Submitted:
18 November 2025
Posted:
19 November 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
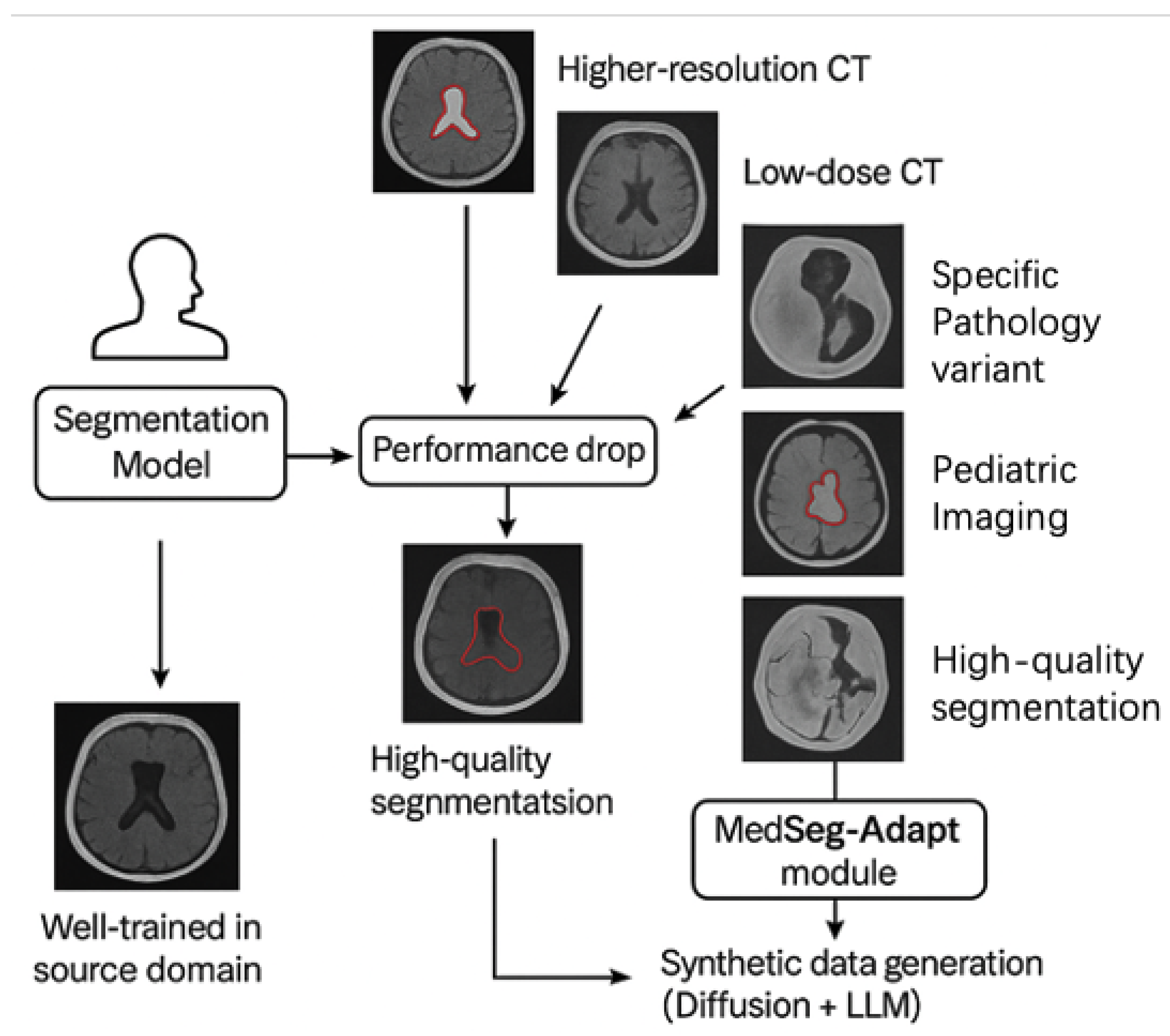
- We propose MedSeg-Adapt, a novel framework for clinical query-guided adaptive medical image segmentation, featuring an autonomous data augmentation module that generates environment-specific data without requiring new reinforcement learning models.
- We introduce MedScanDiff, a comprehensive new benchmark specifically designed to evaluate the generalization and adaptation capabilities of medical image segmentation models across five distinct novel medical imaging environments.
- We demonstrate that fine-tuning existing state-of-the-art medical vision-language models with data generated by MedSeg-Adapt leads to significant and consistent performance improvements (an average of +5.5% DSC) on the MedScanDiff benchmark, validating its efficacy in real-world adaptive scenarios.
2. Related Work
2.1. Medical Vision-Language Models and Query-Guided Segmentation
2.2. Domain Adaptation and Synthetic Data Generation in Medical Imaging
2.3. Advanced Decision-Making and Control in Intelligent Systems
2.4. Broad Applications of AI and Machine Learning
3. Method
3.1. Problem Formulation
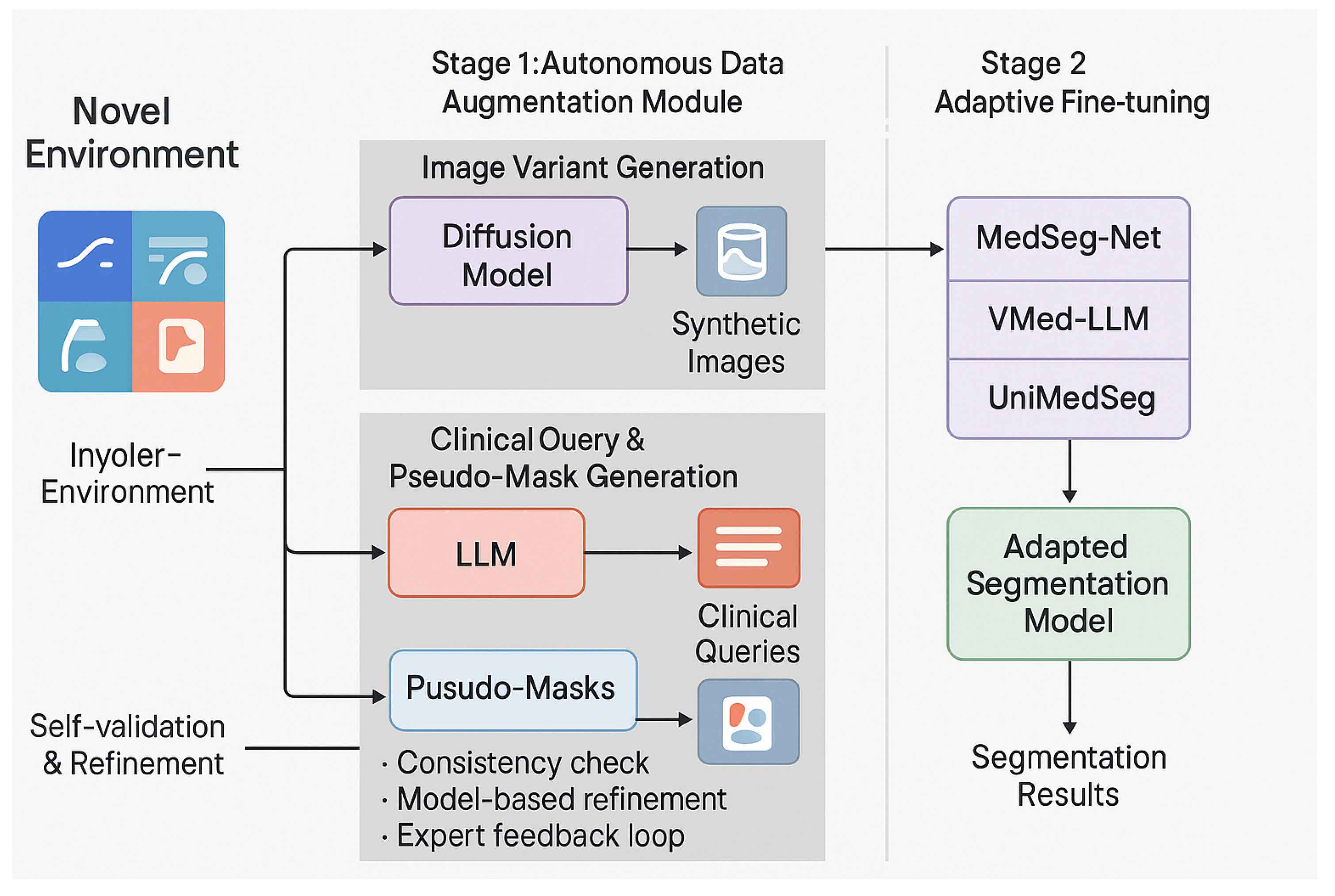
3.2. MedSeg-Adapt Framework Overview
- Autonomous Data Augmentation: For any given novel medical imaging environment , an integral autonomous data augmentation module is activated. This module is responsible for generating a diverse and representative set of synthetic medical images, along with their corresponding natural language clinical queries and preliminary pseudo-segmentation masks. This sophisticated generation process intelligently leverages advanced generative models, particularly medical image Diffusion models, in conjunction with large language models (LLMs), all guided by meticulously designed domain-specific rules and constraints.
- Adaptive Fine-tuning: The synthetic data generated and subsequently self-validated in the first stage are then systematically utilized to fine-tune existing pre-trained medical vision-language models (MVLMs) or specialized medical image segmentation models. This adaptive fine-tuning step is crucial for aligning the base models with the unique characteristics and data distribution of the target novel environment, thereby significantly enhancing their segmentation performance, generalization capabilities, and overall robustness in real-world clinical settings.
3.3. Autonomous Data Augmentation Module
3.3.1. Environment-Specific Data Generation
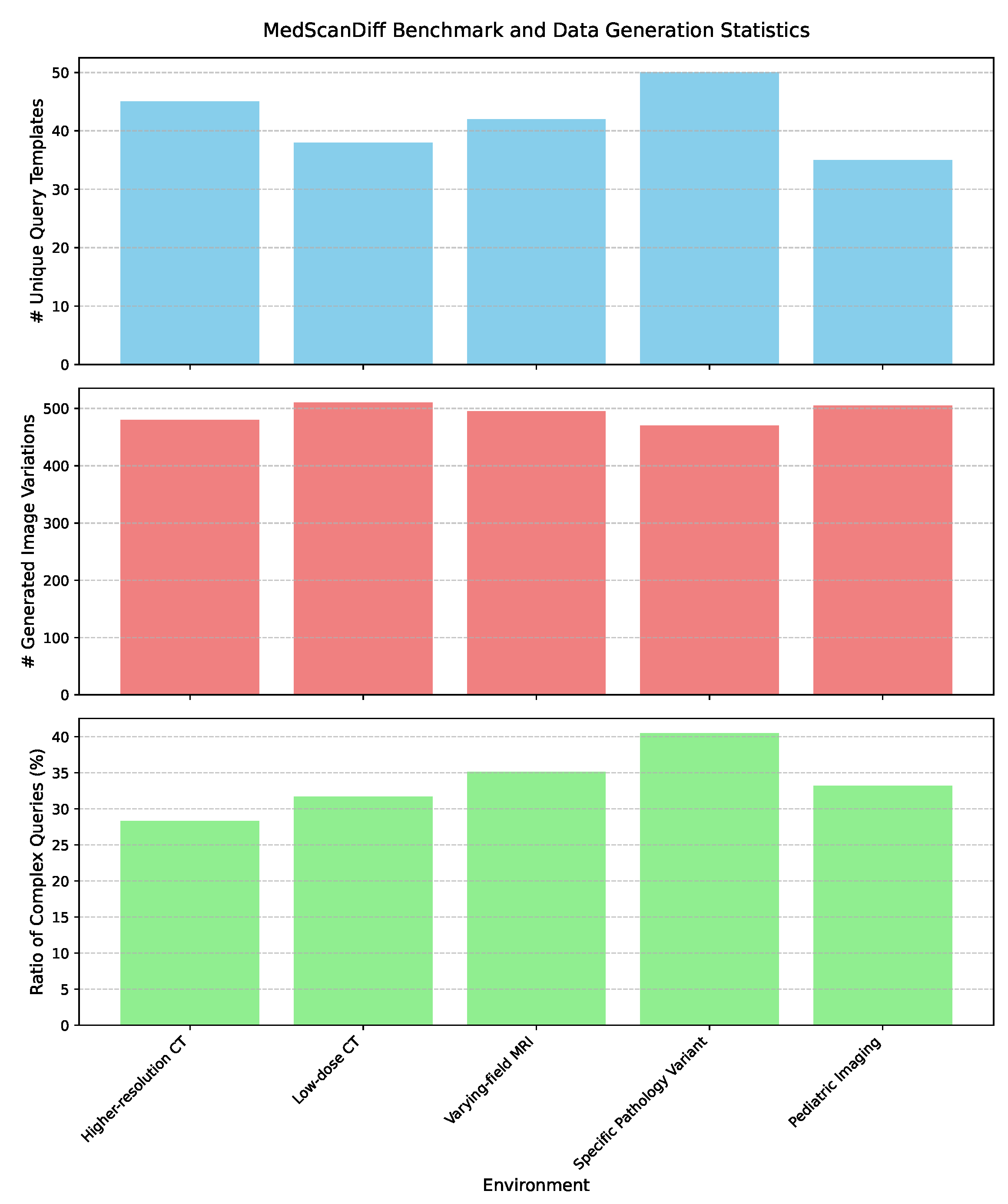
- Image Variant Generation: A medical image Diffusion model, specifically designed for high-fidelity image synthesis, is conditioned on the distinctive characteristics of the target environment . These characteristics can include specific imaging modalities, resolution variations, noise levels, acquisition protocols (e.g., “higher resolution CT”, “low-dose CT”, “pediatric MRI”), or even the prevalence of certain artifacts. This conditional Diffusion model then generates a diverse set of synthetic medical image slices, denoted as . These synthetic images are meticulously crafted to mimic the visual properties, inherent noise patterns, and potential artifacts commonly observed in the target environment . The conditioning information, , can be effectively conveyed through various mechanisms, such as descriptive text prompts, latent image embeddings, or specific control signals that encapsulate the essence of the novel environment. This generative process can be formally conceptualized as:where represents the generative function of the conditional Diffusion model, and denotes the conditioning information pertinent to environment . For each environment, we systematically generate up to 500 unique image variations to ensure comprehensive coverage of the target domain’s visual characteristics.
- Clinical Query and Pseudo-Mask Generation: For each autonomously generated image variant , a large language model (LLM) is subsequently employed to generate a set of 2-3 corresponding natural language clinical queries, denoted as , and preliminary pseudo-segmentation masks, . The LLM is interactively prompted with the generated image (or its extracted salient features, derived via a vision encoder) and the previously specified environment characteristics . This input enables the LLM to formulate clinically relevant and contextually appropriate queries, mimicking how a clinician might inquire about specific structures or pathologies within the image. Simultaneously, the LLM, often augmented with a sophisticated vision encoder to process visual information directly, generates an initial pseudo-mask based on the generated query and the visual content of . This integrated generation process can be formally expressed as:where represents the multimodal capability of the LLM to generate both natural language queries and pixel-level segmentation masks. The generation of queries and masks relies on a sophisticated rule-based and generative model-driven exploration strategy rather than explicit reinforcement learning. This means the LLM actively explores diverse query formulations and segmentation interpretations within the medical domain, guided by pre-defined medical knowledge bases (e.g., anatomical ontologies, disease classifications) and internal consistency checks to ensure clinical plausibility and semantic coherence. This exploration facilitates the creation of a rich and varied synthetic dataset.
3.3.2. Self-Validation and Pseudo-Annotation Refinement
- Consistency Check: The generated clinical query and its corresponding pseudo-mask are meticulously cross-referenced against a comprehensive knowledge base of medical terminology, anatomical relationships, and disease characteristics. An internal consistency score is algorithmically computed to identify any potential discrepancies, ambiguities, or semantic inconsistencies between the textual query and the visual segmentation. Samples failing to meet a predefined consistency threshold are flagged for further scrutiny or regeneration.
- Model-Based Refinement: An ensemble of pre-trained, general-purpose medical segmentation models is strategically employed to independently re-evaluate and re-segment the pseudo-mask , given the synthetic image and the generated query . If a significant divergence is observed between the ensemble’s consensus prediction and the initially generated pseudo-mask (e.g., high Dice dissimilarity or structural disagreement), the pseudo-mask may undergo an automated refinement process (e.g., using weighted averaging or an uncertainty-aware fusion technique) or be flagged for manual review if the divergence is critical. This step leverages the collective knowledge of multiple models to enhance accuracy.
- Expert Feedback Loop: To maintain the highest standards of clinical fidelity and to continuously improve the generative process, a small, representative fraction of the generated data is periodically presented to medical experts for rapid review and feedback. This invaluable human feedback is then systematically incorporated to refine the prompting strategies employed for the LLM and to optimize the conditioning parameters for the Diffusion model. This iterative feedback loop ensures a continuous improvement cycle, progressively enhancing the quality and clinical utility of subsequent data generations. For the MedScanDiff benchmark specifically, the ground-truth annotations against which the adapted models are evaluated are meticulously prepared and validated by experienced medical experts, serving as the ultimate standard.
3.4. Adaptive Fine-tuning
4. Experiments
4.1. Experimental Setup
4.1.1. Models
4.1.2. MedScanDiff Benchmark
4.1.3. Fine-tuning Details
4.1.4. Evaluation Metric
4.2. MedScanDiff Benchmark and Data Generation Analysis
4.3. Main Results on MedScanDiff
4.4. Validation of MedSeg-Adapt’s Adaptive Capabilities
4.5. Qualitative Analysis and Human Evaluation
4.6. Ablation Studies
- Environment-Specific Image Generation: When the Diffusion model’s environment-specific image generation was replaced by generic augmentation techniques (e.g., random affine transformations, intensity shifts) that do not specifically mimic novel domain characteristics, the average DSC dropped from 78.3% to 74.5%. This significant decrease of 3.8 percentage points highlights the critical role of high-fidelity, environment-aware synthetic images in effectively bridging the visual domain gap.
- LLM-driven Query and Pseudo-Mask Generation: Substituting the sophisticated LLM-driven query and pseudo-mask generation with simpler, template-based queries and basic image processing for mask generation (e.g., thresholding or simple edge detection) resulted in an average DSC of 75.9%. This 2.4 percentage point reduction compared to the full framework underscores the importance of the LLM’s ability to create semantically rich, clinically relevant queries and accurate initial pseudo-masks, which are vital for guiding the segmentation model towards the correct anatomical or pathological targets.
- Self-Validation and Refinement: Removing the self-validation and pseudo-annotation refinement step, which ensures the quality and consistency of the generated data, led to a performance drop to 77.0% (a 1.3 percentage point decrease). While less pronounced than the other two components, this still indicates that the rigorous validation process is essential for filtering out low-quality synthetic samples and refining pseudo-masks, thereby preventing the introduction of noise or errors during fine-tuning.
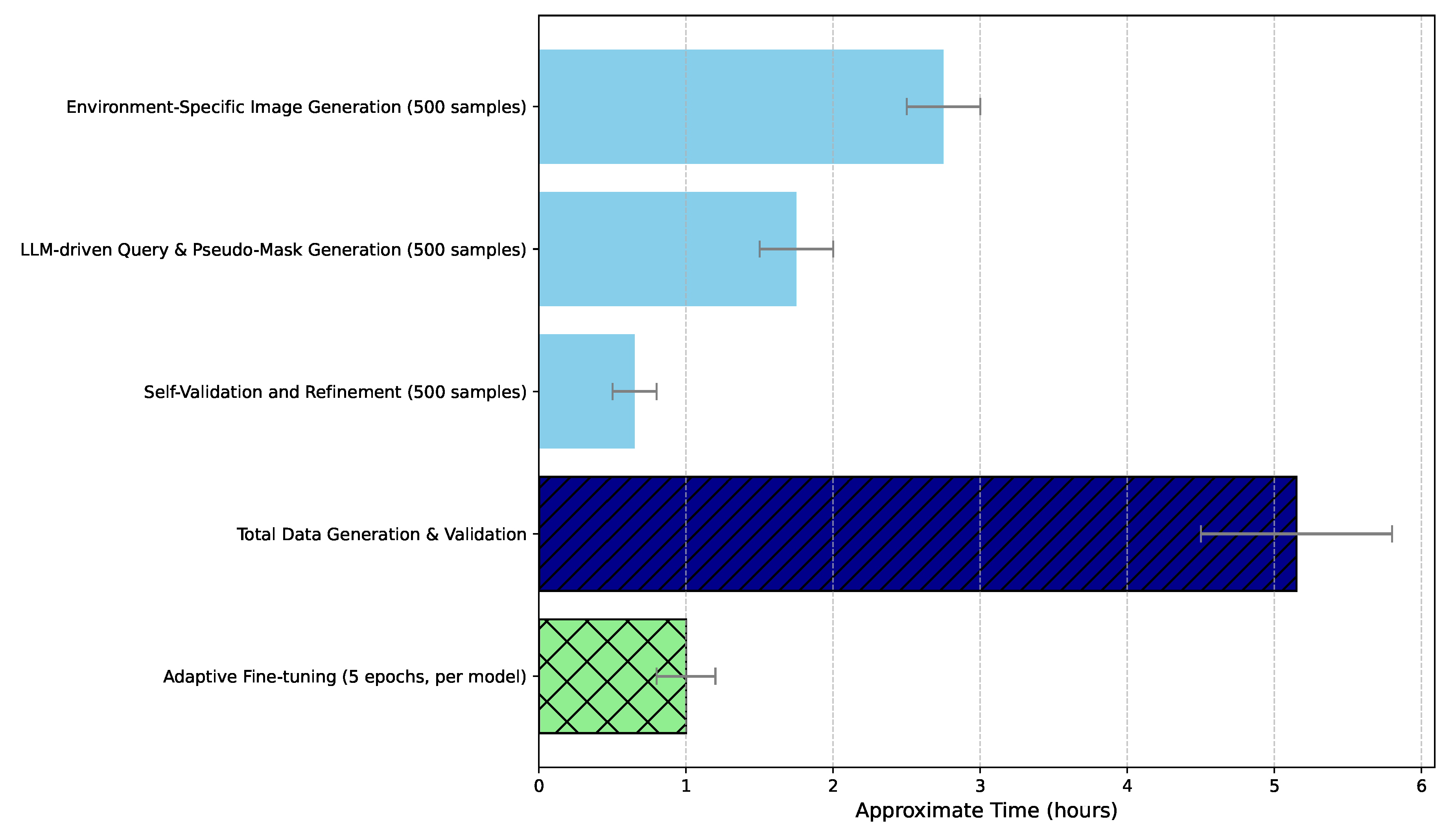
4.7. Efficiency and Scalability Analysis
4.8. Discussion and Future Directions
5. Conclusions
References
- Chang, D.T. Concept-Oriented Deep Learning: Generative Concept Representations. CoRR 2018.
- Clark, H.D.; Reinsberg, S.A.; Moiseenko, V.; Wu, J.; Thomas, S.D. Prefer Nested Segmentation to Compound Segmentation. arXiv preprint arXiv:1705.01643v1 2017.
- Zhou, Y.; Li, X.; Wang, Q.; Shen, J. Visual In-Context Learning for Large Vision-Language Models. In Proceedings of the Findings of the Association for Computational Linguistics, ACL 2024, Bangkok, Thailand and virtual meeting, August 11-16, 2024. Association for Computational Linguistics, 2024, pp. 15890–15902.
- Lin, G. Analyzing PFG anisotropic anomalous diffusions by instantaneous signal attenuation method. arXiv preprint arXiv:1701.00257v2 2017.
- Abdullah.; Huang, T.; Lee, I.; Ahn, E. Computationally Efficient Diffusion Models in Medical Imaging: A Comprehensive Review. CoRR 2025. [CrossRef]
- Zhou, Y.; Geng, X.; Shen, T.; Tao, C.; Long, G.; Lou, J.G.; Shen, J. Thread of thought unraveling chaotic contexts. arXiv preprint arXiv:2311.08734 2023.
- Köhler-Bußmeier, M. Probabilistic Nets-within-Nets. CoRR 2024. [CrossRef]
- Kim, K.; Ailamaki, A. Trustworthy and Efficient LLMs Meet Databases. CoRR 2024. [CrossRef]
- Chen, Q.; Zhao, R.; Wang, S.; Phan, V.M.H.; van den Hengel, A.; Verjans, J.; Liao, Z.; To, M.; Xia, Y.; Chen, J.; et al. A Survey of Medical Vision-and-Language Applications and Their Techniques. CoRR 2024. [CrossRef]
- Liu, C.; Shah, A.; Bai, W.; Arcucci, R. Utilizing Synthetic Data for Medical Vision-Language Pre-training: Bypassing the Need for Real Images. CoRR 2023. [CrossRef]
- Zhang, Y.; He, N.; Yang, J.; Li, Y.; Wei, D.; Huang, Y.; Zhang, Y.; He, Z.; Zheng, Y. mmFormer: Multimodal Medical Transformer for Incomplete Multimodal Learning of Brain Tumor Segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention - MICCAI 2022 - 25th International Conference, Singapore, September 18-22, 2022, Proceedings, Part V. Springer, 2022, pp. 107–117. [CrossRef]
- Yang, R.; Li, H.; Wong, M.Y.H.; Ke, Y.; Li, X.; Yu, K.; Liao, J.; Liew, J.C.K.; Nair, S.V.; Ong, J.C.L.; et al. The Evolving Landscape of Generative Large Language Models and Traditional Natural Language Processing in Medicine. CoRR 2025. [CrossRef]
- Zhou, Y.; Shen, T.; Geng, X.; Long, G.; Jiang, D. ClarET: Pre-training a Correlation-Aware Context-To-Event Transformer for Event-Centric Generation and Classification. In Proceedings of the Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), 2022, pp. 2559–2575.
- Zhou, Y.; Geng, X.; Shen, T.; Long, G.; Jiang, D. Eventbert: A pre-trained model for event correlation reasoning. In Proceedings of the Proceedings of the ACM Web Conference 2022, 2022, pp. 850–859.
- Zhang, Y.; Wang, S.; Zhou, T.; Dou, Q.; Chen, D.Z. SQA-SAM: Segmentation Quality Assessment for Medical Images Utilizing the Segment Anything Model. CoRR 2023. [CrossRef]
- Ouyang, S.; Zhang, J.; Lin, X.; Wang, X.; Chen, Q.; Chen, Y.W.; Lin, L. Language-guided Scale-aware MedSegmentor for Lesion Segmentation in Medical Imaging. arXiv preprint arXiv:2408.17347v3 2024.
- Huang, Y.; Peng, Z.; Zhao, Y.; Yang, P.; Yang, X.; Shen, W. MedSeg-R: Reasoning Segmentation in Medical Images with Multimodal Large Language Models. CoRR 2025. [CrossRef]
- Hu, S.; Liao, Z.; Zhen, L.; Fu, H.; Xia, Y. Cycle Context Verification for In-Context Medical Image Segmentation. CoRR 2025. [CrossRef]
- Guan, H.; Liu, M. DomainATM: Domain adaptation toolbox for medical data analysis. NeuroImage 2023, p. 119863. [CrossRef]
- Xu, Y.; Xie, S.; Reynolds, M.; Ragoza, M.; Gong, M.; Batmanghelich, K. Adversarial Consistency for Single Domain Generalization in Medical Image Segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention - MICCAI 2022 - 25th International Conference, Singapore, September 18-22, 2022, Proceedings, Part VII. Springer, 2022, pp. 671–681. [CrossRef]
- Stolte, S.E.; Volle, K.; Indahlastari, A.; Albizu, A.; Woods, A.J.; Brink, K.M.; Hale, M.T.; Fang, R. DOMINO++: Domain-Aware Loss Regularization for Deep Learning Generalizability. In Proceedings of the Medical Image Computing and Computer Assisted Intervention - MICCAI 2023 - 26th International Conference, Vancouver, BC, Canada, October 8-12, 2023, Proceedings, Part IV. Springer, 2023, pp. 713–723. [CrossRef]
- Mahmood, F.; Chen, R.; Durr, N.J. Unsupervised Reverse Domain Adaptation for Synthetic Medical Images via Adversarial Training. IEEE Trans. Medical Imaging 2018, pp. 2572–2581. [CrossRef]
- Xie, J.; Zhang, Z.; Weng, Z.; Zhu, Y.; Luo, G. MedDiff-FT: Data-Efficient Diffusion Model Fine-tuning with Structural Guidance for Controllable Medical Image Synthesis. CoRR 2025. [CrossRef]
- Feng, Y.; Zhang, B.; Xiao, L.; Yang, Y.; Tana, G.; Chen, Z. Enhancing Medical Imaging with GANs Synthesizing Realistic Images from Limited Data. CoRR 2024. [CrossRef]
- Molino, D.; Feola, F.D.; Shen, L.; Soda, P.; Guarrasi, V. Any-to-Any Vision-Language Model for Multimodal X-ray Imaging and Radiological Report Generation. CoRR 2025. [CrossRef]
- Liang, X.; Hu, X.; Zuo, S.; Gong, Y.; Lou, Q.; Liu, Y.; Huang, S.; Jiao, J. Task Oriented In-Domain Data Augmentation. In Proceedings of the Proceedings of the 2024 Conference on Empirical Methods in Natural Language Processing, EMNLP 2024, Miami, FL, USA, November 12-16, 2024. Association for Computational Linguistics, 2024, pp. 20889–20907. [CrossRef]
- Lin, Z.; Lan, J.; Anagnostopoulos, C.; Tian, Z.; Flynn, D. Safety-Critical Multi-Agent MCTS for Mixed Traffic Coordination at Unsignalized Intersections. IEEE Transactions on Intelligent Transportation Systems 2025, pp. 1–15. [CrossRef]
- Lin, Z.; Lan, J.; Anagnostopoulos, C.; Tian, Z.; Flynn, D. Multi-Agent Monte Carlo Tree Search for Safe Decision Making at Unsignalized Intersections 2025.
- Tian, Z.; Lin, Z.; Zhao, D.; Zhao, W.; Flynn, D.; Ansari, S.; Wei, C. Evaluating scenario-based decision-making for interactive autonomous driving using rational criteria: A survey. arXiv preprint arXiv:2501.01886 2025.
- Yang, Y.; Constantinescu, D.; Shi, Y. Robust four-channel teleoperation through hybrid damping-stiffness adjustment. IEEE Transactions on Control Systems Technology 2019, 28, 920–935.
- Yang, Y.; Constantinescu, D.; Shi, Y. Passive multiuser teleoperation of a multirobot system with connectivity-preserving containment. IEEE Transactions on Robotics 2021, 38, 209–228.
- Wang, P.; Zhu, Z.; Liang, D. Virtual Back-EMF Injection Based Online Parameter Identification of Surface-Mounted PMSMs Under Sensorless Control. IEEE Transactions on Industrial Electronics 2024.
- Wang, P.; Zhu, Z.; Liang, D. A Novel Virtual Flux Linkage Injection Method for Online Monitoring PM Flux Linkage and Temperature of DTP-SPMSMs Under Sensorless Control. IEEE Transactions on Industrial Electronics 2025.
- Wang, P.; Zhu, Z.; Feng, Z. Virtual Back-EMF Injection-based Online Full-Parameter Estimation of DTP-SPMSMs Under Sensorless Control. IEEE Transactions on Transportation Electrification 2025.
- Wang, Z.; Xiong, Y.; Horowitz, R.; Wang, Y.; Han, Y. Hybrid Perception and Equivariant Diffusion for Robust Multi-Node Rebar Tying. In Proceedings of the 2025 IEEE 21st International Conference on Automation Science and Engineering (CASE). IEEE, 2025, pp. 3164–3171.
- Ren, L.; et al. Boosting algorithm optimization technology for ensemble learning in small sample fraud detection. Academic Journal of Engineering and Technology Science 2025, 8, 53–60.
- Ren, L. Reinforcement Learning for Prioritizing Anti-Money Laundering Case Reviews Based on Dynamic Risk Assessment. Journal of Economic Theory and Business Management 2025, 2, 1–6.
- Ren, L. AI-Powered Financial Insights: Using Large Language Models to Improve Government Decision-Making and Policy Execution. Journal of Industrial Engineering and Applied Science 2025, 3, 21–26.
- Wang, Z.; Jiang, W.; Wu, W.; Wang, S. Reconstruction of complex network from time series data based on graph attention network and Gumbel Softmax. International Journal of Modern Physics C 2023, 34, 2350057.
- Wang, Z.; Wen, J.; Han, Y. EP-SAM: An Edge-Detection Prompt SAM Based Efficient Framework for Ultra-Low Light Video Segmentation. In Proceedings of the ICASSP 2025-2025 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). IEEE, 2025, pp. 1–5.
| Model | Higher-res CT | Low-dose CT | Varying-field MRI | Specific Pathology | Pediatric Imaging | Average DSC |
|---|---|---|---|---|---|---|
| MedSeg-Net (Vanilla) | 68.2 | 65.5 | 70.1 | 63.8 | 60.5 | 65.6 |
| MedSeg-Net (MedSeg-Adapt FT) | 73.5 (+5.3) | 71.2 (+5.7) | 74.8 (+4.7) | 69.1 (+5.3) | 66.8 (+6.3) | 71.1 (+5.5) |
| VMed-LLM (Vanilla) | 72.5 | 69.8 | 75.2 | 68.1 | 64.9 | 70.1 |
| VMed-LLM (MedSeg-Adapt FT) | 77.8 (+5.3) | 75.4 (+5.6) | 80.0 (+4.8) | 73.9 (+5.8) | 71.1 (+6.2) | 75.6 (+5.5) |
| UniMedSeg (Vanilla) | 75.1 | 72.3 | 78.5 | 71.0 | 67.2 | 72.8 |
| UniMedSeg (MedSeg-Adapt FT) | 80.3 (+5.2) | 77.9 (+5.6) | 83.1 (+4.6) | 76.8 (+5.8) | 73.5 (+6.3) | 78.3 (+5.5) |
| Model | Average Clinical Score (1-5) | % Cases Requiring Correction |
|---|---|---|
| MedSeg-Net (Vanilla) | 3.2 | 35.0% |
| MedSeg-Net (MedSeg-Adapt FT) | 4.1 | 12.0% |
| VMed-LLM (Vanilla) | 3.5 | 28.0% |
| VMed-LLM (MedSeg-Adapt FT) | 4.3 | 10.0% |
| UniMedSeg (Vanilla) | 3.8 | 22.0% |
| UniMedSeg (MedSeg-Adapt FT) | 4.5 | 8.0% |
| Configuration | Average DSC |
|---|---|
| UniMedSeg (Vanilla) | 72.8 |
| UniMedSeg (MedSeg-Adapt FT - Full Framework) | 78.3 |
| MedSeg-Adapt w/o Env-Specific Image Gen | 74.5 |
| (uses generic image augmentation instead of Diffusion) | |
| MedSeg-Adapt w/o LLM-driven Query/Mask Gen | 75.9 |
| (uses template queries and basic image processing for masks) | |
| MedSeg-Adapt w/o Self-Validation | 77.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




