Submitted:
12 November 2025
Posted:
13 November 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
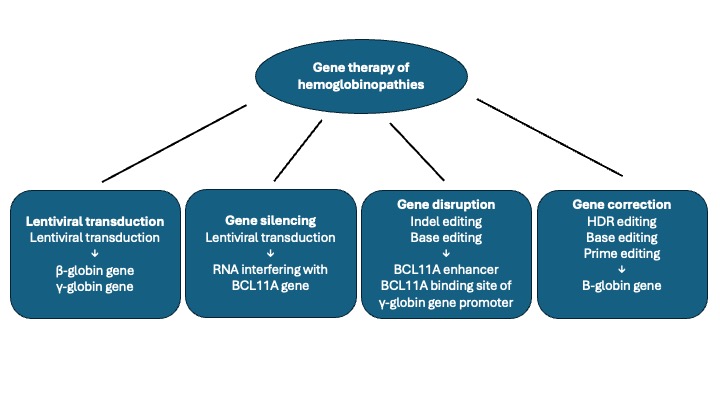
2. Gene Therapy Strategies
3.1. Different Lentiviral Vectors Used in Clinical Trials for Hemoglobinopathies
3.2. Lovo-Cel Therapy for Sickle Cell Disease
3.3. Gene Therapy of SCD Using Lenti/G-βAS3-FB Vector
3.4. Gene Therapy of SCD Using Lentiviral Vectors Encoding HbFG16D
3.5. Lovo-Cel Gene Therapy for β-Thalassemia
3.6. LV-GLOBE Gene-Therapy of β-Thalassemia
3.7. Modified LV Globin Gene Therapy for Pediatric β°/β°
3.8. Lentiviral BCL11A Short Hairpin mRNA
4. Gene Editing Therapy of Hemoglobinopathies
4.1. Gene Editing Based on Double-Strand Break
4.2. Gene Editing Without Double-Strand DNA Break
4.3. Specific Transcriptional and Epigenetic Modulation Using Dead Caspase 9 (dCas9)
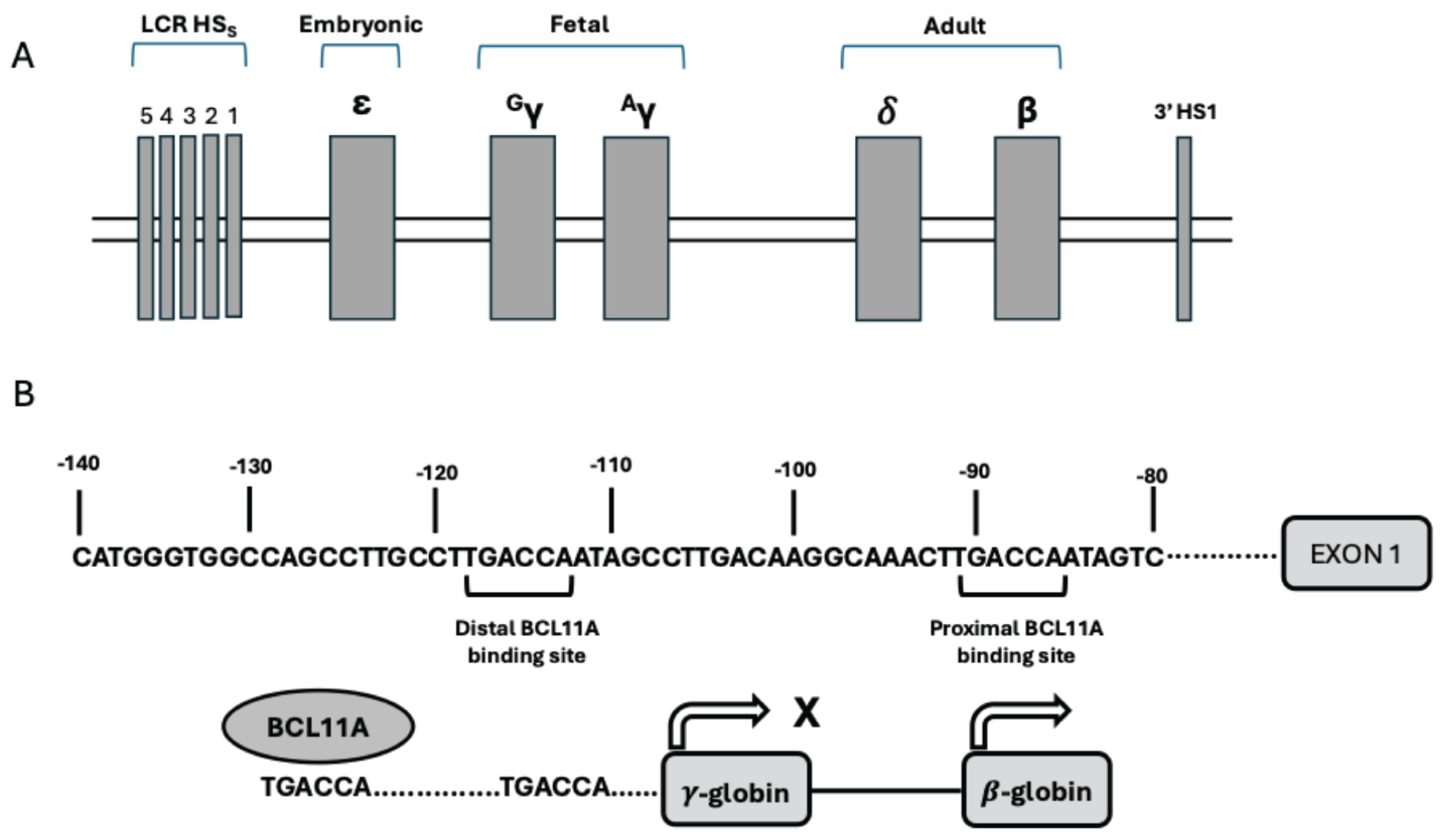
4.4. CRISPR-Cas9 Editing of BCL11A Enhancer: Studies in β-Thalassemia
4.5. CRISPR-Cas9 Editing of BCL11A Enhancer: Studies in Sickle Cell Disease
4.6. Gene Editing of γ-Globin Gene Promoters Using CRISPR-Cas9 Technology
4.7. Base Editing of BCL11A Binding Site in γ-Globin Gene Promoter
4.8. Cas12 Editing of BCL11A Binding Site in γ-Globin Genes Promoter
4.9. Gene Correction Studies Using CRISPR-Cas9 Gene Editing
4.10. Zinc Finger Nuclease-Mediated Gene Editing of BCL11A Erythroid Enhancer in HSCs
5. Gene Therapy of β-Hemoglobinopathies Through In Vivo Gene Editing
6. Affordability of Gene Therapies for Hemoglobinopathies
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Orkin, S.H. The fetal-to-adult hemoglobin switch—mechanisms and therapy. N Engl J Med 2025, 392, 2135–2149. [Google Scholar] [CrossRef]
- Huang, P.; Pesiak, C.A.; Ren, R.; Khandros, E.; Qin, K.; Keller, C.A.; Giardine, B.; Bell, H.W.; Lan, X.; Sharma, M.; et al. HIC2 controls developmental hemoglobin switching by repressing BCL11A transcription. Nat Genet 2022, 54, 1417–1426. [Google Scholar] [CrossRef]
- Bauer, D.E.; Kamran, S.C.; Lessard, S.; Xu, J.; Fujiwara, Y.; Lin, C.; Shao, Z.; Canver, M.C.; Smith, E.C.; Pinello, L.; et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 2013, 342, 2453–2457. [Google Scholar] [CrossRef]
- Masuda, T.; Wang, X.; Maedfa, M.; Canver, M.C.; Sher, F.; Funnel, A.; Fisher, C.; Suclu, M.; Martyn, G.; Norton, L.; et al. Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science 2016, 351, 285–289. [Google Scholar] [CrossRef]
- Piel, F.; Steinberg, M.; Rees, D.C. Sickle cell disease. N Engl J Med 2017, 376, 1561–1573. [Google Scholar] [CrossRef]
- Taher, A.; Musallam, K.; Cappellini, D. β-thalassemias. N Engl J Med 2021, 384, 727–743. [Google Scholar] [CrossRef]
- Jones-Wonni, B. A review of gene therapies for hemoglobinopathies. Hemoglobin 2024, 48, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.; Sathish, S.; London, E.; Johnson, T.L.; Essaewi, K.; Leonard, A.; Tisdale, J.F.; Demirici, S. Genome editing strategies for targeted correction of β-globin mutation in sickle cell disease: from bench to bedside. Mol Ther 2025, 33, 2154–2171. [Google Scholar] [CrossRef] [PubMed]
- Fitzhugh, C.D.; Cordes, S.; Taylor, T.; Cotes, W.; Roskom, K.; Link, M.; Hsieh, M.M.; Tisdale, J.F. At least 20% donor myeloid chimerism is necessary to reverse the sickle phenotype after allogeneic HSCT. Blood 2017, 130, 1946–1948. [Google Scholar] [CrossRef]
- Frangoul, H.; Stuits, A.; Bruce, K.; Domm, J.; Carroll, C.; Aide, S.; Duckworth, M.; Evans, M.; McManus, M. Best practices in gene therapy for sickle cell disease and transfusion-dependent β-thalassemia. Transpl Cell Therapy.
- Poletti, V.; Mavilio, F. Designing lentiviral vectors for gene therapy of genetic diseases. Viruses 2021, 13, 1526. [Google Scholar] [CrossRef] [PubMed]
- Ballantine, J.; Tisdale, J.F. Gene therapy for sickle cell disease: recent advances, clinical trials and future directions. Cytotherapy 2025, 27, 826–834. [Google Scholar] [CrossRef]
- Sadelain, M.; Wang, C.H.; Antoniou, M.; Grosveld, F.; Mulligan, R.C. Generation of a high-titer retroviral vector capable of expressing high levels of the human beta-globin gene.
- Miller, A.D.; Bender, M.A.; Harris, E.; Kalako, M.; Gelinas, R.E. Design of retrovirus vectors for transfer and expression of the human β-globin gene. J Virol 1988, 62, 4337–4345. [Google Scholar] [CrossRef]
- Psatha, N.; Sava, P.; Georgopoulos, G.; Psehaudi, K.; Iwata, M.; Bloom, J.; Ulyanova, T.; Wang, H.; Kiztsau, A.; Vasiloudis, N.I.; et al. Large-scale discovery of potent, compact and erythroid specific enhancers for gene therapy vectors. Nat Commun 2025, 16, 4325. [Google Scholar] [CrossRef]
- Pawliuk, R.; Westerman, K.A.; Fabry, M.E.; Payen, E.; Bouhassira, E.E.; Acharya, S.A.; Ellis, J.; London, I.M.; Eaves, C.J.; Humphries, R.K.; et al. Correction of sickle-cell disease in transgenic mouse models by gene therapy. Science 2001, 294, 2368–2371. [Google Scholar] [CrossRef]
- McCune, S.L.; Reilly, M.P.; Chomo, M.J.; Asakura, T.; Townes, T.M. Recombinant human hemoglobins designed for gene therapy of sickle cell disease. Proc Natl Acad Sci USA 1994, 91, 9852–9856. [Google Scholar] [CrossRef]
- Grimley, M.; Sherestha, A.; Felker, S.; Lutzko, C.; Arumugam, P.I.; Wititng, S.; et al. Safety and efficacy of Aru-1801 in patients with sickle cell disease: early results from the phase 1-2 momentum study of a modified gamma globin gene therapy and reduced intensity conditioning. Blood 2021, 138 (suppl.1), 3970–3972. [Google Scholar] [CrossRef]
- Cabriolu, A.; Odek, A.; Zamparo, L.; Yuan, H.; Leslie, C.D.; Sadelain, M. Globin vector regulatory elements are active in early hematopoietic progenitor cells. Mol Ther 2022, 30, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Marshall, E. Gene editing. Gene genich cures sickle cell in mice. Science 2011, 294, 2268. [Google Scholar]
- Demirci, S.; Gudmundsdottir, B.; Li, Q.; Haro-Mora, J.J.; Nassehi, T.; Drysdale, C.; Yapundich, M.; Gamer, J.; Seifuddin, F.; Tisdale, J.F.; et al. βT87Q-globin gene therapy reduces sickle hemoglobin production, allowing for ex vivo anti-sickling activity in human erythroid cells. Methods Clin Dev 2020, 17, 912–916. [Google Scholar] [CrossRef] [PubMed]
- Ribeil, J.A.; Hacein-Bey-Abina, S.; Payen, E.; Magnani, A.; Semeraro, M.; Magrin, E.; Caccavelli, L.; Neven, B.; Bourget, P.; El Nemer, W.; et al. Gene therapy in a patient with sickle cell disease. N Engl J Med 2017, 376, 848–855. [Google Scholar] [CrossRef]
- Magrin, E.; Semeraro, M.; Hebert, N.; Joseph, L.; Magnani, A.; Chalumeau, A.; Gabrion, A.; Roudaut, C.; Marouene, J.; Lfrere, F.; et al. Long-term outcomes of lentiviral therapy for the β-hemoglobinopathies: the HGB-205 trial. Nat Med 2022, 28, 81–88. [Google Scholar] [CrossRef]
- Kanter, J. Thompson, A.A.; Piercey, F.J.; Hsieh, M.; Uchida, N.; Leboulch, P.; Schmidt, M.; Bonner, M.; Guo, R.; Miller, A.; et al. Loco-cel gene therapy for sickle cell disease: treatment process evolution and outcomes in the initial groups of the HGB-206 study. Am J Hematol 2023, 98, 11–22. [Google Scholar] [CrossRef]
- Kanter, J. Walters, M.C.; Krishnamurti, L.; Mapara, M.Y.; Kwiatkowski, J.L.; Rifkin-Zenenberg, S.; Aygun, B.; Kasow, K.A.; Pierciey, F.J.; Bonner, M.; et al. Biologic and clinical efficacy of LentiGloibin for sickle cell disease. N Engl J Med. 2022, 386, 617‐628.
- Rifkin-Zenenberg, S.; Kanter, J.; Kinney, M.A.; Kwiatkowski, J.L.; Nickel, R.S.; Walters, M.C.; Parikh, S.; Thompson, A.; George, A.P.; Mapara, M.; et al. An update on Lovotibeglogene autotemcel (Lovo-cel) for sickle cell disease (SCD) and analysis of early predictors of response to lovo-cel. Blood 2024, 144 (suppl.1), 511–512. [Google Scholar] [CrossRef]
- Kanter, J.; Chawla, A.; Thompson, A.A.; Kwiatkowski, J.L.; Parikh, S.; Mapara, M.Y.; Rifkin-Zenenberg, S.; Aygun, B.; Kasow, K.A.; Gupta, A.O.; et al. Lovotibeglogene autotemcel gene therapy for sickle cell disease: 60 months follow-up. J Sickle Dis 2024, 1, 3–4. [Google Scholar] [CrossRef]
- Kinney, M.A.; Shestopalov, I.; Christiansen, L.; Jiang, H.; Foos, M.; Elliot, H.; Chawla, A.; Piercey, F.J. Predictors of biologic efficacy with Lovotibeglogene autotemcel (Lovo-cel) gene therapy in patients with sickle cell disease. Transpl Cell Therapy 2024, 30 (supplement 2), 302. [Google Scholar] [CrossRef]
- Heering, W.L.; Gallagher, M.E.; Shah, N.; Morse, K.C.; Mladsi, D.; Dong, O.M.; Chawrla, A.; Leiding, J.; Zhang, L.; Paramore, C.; et al. Cost-effectiveness for Lovotibeglogene autotemcel (Lovo-Cel) gene therapy for patients with sickle cell disease and recurrent vaso-occlusive events in the United States. PharmacoEconomics 2024, 42, 693–714. [Google Scholar] [CrossRef] [PubMed]
- Urbinati, F.; Fernandez, B.C.; Masiuk, K.E.; Poletti, V.; Hollis, R.P.; Koziol, C.; Kaufman, M.L.; Brown, D.; Mavilio, F.; Kohn, D.B. Gene therapy for sickle cell disease: a lentiviral vector comparison study. Human Gene Therapy 2018, 29, 1153–1166. [Google Scholar] [CrossRef] [PubMed]
- Prueksapraopong, C.; Fernandes, A.; Fernandez, B.C.; Roy, S.; Habtemariam, B.; Romero, Z.; Moore, T.M.; Schiller, G.J.; Kohn, D.B. Clinical outcomes of Lenti/G-βAS3-FB lentiviral vector gene therapy for sickle cell disease. Transpl Cell Ther 2025, 31, S255–S256. [Google Scholar] [CrossRef]
- Sobrino, S.; Joseph, L.; Magrin, E.; Chalumeau, A.; Hebert, N.; Corsia, A.; Denis, A.; Roudaut, C.; Aussel, C.; : Leblanc, O.; et al. Severe inflammation and lineage skewing are associated with poor engraftment of engineered hematopoietic stem cells in patients with sickle cell disease. Nat Commun 2025, 16, 3137. [Google Scholar] [CrossRef]
- Perumbeti, A.; Higashimoto, T.; Urbinati, F.; Franco, R.; Meiselman, H.J.; Witte, D.; Lalik, P. A novel human gamma-globin gene vector for genetic correction of sickle cell anemia in a humanized sickle mouse model: critical determinants for successful correction. Blood 2009, 114, 1174–1185. [Google Scholar] [CrossRef]
- Grimley, M.; Davies, S.M.; Shrestha, A.; Shova, A.; Asnani, M.; Kent, M.; Sayani, F.; Quin, C.; Niss, O.; Lutzko, C.; et al. Lentiviral gene therapy with reduced-intensity conditioning for sickle cell disease: a phase 1-2 trial. Nat Med 2025, 31, 2204–2212. [Google Scholar]
- Thompson,A.A.; Walters, M.C.; Kwiatkowski, J.; Rasko, J.; Ribeil,J.A.; Hongeng, S.; Magrin, E.; Schiller, G.J.; Payen, E.; Semeraro, M.; et al. Gene therapy in patients with transfusion-dependent β-thalassemia. N Engl J Med. 2022, 386, 617‐628.
- Locatelli, F.; Thompson, A.A.; Kwiatkowski, J.; Porter, J.B.; Thrasher, A.J.; Hongerng, S.; Sauer, M.G.; Thuret, I.; Lal, A.; Algeri, M.; et al. Betibeglogene autotemcel gene therapy for non-β°/β° genotype β-thalassemia. N Engl J Med 2022, 386, 415–427. [Google Scholar]
- Kwiatkowski, J.; Walters, M.C.; Hongeng, S.; Yannaki, E.; Kulozik, A.E.; Kunz,J.B.; Sauer, M.G.; Tharasher, A.J.; Thuret, I.; Lal, A.; et al. Betibeglogene autotemcel gene therapy in patients with thalassemiadependent, severe genotype β‐thalassemia (HGB‐212): a non‐randomised, multicentre, single‐arm, openlabel, single‐dose, phase 3 trial. Lancet 2024, 404, 2175‐2186 .
- Gibson, N.M.; Friedman, D.F.; Elgarten, C.W.; Haimed, A.; Khandros, E.; Worster, E.; Bardahl, J.; Wray, L.; Wang, Y.; Thompson, A.A.; et al. Post-approval, real-world experience with Betibeglogene Autotemcel for transfusion-dependent beta thalassemia. Transpl Cell Therapy 2025, 31 (suppl. 1). S254.. [Google Scholar] [CrossRef]
- Mizza, A.; Ritsert, M.L.; Tao, G.; Thakar, H.; Labitz, S.; Heine, S.; Kosher, L.; Durken, M.; Schmitt, A.; Pavel, P.; et al. Gene therapy in transfusion-dependent non-β°/β° genotype β-thalassemia: first real-world experience of beti-cel. Blood Adv 2025, 9, 29–38. [Google Scholar]
- Marktel, S.; Scaramuzza, S.; Cicalese, M.P.; Giglio, F.; Galimberti, S.; Lidonnici, M.R.; Calbi, V.; Assanelli, A.; Bernardo, M.E.; Rossi, C.; et al. Intrabone hematopoietic stem cell gene therapy for adult and pediatric patients affected by transfusion-dependent β-thalassemia. Nat Med 2019, 25, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, Z.; Chen, S.; Huang, Y.; Ling, S.; Wang, Q.; Wang, Q. Preclinical efficacy and safety evaluation of BD211 autologous CD34+ hematopoietic stem cell injection for transfusion-dependent β-thalassemia in NCG-X mice. Front Cell Dev Biology 2025, 13, 1607707. [Google Scholar]
- Li, S.; Ling, S.; Wang, D.; Wang, X.; Hao, F.; Yin, L.; Yuan, Z.; Liu, L.; Zhang, L.; Li, Y.; et al. Modified lentiviral globin gene therapy for pediatric β°/β° transfusion-dependent β-thalassemia: a single-center, single-arm pilot trial. Cell Stem Cell 2024, 31, 961–973. [Google Scholar]
- Brendel, C.; Negre, O.; Rothe, M.; Guda, S.; Parson, G.; Harris, C.; McGiuness, M.; Abriss, D.; Tsytsykova, A.; Klatt, D.; Bentler, M.; et al. Preclinical evaluation of a novel lentiviral vector driving lineage-specific BCL11A knockdown fro sickle cell gene therapy. Mol Therapy Methods Clin Dev 2020, 17, 589–595. [Google Scholar] [CrossRef]
- Esrick, E.B.; Lehmann, L.E.; Biffi, A.; Achebe, M.; Brendel, C.; Ciuculescu, M.F.; Daley, H.; MacKinnon, B.; Morris, E.; Federico, A.; et al. Post-transcriptional genetic silencing of BCL11A to treat sickle cell disease. N Engl J Med 2021, 384, 205–215. [Google Scholar]
- Esrick, E.B.; Federico, A.; Abriss, D.; Armant, M.; Boardman, K.; Brendel, C.; Ciuculescu, M.F.; Daley, H.; Dansereau, C.; Fernandes, A.; et al. Induction of fetal hemoglobin and reduction of clinical manifestations in patients with severe Ahmir-base lentiviral gene therapy for post-transcriptional gene editing of BCL11A: updated results from pilot and feasibility trial. Blood 2022, 140 (suppl. 1), 10665–10667. [Google Scholar]
- Esrick, E.B.; Lehmann, L.E.; Federico, A.; Daley, H.; Dansereau, C.; DeOliveira, S.; Everett, J.; Kao, P.C.; Liu, B.; Moore, T.; et al. Long-term follow-up of the firsty in human post-transcriptional genetic silencing of BCL11A in sickle cell disease in a phase 1 pilot and feasibility study. Blood 2025, 146 (suppl.1), abs25–2853. [Google Scholar]
- Jimenez-Kurlander, L.; Kao, P.C.; Morris, E.; Federico, A.; Moore, T.; DeOliveira, S.; Fernandes, A.; Kohn, D.B.; London, W.B.; Williams, D.A.; et al. Patient and parent-reported outcomes post-treatment with Shmir-based lentiviral gene therapy for sickle cell disease. Blood 2023, 142 (suppl.1), 3750. [Google Scholar] [CrossRef]
- De Souza, D.C.; Hebert, N.; Esrick, E.B.; Ciulescu, M.F.; Archer, N.A.; Armant, M.; Audereau, E.; Brendel, C.; Di Caprio, G.; et al. Genetic reversal of the globin switch concurrently modulates both fetal and sickle hemoglobin and reduces red cell sickling. Nat Commun 2023, 14, 5850. [Google Scholar] [CrossRef] [PubMed]
- Spencer Chapman, M.S.; Cull, A.H.; Ciuculescu, M.F.; Esrick, E.B.; Mitchell, E.; Jung, H.; O’Neill, L.; Roberts, K.; Fabre, M.A.; Williams, N.; et al. Clonal selection of hematopoietic stem cells after gene therapy for sickle cell disease. Nature 2023, 29, 3175–3183. [Google Scholar] [CrossRef]
- Xue, C.; Greene, E.C. DNA repair pathway choices in CRISPR-Cas9-mediated genome editing. Trends Genet 2021, 37, 639–656. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Kostamo, Z.; Ortega, M. A: Xu, C.; Feliciano, P.R.; Budak, E.; Lam, D.; Winton, V.; Jenkins, R.; Venugopal, A.; Zhang, M.; et al. Base editing HbS to HbG-Makassar improves hemoglobin function supporting its use in sicke cell disease. Nat Commun. 2025, 16, 1441.
- Radtke, S.; Fields, E.; Swing, K.; Kanestrom, G.; Yen, J.S.; Pande, D.; Enstrom, M.R.; Humbert, O.; Weiss, M.J.; Liu, D.R.; et al. Engraftment and persistence of HBB base-edited hematopoietic stem cells in nonhuman primates. Sci Transl Med 2025, 17, eadn2601. [Google Scholar] [CrossRef]
- Mayuranathan, T.; Newby, G.A.; Feno, R.; Yao, Y.; Mayberry, K.D.; Lazzarotto, C.R.; LI, Y.; Levine, R.M.; Nimmagadda, N.; Dempsey, E.; Kang, G.; et al. Potent and uniform fetal hemoglobin induction via base editing. Nat Genet 2023, 55, 1210–1220. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Qiu, H.Y.; Sun, S.; Fu, Z.C.; Wang, Q.Q.; Qian, X.; Wang, L.; Zhai, X.; Wei, J.; Wang, Y.; et al. Base editing of the HBG promoter induces potent fetal hemoglobin expression with no detectable off-target mutations in human HSCs. Cell Stem Cell 2023, 30, 1624–1639. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Martinucci, P.; Amistadi, S.; Felix, T.; Mombled, M.; Tachtsidi, A.; Corre, G.; Chalumeau, A.; Hardouin, G.; Martin, J.; et al. Multiplex base editing of BCL11A regulatory elements to treat sickle cell disease. Cell Rep Med 2025, 26, 102376. [Google Scholar] [CrossRef]
- Rajendiran, V.; Devaraju, N.; Haddad, M.; Ravi, N.S.; Panigrahi, L.; Paul, J.; Gopalakrishnan, C.; Wyman, S.; Ariudainambi, K.; Mahalingam, G.; et al. Base editing of key residues in the BCL11A-XL-specific zinc finger domains derepresses fetal globin expression. Mol Ther 2024, 32, 663–677. [Google Scholar] [CrossRef]
- Chen, P.J.; Liu, D.R. Prime editing for precise and highly versatile genome manipulation. Nat Rev Genet 2023, 24, 161–177. [Google Scholar] [CrossRef]
- Chauhan, V.; Sharp, P.A.; Langer, R. Engineered prime editors with minimal genomic errors. Nature 2025, in press. [Google Scholar] [CrossRef] [PubMed]
- Everette, K.A. ;Newby GA, Levine, R.M.; Mayberry, K.; Jang, Y.; Mayuranathan, T.; Nimmagadda, N.; Dempsey, E.; Li, Y.; Bhoopala, N.; et al. Ex vivo prime editing of patient haematopoietic stem cells rescues sickle-cell disease phenotypes after engraftment in mice. Nat Biomed Eng. 2023, 7, 616‐626.
- Chalumeau, A.; Dames, M.B.; Fontana, L.; Amistadi, S.; Anatoniou, P.; Loganathan, P.; Mombled, M.; Corre, G.; Peterka, M.; Amendola, M.; et al. A prime editing strategy to rewrite the γ-globin promoters and reactivate fetal hemoglobin for sickle cell disease. Blood 2025, in press. [Google Scholar] [CrossRef]
- Fiumara, M.; Ferrari, S.; Omer-Javed, A.; Beretta, S.; Albano, L.; Canarutto, D.; Varesi, A.; Gaddoni, C.; Brombin, C.; Cuganta, F.; et al. Genotoxic effects of base and prime editing in human haematopoietic stem cells. Nature Biotechnol 2024, 42, 877–891. [Google Scholar] [CrossRef]
- Hwang, G.H.; Lee, S.H.; Oh, M.; Kim, S.; Habib, O.; Jang, H.K.; Kim, H.S.; Kim, Y.; Kim, C.H.; Kim, S.; et al. Large DNA deletions occur during DNA repair at 20-fold lower frequency for base editors and prime editors than for Cas9 nucleases. Nature Biomed Eng 2025, 9, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, A.A.; Lim, W.A.; Qi, L.S. Beyond editing: Repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016, 17, 5–15. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2021, 184, 844. [Google Scholar] [CrossRef]
- Bell, H.W.; Feng, R.; Shah, M.; Yao, Y.; Douglas, J.; Doerfler, P.A.; Mayuranathan, T.; O’Dea, M.F.; Li, Y.; Wang, Y.D.; et al. Removal of promoter CpG methylation by epigenome editing reverses HBG silencing. Nat Commun 2025, 16, 6919. [Google Scholar] [CrossRef]
- Ye, L.; Wang, J.; Tan, Y.; Beyer, A.I.; Zie, F.; Muench, M.O.; Kan, Y.W. Genome editing using CRISPR-Cas9 to create the HPFH genotype in HSPCs: an approach for treating sickle cell disease and β-thalassemia. Proc Natl Acad Sci USA 2016, 113, 10661–10665. [Google Scholar] [CrossRef]
- We,Y. ; Zeng, J.; Roscoe, B.P.; Liu, P.; Yao, Q.; Lazzarotto, C.; Clement, K.; Cole, M.A.; Luk, K.; Baricordi, C.; et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med 2019, 25, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Frangoul, H.; Althshuler, D.; Cappellini, M.D.; Chen, Y.S.; Domm, J.; Eustace, B.K.; Foeli, J.; de la Fuente, J.; Grupp, S.; Handgretinger, R.; et al. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N Engl J Med 2021, 384, 252–260. [Google Scholar] [CrossRef]
- Locatelli, F.; Lang, P.; Wall, D.; Meisel, R.; Carbacioglu, S.; Li, A.M.; d e la Fuente, J.; shah, A.J.; Carpentier, B.; Kwiatkowski, J.L.; et al. Examglogene autotemcel for transfusion-dependent β-thalassemia. N Engl J Med 2024, 390, 1663–1676. [Google Scholar] [CrossRef]
- Frangoul, H.; Locatelli, F.; Lang, P.; Meisel, R.; Wall, D.A.; Carbacioglu, S.; Li, A.; de la Fuente, J.; Shah, A.J.; Carpenter, B.; et al. Durable clinical benefits in transfusion-dependent β-thalassemia with Examglogene Autotemcel. Transpl Cel Ther. 2023, 31 (suppl.1) S252.
- Locatelli, F.; Meisel, R.; Carbacioglu, S.; de la Fuente, J.; Algeri, M.; Ruppechjt, J.; Kuo, K.; Shah, A.; Lang, P.; Merkeley, H.; et al. Correction of ineffective erythropoiesis and durable clinical benefit with exagamglogene autotemcel for transfusion-dependent β-thalassemia. Blood 2025, 146 (suppl.1), abs25–8312. [Google Scholar]
- Fu, B.; Liao, J.; Chen, S.; Li, W.; Wang, Q.; Hu, J.; Yang, F.; Hsiao, S.; Jiang, Y.; Wang, L.; et al. CRISPR-Cas9-mediated gene editing of the BCL11A enhancer for pediatric β°/β° transfusion-dependent β-thalassemia. Nat Med 2022, 28, 1573–1580. [Google Scholar] [CrossRef]
- Zheng, B.; Liu, R.; Zhang, X.; Fu, B.; Xu, Y.; Shi, J.; Feng, X.; Wang, L.; Wang, C.; Liang, R.; Tan, L.; et al. Efficacy and safety of BRL-101, CRISPR-Cas9-mediated gene editing of the Bcl11A enhancer in transfusion-dependent β-thalassemia. Blood 2023, 142, 4995–4996. [Google Scholar] [CrossRef]
- Zheng, B.; Liu, R.; Zhang, X.; Fu, B.; Xu, Y.; Shi, J.; Feng, X.; Li, D.; Wu, Y.; Liu, M.; et al. Efficacy and safety of BRL-101, CRISPR-Cas9-mediated gene editing of the Bcl11A enhancer in transfusion-dependent beta-thalassemia. EHA Library. 2024, p1514.
- Frangoul,H. ; Locatelli, F.; Sharma, A.; Bhatia, M.; Mapara, M.; Liem, R.I.; Telfer, P.; Shah, A.J.; Cavazzana, M.; Corbacioglu, S.; et al. Examglogene autotemcel for severe sickle cell disease. N Engl J Med 2024, 390, 16749–1662. [Google Scholar]
- Grupp, S.A.; Locatelli, F.; Sharma, A.; Bhatia, M.; Mapara, M.; Liem, R.I.; Wall, D.A.; Molinari, L.; Dedeken, L.; Kuo, K.; et al. Durable clinical benefits in severe sickle cell disease with exagamglogene autotemcel. Transl Cell Therapy. 2025, 31 (suppl. 1) S20‐S21.
- Grupp, S.; Locatelli, F.; Sharma, A.; Bhatia, M.; Mapara, M.; Liem, R.; Dedeken, L.; Molinari, L.; Eckrich, M.; Kuo, K.; et al. Long-term follow-up demonstrates durable clinical benefits of examglogene autotemcel for sickle cell disease with recurrent vaso-occlusive crises: final results of Climb SCD-121. Blood 2025, 146 (suppl.1), abs25–801. [Google Scholar]
- Sharma, S.A.; Locatelli, F.; Bhatia, M.; Molinari, L.; Mapara, M.; Liem, R.; Wall, D.A.; Molinari, L.; Dedekan, L.; Wall, D.; et al. Improvements in health-reklated quality of life in patients with severe sickle cell disease after examglogene autotemcel. Blood Adv 2025, in press. [Google Scholar] [CrossRef] [PubMed]
- Frangoul, H.; de la Fuente, J.; Algeri, M.; Chopra, Y.; Amrolia, P.; Sharma, A.; Meisel, R.; Cappellini, M.D.; Carbacioglu, S.; Kattamis, A.; et al. First results of examglogene autotemcel in pediatric patients aged 5-11 years with transfusion-dependent β-thalassemia or sickle cell disease with recurrent severe vaso-occlusive crises. Blood 2025, 146 (suppl.1), abs25–8416. [Google Scholar]
- Ye, D.; Chen, M.; Zhu, Y.; Feng, X.; Xu, L.; Huang, H. Comprehensive regulation of γ-globin expression by epigenetic modification s and protein post-translational modifications. Clin Epigenetics 2025, 17, 173. [Google Scholar] [CrossRef]
- Wongabirisuth, C.; Innachai, P.; Saisawang, C.; Tabsuwan, A.; Jearawirslyaparisan, N.; Kaewprommal, P.; Piryapoagsa, J.; Chiangyong, W.; Anurathapani, U.; Sandeis, D.; et al. Disrupting ZBTB7A or BCL11A binding sites reactivates fetal hemoglobin in erythroblasts from healthy and β°/HbE individuals. Scient Rep 2025, 15, 25580. [Google Scholar] [CrossRef] [PubMed]
- Traxler, E.A.; Yao, Y.; Woodard, K.J.; Kurita, R.; Nakamura, Y.; Hughes, J.R.; Hardison, R.C.; Blobel, G.A.; Li, C.; et al. A genome editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med 2016, 22, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Métais, J.Y.; Doerfler, P.A.; Mayuranathan, T.; Bauer, D.E.; Fowler, S.C.; Hsieh, M.M.; Katta, V.; Kerwala, S.; Lazzarotto, C.R.; Luk, K.; et al. Genome editing of HBG1 and HBG2 to induce fetal hemoglobin. Blood Adv 2019, 3, 3379–3392. [Google Scholar] [CrossRef]
- Sharma, A.; Boelens, J.J.; Cancio, M.; Hanking, J.S.; Bhad, P.; Azizy, M.; Lewandoski, A.; Zhao, X.; Chitnis, S.; Peddinti, R.; et al. CRISPR-Cas9 editing of the HBBG1 and HBBG2 promoters to treat sickle cell disease. N Engl J Mded 2023, 389, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.O.; Sharma, A.; Frangoul, H.; Dalai, J.; Kanter, J.; Alavi, A.; DiPersio, J.; Eapen, M.; Jaroscak, J.J.; Ayala, E.; et al. Initial results from the BEACON clinical study: a phase 1-2 study evaluating the safety and efficacy of a single dose of autologous CD34+ base edited hematopoietic stem cells (BEAM-101) in patients with sickle cell disease with severe vaso-occlusive crises. Blood 2024, 144 (suppl.1), 513. [Google Scholar] [CrossRef]
- Chockalingram, P.S.; Chen, G.; Minella, A.C.; Chen, Y.; Shehan, V.; Zhang, N.; Armant, M.; Zaidi, A.U.; Goodrich, R.; Hines, P.C.; et al. Impact of BEAM-101 treatemnt on red blood cell hemoglobin expression, rheology and sickling properties: initial data from the BEACON phase 1-2 study of autologous CD34+ base edited hematopoietic stem cells in sickle cell disease. Blood 2024, 144 (suppl.1), 4957–4958. [Google Scholar] [CrossRef]
- Gupta, A.O.; Sharma, A.; Frangoul, H.; Dalai, J.; Kanter, J.; Alavi, A.; DiPersio, J.; Eapen, M.; Jaroscak, J.J.; Ayala, E.; et al. Base editing for sickle cell disease: ongoing results from the Beacon study evaluating the safety and efficacy of BEAM-101, the first base-edited autologous CD34+ HSPC one-time cell therapy. EHA Meeting 2025. abst. PF 1151.
- Gupta, A.; Sharma, A.; Frangoul, H.; Kanter, J.; Mapara, M.; Dalai, J.; Alavi, A.; Jaroscak, J.; Ayala, E.; DiPersio, J.; et al. Robust HbF induction and improvement of anemia and hemolysis with base editing in sickle cell disease: safety and efficacy findings from the ongoing BEACON study. Blood 2025, 146 (suppl.1), abs25–2531. [Google Scholar]
- Wang, L.; Xue, W.; Zhang, H.; Gao, R.; Qiu, H.; Wei, J.; Zhou, L.; Lei, Y.N.; Wu, X.; Li, X.; et al. Eliminating base-editor-induced genome-wide and transcriptome-wide off-target mutations. Nat Cell Biol 2021, 23, 552–563. [Google Scholar] [CrossRef]
- Wang, L.; Li, P.; Wang, Y.; Lan, K.; Zheng, H.; Zhu, D.; Zhang, Y.; Guo, R.; Ma, H.; He, J.; et al. Development of best-in-class gene editing therapy for β-hemoglobinopathies using innovative transformer base editor (tBE). Blood 2023, 142 (suppl.1), 5243. [Google Scholar] [CrossRef]
- Lai, Y.; Liu, R.; Wang, L.; Ma, X.K.; Li, Y.; Yang, G.; Shi, L.; Wei, J.; Wei, Z.; Zhou, X.; et al. Rapid, efficient and durable fetal hemoglobin production following CS-101 treatment in transfusion-dependent β-thalassemia participants: an autologous, ex vivo edited CD34+ stem cell product using the innovative transformer base editor (tBE). Blood 2024, 144 (suppl.1), 4962–4963. [Google Scholar] [CrossRef]
- Lai, Y.; Liu, R.; Wang, L.; Ma, X.K.; Li, Y.; Yang, G.; Shi, L.; Wei, J.; Wei, Z.; Zhou, X.; et al. Treatment of patients with severe transfusion-dependent β-thalassemia with CS-101, an autologous, ex vivo edited, CD34+ hematopoietic stem cell product using innovative transformer base editor (TBE). 2025, EHA Meeting abst. S291.
- Chen, J.; Lai, Y.; Zhai, X.; Zhao, W.L.; Liu, R.; Qian, X.; Tang, W.; Wang, L.; Ma, X.K.; LI, Y.; et al. Rapid, efficient and durable fetal hemoglobin production following CS-101 treatment in transfusion-dependent β-thalassemia participants; an autologous, ex vivo edited CD34+ stem cell product using the innovative transformer base editor (tBE). Blood 2025, 146 (suppl.1), abs25–8614. [Google Scholar]
- Paul, B.; Montoya, G. CRISPR-Cas12a: functional overview and applications. Biochem J 2020, 43, 8–17. [Google Scholar] [CrossRef]
- De Dreuzy, E.; Haeth, G.; Zuris, J.A.; Sousa, P.; Viswanathan, R.; Scott, S.; Da Silva, J.; Ta, T.; Copehart, S.; Wang, T.; et al. EDIT-301: an experimental autologous cell therapy comprising Cas12a-RNP modified mPB-CD34+ cells for the potential treatment of SCD. Blood. 2019;134(suppl 1):4636.
- Hanna, R.; Frangoul, H.; McKinney, C.; Pineiro, L.; Mapara, M.; Chang, K.H.; Jaskolka, M.; Kim, K.; Farrington, D.L.; Wally. M.; et al. EDIT-301: an experimental autologous cell therapy comprising Cas12a-RNP-modified mPB-CD34+ cells for the potential treatment of SCD. Blood 2019, 134 (suppl.1), 4636. [Google Scholar]
- Hanna, R.; Frangoul, H.; McKinney, C.; Pineiro, L.; Mapara, M.; Dalal, J.; Rangarajan, H.; Atkins, H.; Chang, K.H.; Mei, B.; et al. As Cas12a gene editing of HBG1/2 promoters with EDIT-301 results in rapid and sustained normalization of hemoglobin with increased fetal hemoglobin in patients with severe sickle cell disease and transfusion-dependent beta-thalassemia. Blood 2023, 142 (suppl.1), 4996. [Google Scholar] [CrossRef]
- Hanna, R.; Frangoul, H.; Pineiro, L.; McKinney, C.; Mapara, M.; Dalai, J.; Rangarajan, H.; Atkins, H. ; Bhatia, Chellapandian, D.; et al. CRISPR-Cas12a gene editing of the HBG 1-2 promoters leads to sustained normalization of total hemoglobin and increased fetal hemoglobin in patients with severe sickle cell disease: updated results from the RUBY study. Blood 2025, 146 (suppl.1), abs25-8372. [Google Scholar]
- Frangoul, H.; Hanna, R.; Walters, >M. C.; Chang, K.H.; Jaskolka, M.; Kim, K.; Mei, B.; Afonja, O.; Thompson, A. Reni-Cel, an investigational As Cas12a gene-edited cell medicine, led to substantial engraftment, increased hemoglobin, and reduced transfusion dependence in patients with transfusion-dependent beta-thalassemia treated in the Edithal trial. Blood 2024, 144 (suppl.1), 7456–7457. [Google Scholar]
- Uchida, N.; Li, L.; Nassehi, T.; Drysdale, C.M.; Yapundich, M.; Gasner, J.; Haro-Mora, J.; Demirci, S.; Leonard, A.; Bonifacino, A.C.; et al. Preclinical evaluation for engraftment of CD34+ cells gene-edited at the sickle disease locus in xenograft mouse and non-human primate. Cell Rep Med 2021, 2, 100247. [Google Scholar] [CrossRef] [PubMed]
- Kanter, J.; DiPersio, J.F.; Leavey, P.; Shyr, D.C.; Thompson, A.A.; Porteus, M.H.; Intondi, A.; Lahiri, P.; Dever, D.P.; Petrusich, A.; et al. Cedar trial in progress: a first in human, phase 1-2 study of the correction of a single nucleotide mutation in autologous HSCs (GPH 101) to convert HbS to HbA for treating severe SCD. Blood 2021, 138 (suppl.1), 1864–1865. [Google Scholar] [CrossRef]
- Shyr, D.C.; Lowsky, R.; Miller, W.; Schroeder, M.A‐; Bucholz, T.; Dougall, K.; Intondi, A.; Charles, A.; Leher, J.; Lehrer, J.; et al. One year follow‐up on the first patient treated with Nula‐Cel: an autologous CRISPR/Cas9 gene corrected CD34+ cell product to treat sickle cell disease. Blood 2023, 142 (suppl.1), 5000.
- Lee, B.; Gin, A.; Wu, C.; Singh, K.; Grice, M.; Mortlock, R.; Abraham, D.; Fan, X.; Zhou, Y.; Aljanahi, A.; et al. Impact of CRISPR/HDR editing versus lentiviral transduction on long-term engraftment and clonal dynamics of HSPCs in rhesus macaques. Cell Stem Cell 2024, 31, 455–466. [Google Scholar] [CrossRef]
- Lessard, S.; Rimmelé, P.; Ling, H.; Moran, K.; Vieira, B.; Lin, Y.D.; Rajani, G.M.; Hong, V.; Reik, A.; Boismenu, R.; et al. Zinc finger nuclease-mediated gene editing in hematopoietic stem cells results in reactivation of fetal hemoglobin in sickle cell disease. Scient Rep 2024, 14, 24298. [Google Scholar] [CrossRef]
- Li, C.; Georgakopoulou, A.; Newly, G.A.; Chen, P.J.; Everette, K.A.; Paschoudi, K.; Vlachaki, E.; Gil, S.; Anderson, A.K.; Koob, T.; et al. In vivo HSC prime editing rescues sickle cell disease in a mouse model. Blood 2023, 141, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- LiC.; C.; Anderson, A.K.; Ruminski, P.; Kapova, D.; Kidem, H.P.; DiPersio, J.F.; Lieber, A. A simplified GCSF‐ free procedure allows for in vivo HSC gene therapy of sickle cell disease in a mouse model. Blood Adv 2024, 8, 4089‐4095.
- Li, C.; Georgakopoulou, A.; Paschoudi, K.; Anderson, A.K.; Huang, L.; Gil, S.; Giannaki, M.; Vlachaki, E.; Newby, G.A.; Liu, D.R.; et al. Introducing a hemoglobin G-Makassar variant in HSCs by in vivo base editing treats sickle cell disease in mice. Mol Ther 2024, 32, 4353–4371. [Google Scholar] [CrossRef] [PubMed]
- Breda, L.; Papp, T.E.; Triebwasser, M.P.; Yadegari, A.; Fedorky, M.T.; Tanaka, N.; Abdulmalik, O.; Pavani, G.; Wang, Y.; et al. In vivo hematopoietic stem cell modification by mRNA delivery. Science 2023, 381, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Chatterjee, S.; Dilliard, S.A.; Moore, S.; Xiao, Y.; Bian, X.; Yamada, K.; Sung, Y.C.; Levine, R.M.; Mayberry, K.; et al. Bone-marrow-homing lipid nanoparticles for genome editing in diseased and malignant hematopoietic stem cells. Nat Biotechnol 2024, 19, 1409–1417. [Google Scholar]
- Xu, S.; Liang, D.; Wang, Q.; Cheng, Y.; Xie, D.; Gui, Y.; Zhang, H.; Feng, C.; Zhao, F.; Ren, W.; et al. In vivo genome editing of human hematopoietic stem cells for treatment of blood disorders using mRNA delivery. Nat Biomed Eng 2025, in press. [Google Scholar] [CrossRef]
- Liao, J.; Chen, S.; Hsiao, S.; Jiang, Y.; Yang, Y.; Zhang, Y.; Wang, X.; Lai, Y.; Bauer, D.E.; Wu, Y. Therapeutic base adenine editing of human hematopoietic stem cells. Nat Commun 2023, 14, 207. [Google Scholar] [CrossRef]
- Milani, M.; Fabiano, A.; Perez-Rodriguez, M.; Hernandez, R.J.; Zecchillo, A.; Zonari, E.; Ottonello, S.; Basso-Ricci, L.; Canepari, C.; Volpin, M.; et al. In vivo hematopoietic stem cell gene therapy enabled by postnatal trafficking. Nature 2025, 643, 1097–1106. [Google Scholar] [CrossRef]
- Milani, M.; Annoni, A.; Moalli, F.; Liu, T.; Cesana, D.; Calabria, A.; Bartolaccini, S.; Biffi, M.; Russo, F.; Visigalli, I.; et al. Phagocytosis-shielded lentiviral vectors improve liver gene therapy in nonhuman primates. Soc Transl Med 2019, 11, eaay7325. [Google Scholar] [CrossRef]
- Charlesworth, C.T.; Homma, S.; Amaya, A.K.; Dib, C.; Vaidyanathan, S.; Tan, T.K.; Miyauchi, M.; Nakauchi, Y.; Suchy, F.P.; Wang, S.; et al. Highly efficient in vivo hematopoietic stem cell transduction using an optimized self-complementary adeno-associated virus. Mol Ther Methods Clin Dev 2025, 33, 1–10. [Google Scholar] [CrossRef]
- Karimzadeh, A.; Kim, R.; Garcia, V.; Fiorea, M.; Peacker, B.L.; Kobayashi, S.; Watkins, D.; Messemer, K.; Zheng, J.; Bauer, D.E.; et al. In situ gene editing of hematopoietic stem cells via AAV-delivered CRISPR guide RNAs. Blodd Adv 2025, in press. [Google Scholar] [CrossRef] [PubMed]
- Bhoorasingh, M.; Vail, E.; Yoon, Y.; Black, R.; Reddy, M.; Jeong, S. Engineered extracellular vesicles for in vivo therapy in sickle cell disease. Blood 2025, 146 (suppl.1), abs25–2915. [Google Scholar]
- Tozzi, L.; Schiroli, G.; Heshmati, Y.; Cao, Y.; Monte, T.; Wangweerawong, A.; Rottman, J.B.; Palchaudhuri, R.; Ribeil, J.A.; Manis, J.; et al. In vivo HSC gene editing for correction of the sickle cell mutation using RNA gene writers. Blood 2024, 144 (suppl.1), 515. [Google Scholar] [CrossRef]
- Heshmati, Y.; Schiroli, G.; Tozzi, L.; Monte, M.; Beytour, N.; Wangweerawong, A.; Rottman, J.; Salomon, W.E.; Palchaudhuri, R.; Wang, J.; et al. In vivo correction of the sickle cell disease mutation in hematopoietic stem cells using RNA gene writers. Blood 2025, 146 (suppl.1), abs25–7314. [Google Scholar]
- Raguram, A.; Banskota, S.; Liu, D.R. Therapeutic in vivo delivery of gene editing agents. Cell 2022, 185, 2806–2827. [Google Scholar] [CrossRef]
- Jones, R.J.; Kassim,A:; Brodsky, R. A.; De Baun, M. Is allogeneic transplantation for sickle cell disease still relevant in the era of gene therapy? Blood Adv 2025, 9, 877–883. [Google Scholar] [CrossRef]
- Kassim,A. A.; de la Funte, J.; Wilkerson, K.L.; Alahmari, A.; Seber, A.; Bonfim, C.; Simoes, B.P.; Alzahrani, M.; Eckrich, M.J.; Horn, B.; et al. An international learning collaborative phase 2 trial for haploidentical bone marrow transplant in sickle cell disease. Blood 2024, 143, 2654–2663. [Google Scholar] [CrossRef]
- Kassim, A.A.; Walters, M.C.; Eapen, M.; Smith, M.; Logan, B.R.; Solh, M.; McKinney, C.; Nieder, M.; Ross, M.; Kent, M.; et al. Haploidentical bone marrow transplantation for sickle cell disease. NEJM Evidence 2025, 4, EVIDoa2400192. [Google Scholar] [CrossRef] [PubMed]
- Santorone, S.; Angelini, S.; Natale, A.; Vaddinelli, D.; Spadano, R.; Casciani, P.; Papola, F.; DiLembo, E.; Iannetti, G.; Di Bartolomeo, P. survival and late effects of hematopoietic cell transplantation in patients with thalassemia major. Bone Marrow Trasplant 2022, 57, 1689–1697. [Google Scholar] [CrossRef] [PubMed]
- Santorone, S.; Pepe, A.; Meloni, A.; Natale, A.; Pistoia, L.; Olioso, P.; Papalinetti, G.; Cuccia, L.; Spasiano, A.; Lisi, R.; et al. Secondary solid cancer following hematopoietic transplantation in patients with thalassemia major. Bone Marrow Transpl 2018, 53, 39–43. [Google Scholar] [CrossRef]
- De Franceschi, L.; Locatelli, F.; Rees, D.; Chabannon, C.; Dalle, J.H.; Rivella, S.; Iolascon, A.; Lobitz, S.; Abboud, M.R.; de la Fuente, J.; et al. Selecting patients with sickle cell disease for gene addition or gene editing-based therapeutic approaches: report on behalf of a joint EHA specialized working group and EBMT hemoglobinopathies working party consensus conference. HemaSphere 2025, 9, e70089. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; John, T. To pursue gene therapy or not? Is it feasible after graft failure in allogeneic hematopoietic cell transplant recipients? Blood Adv. 2025, 9, 3845‐3849.
- Kelkar, A.H.; Achebe, M.O.; Hantel, A. An ethical allocation scheme for scarce gene therapies in sickle cell disease and transfusion-dependent β-thalassemia. Blood Adv 2025, 9, 4502–4510. [Google Scholar] [CrossRef]
- John, T.; Czechowiez, A. Clinical hematopoietic stem cell-based gene therapy. Mol Therapy 2025 33, 2663–2683. [CrossRef] [PubMed]
- Boulad, F.; Maggio, A.; Wang, X.; Moi, P.; Acuto, S.; Kogel, F.; Takpradit, C.; Mansilla-Soto, J.; Cabriolu, A.; Odak, A.; et al. Lentiviral globin gene therapy with reduced-intensity conditioning in adults with β-thalassemia: a phase 1 trial. Nat Med 2022, 28, 63–70. [Google Scholar] [CrossRef]
- Hsieh, M.M.; Bonner, M.; Pierciey, F.J.; Uchida, N.; Rottman, J.; Demopoulos, L.; et al. Myelodysplastic syndrome unrelated to lentiviral vector in patient treated with gene therapy for sickle cell disease. Blood Adv 2020, 4, 2058–2063. [Google Scholar] [CrossRef] [PubMed]
- Goyal, S.; Tisdale, J.; Schmidt, M. , Kanter, J.; Jaroscak, J.; Whitney, D.; et al. Acute myeloid leukemia case after gene therapy for sickle cell disease. N Engl J Med 2022, 386, 138–147. [Google Scholar] [CrossRef]
- Liggett, L.A.; Cato, L.D.; Weinstock, J.S.; Zhang, Y.; Nourale, S.M.; Gladwin, M.T.; Garrett, M.E.; Ashley-Koch, A.; Telen, M.J.; Custer, S.; et al. Clonal hematopoiesis in sickle cell disease. J Clin Invest 2022, 132, e156060. [Google Scholar] [CrossRef]
- Pincez, T.; Lee, S.; Ilboudo, Y.; Preuss, M.; Hung d’Alexandry d’Orengiani, A.L.; Bartolucci, P.; Galacteros, F.; Loly, P.; Bauer, D.E.; Loos, R.; et al. Clonal hematopoiesis in sickle cell disease. Blood 2021, 138, 2148–2152. [Google Scholar] [CrossRef]
- Solomou, E.E.; Delaporta, P.; Stamatia, L.; Katsika, E.V.; Catzieleftheriou, M.; Toutoudaki, K.; Glentis, S.; Stamatopoulos, K.; Catzidimitriou, A.; Kattamis, A. High incidence of clonal hematopoiesis in transfusion-dependent thalassemia patients. Blood 2022, 140 (suppl.1), 8213–8214. [Google Scholar] [CrossRef]
- De Luna, G.; Cretin, J.; Redjoul, R.; Beckerich, F.; Habiobi, A.; Menouche, D.; Hebert, N.; Giovannangeli, C.; Maury, S.; Bartolucci, P.; Sloma, I. Screening for clonal hematopoiesis in patients with β-hemoglobinopathies who are candidates to transplant approaches. Blood 2025, 146 (suppl.1), abs25–2407. [Google Scholar]
- Weeks, L.; Fitzhugh, C.; Pollock, S.; Osei, M.; Rickles-Young, M.; Murdock, M.; Townsend, M.; Reilly, C.; Dinardo, C.; Sabino, E.; et al. Sickle cell disease is associated with early onset clonal hematopoiesis involving DNA damage response pathway mutations. Blood 2025, 146 (suppl.1), abs25–7115. [Google Scholar]
- Zeng, J.; Nguyen, M.A.; Liu, P.; Ferreira da Silva, L.; Levesque, S.; Lin, L.Y.; Justus, D.G.; Petri, K.; Clement, K.; Porter, S.N. Gene editing without ex vivo culture evades genetoxicity in human hematopoietic cells. Cell Stem Cell 2025, 32, 191–208. [Google Scholar] [CrossRef]
- Demirci, S.; Zeng, J.; Pelchaudhuri, R.; Wu, C.; Abraham, D.M.; Hayal, T.B.; Essawi, K.; Nguyen, M.A.; Stasula, U.; Chu, R.; et al. BCL11A +58/+55 enhancer-editing facilitates HSPC engraftment and HbF induction in rhesus macaques conditioned with a CD45 antibody-drug conjugate. Cell Stem Cell 2025, 32, 209–226. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.; Sathish, S.; London, E.; Le, A.; Li, Q.; Gudmundsdottir, B.; Lee, D.Y.; Burke, E.V.; Yates, B.P.; Liu, D.R.; et al. Comparative analysis of CRISPR-Cas9, lentiviral transduction, and base editing for sickle cell disease in a murine model. Blood Adv 2025, in press. [Google Scholar] [CrossRef] [PubMed]
| Initial evaluation: patient selection and preparation |
| HSC mobilization with G-CSF and Plexifor, apheresis and HSCs/HPCs collection |
| CD34+ HSC/HPC purification and genetic manipulation (lentiviral transfection or gene editor electroporation) |
| Patient hospitalization and myeloablative conditioning |
| Infusion of engineered HSCs/HPCs; post-transplantation hospitalization and supportive care |
| Clinical support, monitoring and follow-up (including post-transplantation support, evaluation of hematological outcomes of gene-modified hematopoiesis, evaluation of late effects) |
| Main genetic components of lentiviral vectors used for gene therapy of hemoglobinopathies |
|
|
|
|
| Therapy and Clinical trial-gov. No. |
Disease | Gene therapy approach | Status and number of patients |
|---|---|---|---|
| Lov-Cel (Blue Bird Bio) NCT 02140554 HGB-205 and HGB-206 |
Sickle Cell Disease | Lentiviral globin gene addition (modified β-globin βT87Q) |
Approved (FDA 2023) 55 patients |
| Beti-Cel (Blue Bird Bio) NCT 01745120 HGB-204, HGB-205, HGB-207, HGB-212 |
β°/β°, β+/β° thalassemia |
Lentiviral globin gene addition (modified β-globin βT87Q) |
Approved (EMA 2019, FDA 2022) 40 patients |
| BCH-BB694 (Boston Children Hospital) NCT 03282656 |
Sickle Cell Disease | Lentiviral BCL11A-shmiR addition | Trial in progress 10 patients |
| LVV GbGM NCT 02186418 |
Sickle Cell Disease | Lentiviral globin gene addition (modified γ-globin γG16D) |
Trial in progress 7 patients |
| Exa-Cel (CRISPR Therapeutics/Vertex Pharmaceuticals) NCT 03745287 CLIMB SCD-121/131 |
Sickle Cell Disease | CRISPR-Cas9 editing of BCL11A enhancer | Approved (EMA 2023, FDA 2023/2024) 44 patients |
| Exa-Cel (CRISPR Therapeutics/Vertex Pharmaceuticals) NCT 03655678 CLIMB THAL-111 |
β°/β°, β+/β° thalassemia |
CRISPR-Cas9 editing of BCL11A enhancer | Approved 52 patients |
| Reni-Cel (Editas Medicine) NCT 05444894 EdiThal |
β°/β°, β+/β°like thalassemia |
Cas 12 editing of BCL11A binding sited in γ-globin promoter | Trial in progress 7 patients |
| Reni-Cel (Editas Medicine) NCT 04853576 Ruby |
Sickle Cell Disease | Cas 12 editing of BCL11A binding sited in γ-globin promoter | Trial in progress 21 patients |
| BEAM-101 (Beam Therapeutics) NCT 05456880 |
Sickle Cell Disease | Base editing of BCL11A binding site in -globin promoter | Trial in progress 5 patients |
| BRL-101 (BRL Medicine Inc) NCT 04211480 NCT 04205435 |
β°/β°, β+/β°, β+/β+ thalassemia |
CRISPR Cas 9 editing of BCL11A enhancer | Trial in progress (15 patients) |
| Gene editing strategy Editing efficacy |
Specifity | Main advantage | Main limitations | Translational applications |
|---|---|---|---|---|
| CRISPR/Cas9 High |
Moderate | Cost effectiveness, speed, efficiency, ease of use, flexibility and versatility, multiplexing | Off-target effects, double-strand breaks induced genotoxicity Active only in proliferating cells (HDR repair) |
Disruption of regulators elements regulating HbF synthesis (NHEJ repair) Correction of point mutations (HDR repair) Approved for clinical use. |
| CRISPR/Cas12 High |
High | Efficiency in HDR repair, multiplexing, fewer off-target effects, more precision in gene targeting than CRISPR/Cas9 | Double-strand breaks induced genotoxicity, possible off-target effects | Disruption of regulators elements regulating HbF synthesis Clinical trials in progress |
| Base editing Very high |
High | Efficient editing No donor template needed, versatile applications, no double-strand breaks, precise correction of single-base changes |
Bystander effects, PAM dependency, limited conversion for some base pairs | Base-editing of regulatory elements in HBG1 and HBG2 gene promoters Clinical trials in progress |
| Prime editing Moderate |
Very high | High versatility (base substitutions, small insertions and deletions), precision, reduced off-target effects | Low efficiency (particularly for some DNA sequences), some possible off-target effects Difficult delivery for in vivo studies |
Pre-clinical studies |
| CRISPR a/i | Very high | Efficient activation or inhibition of specific genes | Effect on gene expression insufficient for therapeutic purposes | Pre-clinical studies |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





