1. Introduction
Liver diseases such as fibrosis, liver cancer, and nonalcoholic fatty liver disease are major global health concerns. Traditional diagnostic methods like liver biopsy are risky and not effective for early detection, which is why noninvasive and accurate diagnostic tools are urgently needed [
15]. AI offer new methods to analyze complex data and detect patterns that traditional techniques often miss [
16]. Research studies have shown that AI-assisted imaging, gene expression analysis, and gut microbiome tools can significantly improve diagnostic accuracy [
17]. Frameworks like MaLLiDD and advanced imaging techniques also contribute to better diagnostic outcomes [
18,
19]. But still, challenges remain, such as data imbalance, inconsistent reporting standards, and the need for large, high-quality datasets [
20,
21]. These findings suggest that with proper validation and collaboration, AI and ML can greatly enhance their clinical applicability [
22,
23,
24].
2. Literature Review
Decharatanachart et al. [
1] discussed the limitations of current liver diagnostic methods, stating that while liver biopsy is the standard, it is invasive and noninvasive tests lack accuracy. Their study showed that AI-based methods outperformed traditional diagnostics. Similarly, Sowa et al. [
4] analyzed liver fibrosis in NAFLD patients and emphasized the risks of liver biopsy and the inadequacy of noninvasive tests, especially in early stages. They proposed a new machine learning model using blood markers, achieving 79% accuracy.
Peng et al. [
2] highlighted the limitations of the MELD score in predicting liver dysfunction. They used machine learning models such as ANN, CART, and SVM with 15 features including INR and sodium. ANN achieved the highest accuracy of 91.2%, outperforming MELD’s 67%. Sorino et al. [
3] evaluated eight machine learning algorithms for NAFLD diagnosis using routine clinical and lab data, finding that SVM was most effective.
Yu et al. [
6] addressed the difficulty in diagnosing early-stage NAFLD due to the lack of biomarkers. Using machine learning on gene data, they identified five key genes (BBOX1, FOSB, NR4A2, RAB26, SOCS2) that improved diagnostic accuracy. Azam et al. [
6] tested five algorithms on Indian liver disease data, selecting key features that improved KNN accuracy to 74.15%.
Rehman et al. [
5] proposed the MaLLiDD framework to address imbalanced data and feature selection issues. Their model achieved 99.56% accuracy using random forest. Radiya et al. [
5] used machine learning on liver CT scans, emphasizing the lack of standardized reporting and the need for clinical collaboration [
26,
27,
28].
Liu et al. [
7] reviewed 11 studies using gut microbiome data for diagnosing liver fibrosis and cirrhosis, reporting sensitivity of 0.81 and specificity of 0.85. Drozdz et al. [
8] identified cardiovascular risk in MAFLD patients with machine learning models, achieving 84% accuracy.
Hou et al. [
5] developed a model to predict bleeding risk in liver cirrhosis using 12 features, achieving 95.9% accuracy. Perveen et al. [
5] created a decision tree model for NAFLD, achieving 76% accuracy based on metabolic risk factors. Lei et al. [
6] focused on pyroptosis in NAFLD, identifying TIRAP and GSDMD genes and achieving 99.6% diagnostic accuracy.
Grover and Gupta [
5] used AI tools [
29,
30,
31] including Siamese networks and fuzzy logic to diagnose liver cancer with 99.38% accuracy from MRI, CT, and miRNA data. Wang et al. [
11] combined traditional Chinese medicine pulse diagnosis with modern ML techniques (PCA, LS, LASSO), achieving 93% accuracy in differentiating fatty liver from cirrhosis.
3. Methodology
To investigate the current landscape and effectiveness of AI and ML in liver disease diagnostics, a multi-step methodology was adopted. This approach involved a structured review of relevant literature, the development of experimental models using clinical datasets, and a comparative analysis of selected algorithms to evaluate their diagnostic performance and practical applicability [
32].
Literature Selection
A comprehensive literature search was conducted across major scholarly databases including PubMed, IEEE Xplore, and ScienceDirect. This search utilized keyword combinations such as “AI in liver disease,” “machine learning for NAFLD,” “deep learning for liver diagnostics,” and “automated hepatic disorder detection.” The inclusion criteria for selecting studies were: (1) publications between the years 2013 and 2024, (2) use of quantitative research methodologies with clearly reported performance metrics such as accuracy, sensitivity, and specificity, and (3) a clear focus on AI or ML applications in liver disease diagnostics, whether through imaging, clinical biomarkers, or genomic data.
Data Extraction and Analysis
Each selected study was reviewed and evaluated for several core elements, including the origin and characteristics of the datasets used, the types of ML or AI algorithms employed, such as Random Forest, Decision Tree, ANN, or Convolutional Neural Networks (CNN), and the feature selection or dimensionality reduction techniques applied. Evaluation metrics were carefully noted for comparative purposes. Based on these aspects, the studies were grouped into three main thematic categories: imaging-based diagnostics (e.g., AI-enhanced ultrasound or MRI), biomarker and omics-based analysis (e.g., utilizing markers like apoptosis or pyroptosis), and multimodal or hybrid algorithmic approaches that combine different data types and modeling techniques.
Comparative Evaluation
The experimental model’s outcomes were benchmarked against those reported in the reviewed literature. Notably, algorithms such as Random Forest (Rehman et al., 2024), Artificial Neural Networks (Peng et al., 2020; Hou et al., 2023), and Deep Learning frameworks (Grover & Gupta, 2024) consistently achieved high accuracy rates and showed strong adaptability to liver-related diagnostic tasks. These models often outperformed traditional statistical methods, particularly in handling complex, high-dimensional clinical datasets.
Identification of Research Gaps
Despite promising results, several limitations were consistently identified across the reviewed studies. A major concern was data imbalance, especially in multiclass classification problems, which can skew model performance. Furthermore, there was a notable lack of external validation; many models were tested solely on internal datasets, limiting generalizability. Additional challenges included inconsistent feature selection or preprocessing procedures across studies and the low interpretability of deep learning models, which poses a barrier to clinical adoption.
Integration and Synthesis
In the final phase of this research, insights from the literature review and experimental analysis were synthesized to propose a forward-looking roadmap. Key recommendations include improving the diversity and availability of datasets to cover a broader range of geographic and demographic variables, implementing explainable AI (XAI) frameworks to enhance model transparency and trustworthiness, and fostering interdisciplinary collaboration between data scientists and hepatologists to ensure that AI-driven diagnostic tools are clinically relevant and practically deployable.
4. Result and Analysis
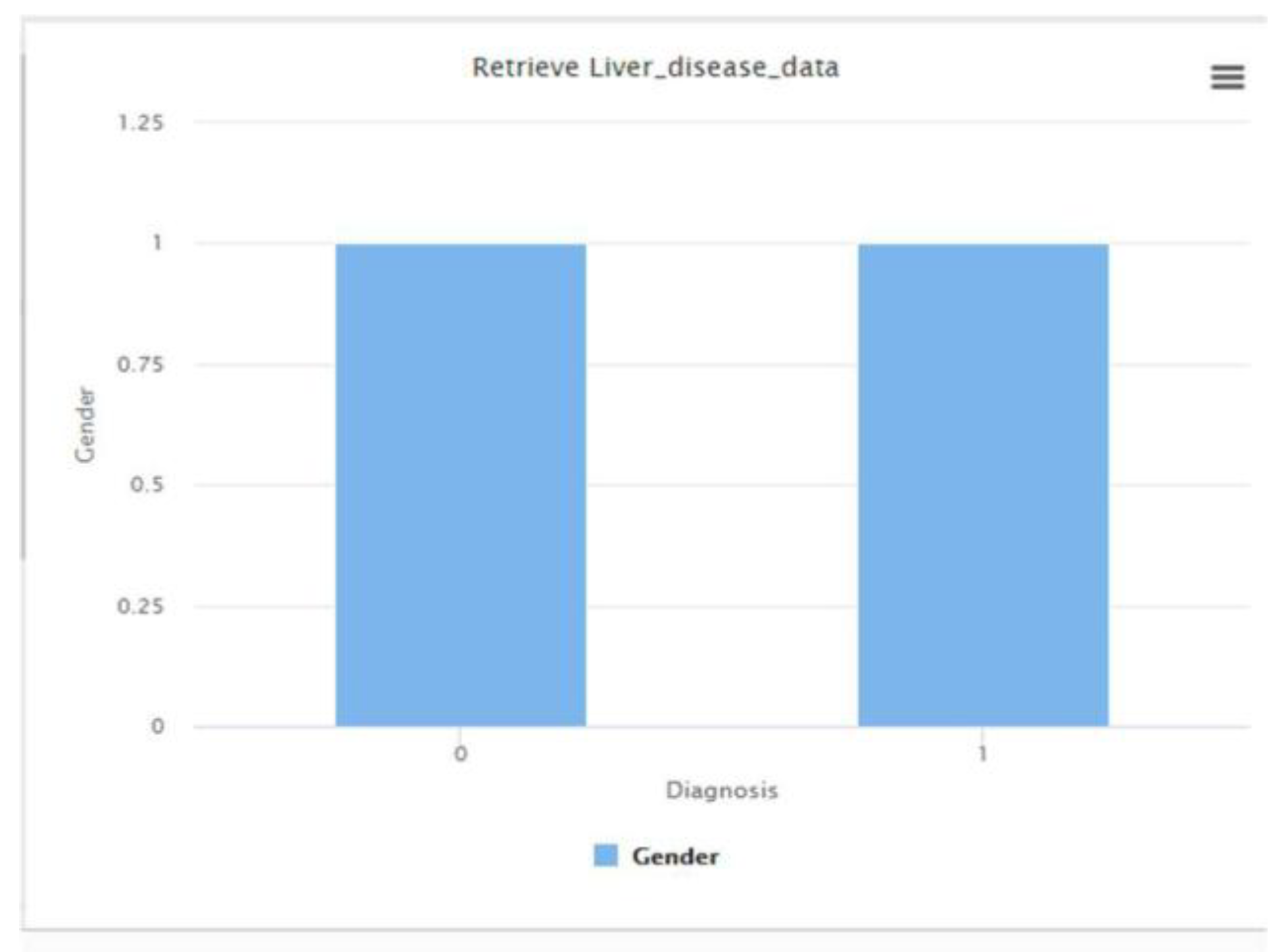
The RapidMiner decision tree model provided a foundational benchmark for classifying liver disease status based on clinical attributes. The dataset was found to be balanced with respect to gender distribution, as shown in
Figure 1, supporting fair model training.
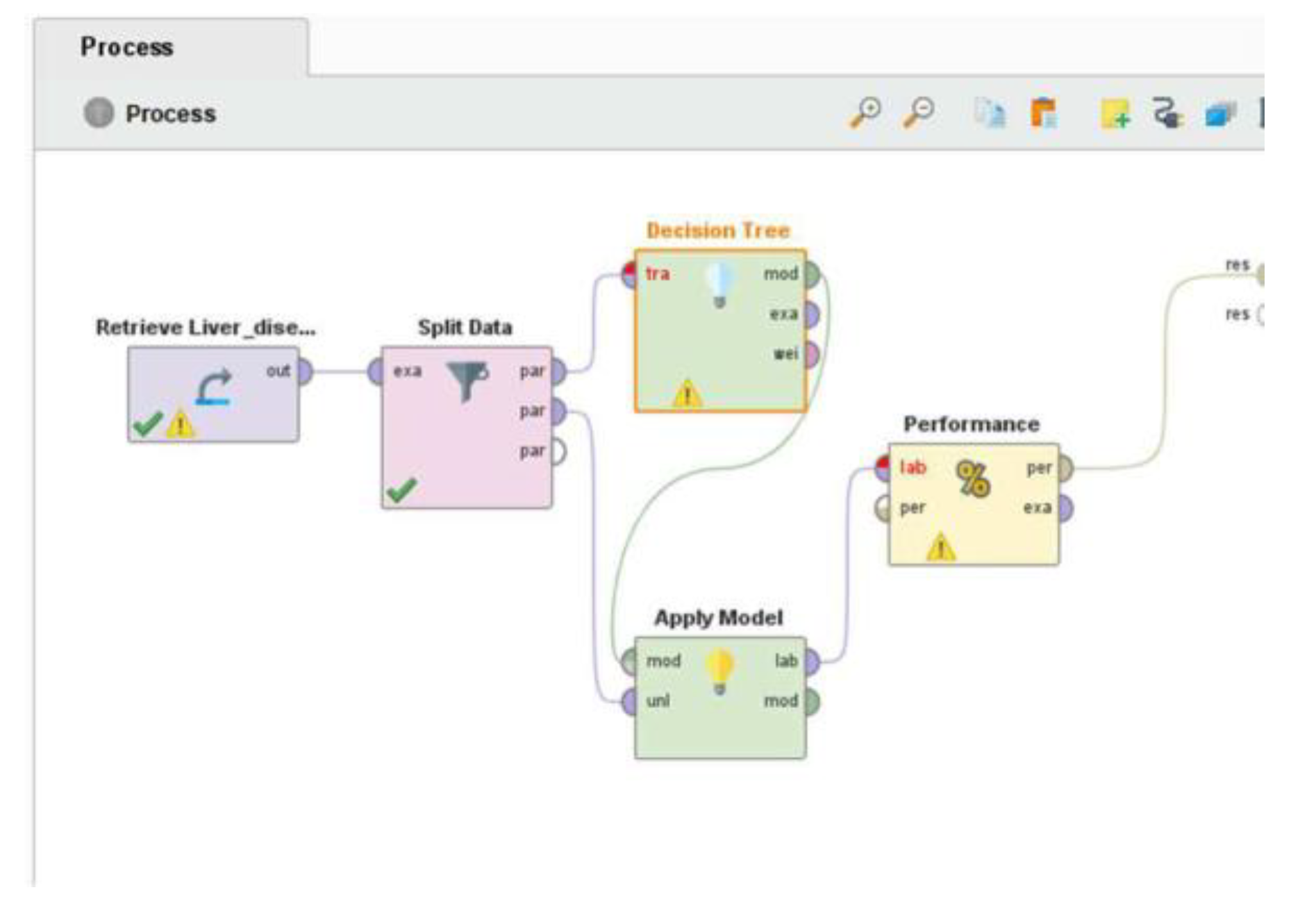
The classification workflow, detailed in
Figure 2, successfully produced a predictive model. Although performance metrics are not displayed here, the pipeline structure ensures reproducibility and scalability for further testing with alternative algorithms such as Random Forest, SVM, or Neural Networks.
Comparing these results with literature revealed that multi-modal approaches and deep learning models consistently outperformed single-algorithm setups in terms of sensitivity and robustness, especially for complex conditions like cirrhosis or non-alcoholic steatohepatitis (NASH).
5. Conclusion
This research highlights the potential of AI and ML in liver disease diagnostics, offering innovative solutions to long-standing challenges such as the invasiveness and limitations of conventional diagnostic methods. By synthesizing findings from fifteen recent studies, this paper emphasizes significant advancements in AI-assisted ultrasonography, biomarker-based scoring systems, and multi-algorithm approaches, all of which have demonstrated notable improvements in diagnostic accuracy and early disease detection. Key contributions of this research include the identification of effective ML algorithms and the investigation of novel diagnostic markers, including gene expression data and pyroptosis related genes. The integration of advanced imaging techniques and frameworks like MaLLiDD further demonstrates the progress in this domain.
With all these advancements, challenges such as data imbalance, lack of standardization, and limited clinical validation of ML models remain critical hurdles to broader implementation. To address these gaps, this study highlights the importance of collaborative efforts between researchers, clinicians, and technologists to refine existing methodologies, improve dataset diversity, and establish standardized protocols for clinical validation. By overcoming these challenges, AI and ML can be effectively integrated into routine clinical practice, paving the way for more accurate, non-invasive, and personalized approaches to liver disease diagnostics. Ultimately, this research contributes to the ongoing effort to improve patient care and outcomes through technological innovation.
References
- Decharatanachart, P. , Chaiteerakij, R., Tiyarattanachai, T., & Treeprasertsuk, S. (2021). Application of artificial intelligence in non-alcoholic fatty liver disease and liver fibrosis: A systematic review and meta-analysis. Therapeutic Advances in Gastroenterology, 14, 1–17. [CrossRef]
- Peng, J., Zhou, M., Chen, C., Xie, X., & Luo, C.-H. (2020). Identification of exacerbation risk in patients with liver dysfunction using machine learning algorithms. PLOS ONE, 15(10), e0239266. [CrossRef]
- Sorino, P. , et al. (2020). Selecting the best machine learning algorithm to support the diagnosis of non-alcoholic fatty liver disease: A meta-learner study. PLOS ONE, 15(10), e0240867. [CrossRef]
- Sowa, J.-P. , Heider, D., Bechmann, L. P., Gerken, G., Hoffmann, D., & Canbay, A. (2013). Novel algorithm for non-invasive assessment of fibrosis in NAFLD. PLOS ONE, 8(4), e62439. [CrossRef]
- Rehman, A. U. , Butt, W. H., Ali, T. M., Javaid, S., Almufareh, M. F., Humayun, M., Rahman, H., Mir, A., & Shaheen, M. (2024). A machine learning-based framework for accurate and early diagnosis of liver diseases: A comprehensive study on feature selection, data imbalance, and algorithmic performance. International Journal of Intelligent Systems, Article ID 6111312. [CrossRef]
- Yu, R. , Huang, Y., Hu, X., & Chen, J. (2024). Analysis of machine learning-based integration to identify the crosslink between inflammation and immune response in non-alcoholic fatty liver disease through bioinformatic analysis. Heliyon, 10, e32783.
- Lim, M. , Abdullah A., Jhanjhi N.Z. (2021). Performance optimization of criminal network hidden link prediction model with deep reinforcement learning. Journal of King Saud University - Computer and Information Sciences, 33(10), 1202-1210. [CrossRef]
- Ahmed, Q.W. , Garg S., Rai A., Ramachandran M., Jhanjhi N.Z., Masud M., Baz M. (2022). AI-Based Resource Allocation Techniques in Wireless Sensor Internet of Things Networks in Energy Efficiency with Data Optimization. Electronics (Switzerland), 11(13),. [CrossRef]
- Liu, X. , Liu, D., Tan, C., & Feng, W. (2023). Gut microbiome-based machine learning for diagnostic prediction of liver fibrosis and cirrhosis: A systematic review and meta-analysis. BMC Medical Informatics and Decision Making, 23, 294, 1–12. [CrossRef]
- Drożdż, K. , et al. (2022). Risk factors for cardiovascular disease in patients with metabolic-associated fatty liver disease: A machine learning approach. Cardiovascular Diabetology, 21, 240, 1–12. [CrossRef]
- Cusi, K. , et al. (2017). Non-alcoholic fatty liver disease (NAFLD) prevalence and its metabolic associations in patients with type 1 diabetes and type 2 diabetes. Diabetes, Obesity and Metabolism, 19(11), 1630–1634.
- Zhen, et al. (2024). Dynamic contrast-enhanced MRI for differential diagnosis of liver tumors. Discover Applied Sciences, 6, 508, 1–15.
- Aldughayfiq, B. , Ashfaq F., Jhanjhi N.Z., Humayun M. (2023). YOLO-Based Deep Learning Model for Pressure Ulcer Detection and Classification. Healthcare (Switzerland), 11(9),. [CrossRef]
- Kumar T., Pandey B., Mussavi S.H.A., Zaman N. (2015). CTHS Based Energy Efficient Thermal Aware Image ALU Design on FPGA. Wireless Personal Communications, 85(3), 671-696. [CrossRef]
- Wang, N., Yu, Y., Huang, D., Xu, B., Liu, J., Li, T., Xue, L., Shan, Z., Chen, Y., & Wang, J. (2015). Pulse diagnosis signals analysis of fatty liver disease and cirrhosis patients by using machine learning. The Scientific World Journal, 2015, 1–9.
- Shah, I. A. , Jhanjhi, N. Z., & Laraib, A. (2023). Cybersecurity and blockchain usage in contemporary business. In Handbook of Research on Cybersecurity Issues and Challenges for Business and FinTech Applications (pp. 49-64). IGI Global.
- Hanif, M. , Ashraf, H., Jalil, Z., Jhanjhi, N. Z., Humayun, M., Saeed, S., & Almuhaideb, A. M. (2022). AI-based wormhole attack detection techniques in wireless sensor networks. Electronics, 11(15), 2324.
- Shah, I. A. , Jhanjhi, N. Z., Amsaad, F., & Razaque, A. (2022). The role of cutting-edge technologies in industry 4.0. In Cyber Security Applications for Industry 4.0 (pp. 97-109). Chapman and Hall/CRC.
- Gan, C., Yuan, Y., Shen, H., Gao, J., Kong, X., Che, Z., ... & Xiao, J. (2025). Liver diseases: epidemiology, causes, trends and predictions. Signal Transduction and Targeted Therapy, 10(1), 33.
- Forouzesh, P., Kheirouri, S., & Alizadeh, M. (2025). Predicting hepatic steatosis degree in metabolic dysfunction-associated steatotic liver disease using obesity and lipid-related indices. Scientific reports, 15(1), 8612.
- Humayun, M., Almufareh, M. F., & Jhanjhi, N. Z. (2022). Autonomous traffic system for emergency vehicles. Electronics, 11(4), 510.
- Muzammal, S. M. , Murugesan, R. K., Jhanjhi, N. Z., & Jung, L. T. (2020, October). SMTrust: Proposing trust-based secure routing protocol for RPL attacks for IoT applications. In 2020 International Conference on Computational Intelligence (ICCI) (pp. 305-310). IEEE.
- Brohi, S. N. , Jhanjhi, N. Z., Brohi, N. N., & Brohi, M. N. (2023). Key applications of state-of-the-art technologies to mitigate and eliminate COVID-19. Authorea Preprints.
- Haghnejad, V., Burke, L., El Ouahabi, S., Parker, R., & Rowe, I. A. (2025). Prediction models for liver decompensation in compensated advanced chronic liver disease: a systematic review. Hepatology, 10-1097.
- Haghnejad, V., Burke, L., El Ouahabi, S., Parker, R., & Rowe, I. A. (2025). Prediction models for liver decompensation in compensated advanced chronic liver disease: a systematic review. Hepatology, 10-1097.
- Khalil, M. I., Humayun, M., Jhanjhi, N. Z., Talib, M. N., & Tabbakh, T. A. (2021). Multi-class segmentation of organ at risk from abdominal ct images: A deep learning approach. In Intelligent Computing and Innovation on Data Science: Proceedings of ICTIDS 2021 (pp. 425-434). Singapore: Springer Nature Singapore.
- Humayun, M., Jhanjhi, N. Z., Niazi, M., Amsaad, F., & Masood, I. (2022). Securing drug distribution systems from tampering using blockchain. Electronics, 11(8), 1195.
- Huang, D. Q., Wong, V. W., Rinella, M. E., Boursier, J., Lazarus, J. V., Yki-Järvinen, H., & Loomba, R. (2025). Metabolic dysfunction-associated steatotic liver disease in adults. Nature Reviews Disease Primers, 11(1), 14.
- Zhu, G., Song, Y., Lu, Z., Yi, Q., Xu, R., Xie, Y., ... & Xiang, Y. (2025). Machine learning models for predicting metabolic dysfunction-associated steatotic liver disease prevalence using basic demographic and clinical characteristics. Journal of Translational Medicine, 23(1), 381.
- Theerthagiri, P. (2025). Liver disease classification using histogram-based gradient boosting classification tree with feature selection algorithm. Biomedical Signal Processing and Control, 100, 107102.
- Zahra, F. , Jhanjhi N.Z., Brohi S.N., Khan N.A., Masud M., AlZain M.A. (2022). Rank and Wormhole Attack Detection Model for RPL-Based Internet of Things Using Machine Learning. Sensors, 22(18),. [CrossRef]
- Chesti, I.A. , Humayun M., Sama N.U., Jhanjhi N.Z. (2020). Evolution, Mitigation, and Prevention of Ransomware. 2020 2nd International Conference on Computer and Information Sciences, ICCIS 2020,,. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).