0.0692 |
| RCu15 | 2 | 0.000 | 0.485 | 0.356 | 0.9989 |
| RCu16 | 2 | 0.364 | 0.312 | 0.253 | -0.0981 |
| RCu17 | 4 | 0.364 | 0.333 | 0.302 | -0.0912 |
| RCu18 | 2 | 0.000 | 0.485 | 0.356 | 0.9989 |
| RCu19 | 2 | 0.000 | 0.485 | 0.356 | 0.9989 |
| RCu20 | 2 | 0.000 | 0.485 | 0.356 | 0.9989 |
| RCu21 | 3 | 0.182 | 0.177 | 0.163 | -0.0392 |
| RCu22 | 2 | 0.182 | 0.173 | 0.152 | -0.0405 |
| RCu23 | 2 | 0.182 | 0.173 | 0.152 | -0.0405 |
| Mean ± SEM | 2.33 ± 1.18 | 0.07 ± 0.13 | 0.41 ± 0.23 | 0.34 ± 0.20 | 0.60 ± 0.49 |
| 2,4-D | Marker | p-value | Marker R2 |
| 0.5 mg/L | RM21 | 0.012676 | 0.516757* |
| RM411 | 0.023644 | 0.450997* | |
| RM569 | 0.023644 | 0.450997* | |
| RCu4 | 0.004862 | 0.604351** | |
| RCu5 | 0.004862 | 0.604351** | |
| 2.0 mg/L | RCu17 | 0.029544 | 0.42575* |
| 4.0 mg/L | RM414 | 0.024497 | 0.447042* |
| RCu2 | 0.010716 | 0.533261* | |
| RCu7 | 0.024497 | 0.447042* | |
| RCu10 | 0.024497 | 0.447042* | |
| RCu13 | 0.036366 | 0.401376* | |
| RCu15 | 0.024497 | 0.447042* | |
| RCu18 | 0.024497 | 0.447042* | |
| RCu19 | 0.024497 | 0.447042* | |
| RCu20 | 0.024497 | 0.447042* |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





