Submitted:
29 October 2025
Posted:
30 October 2025
You are already at the latest version
Abstract
Keywords:
Background
Methods
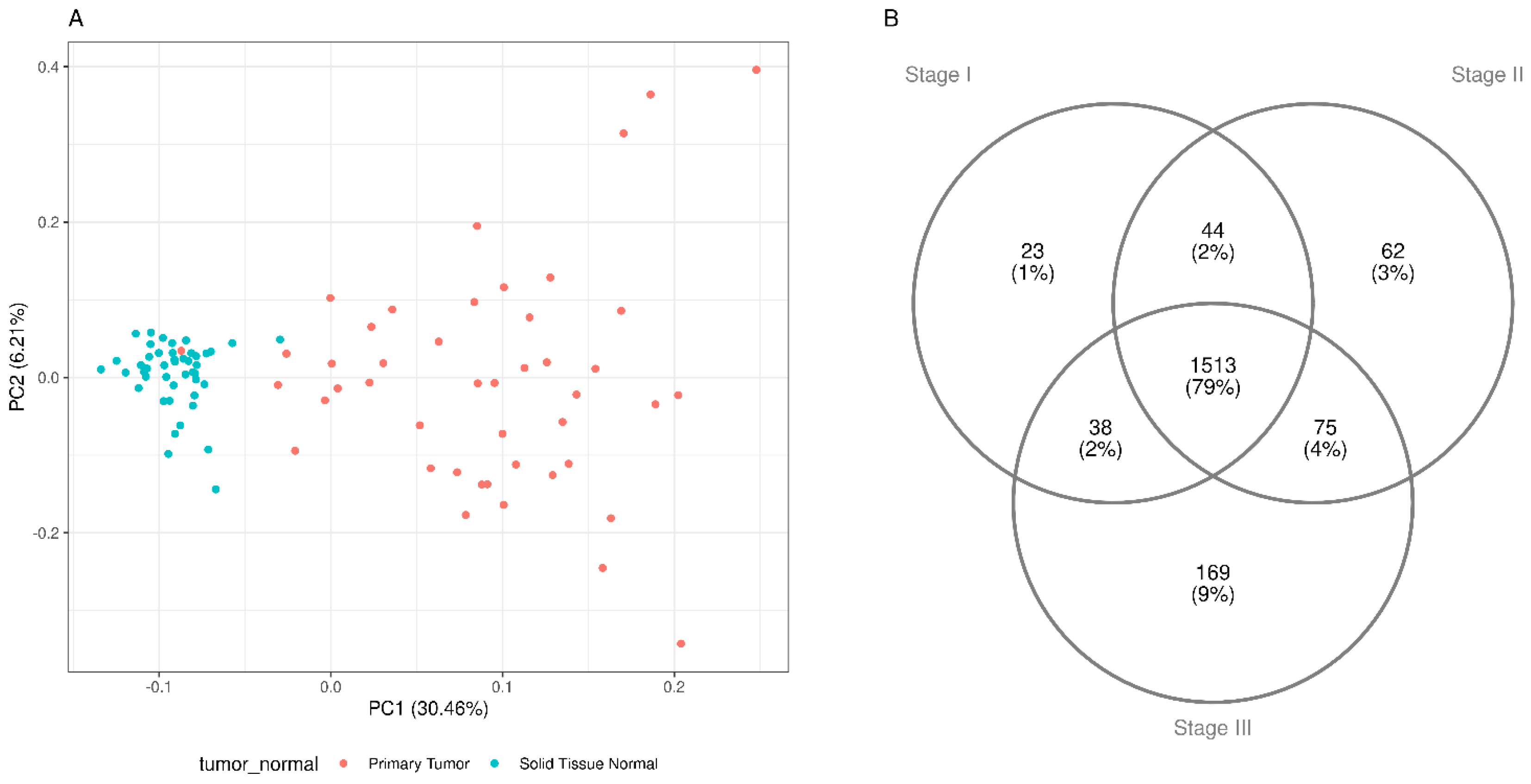
Results
| Normal | Tumor | ||||||
|---|---|---|---|---|---|---|---|
| # | GS1 | Av2 | SD3 | AV | SD | FC | Citation |
| 1 | BRCA1 | 2,39 | 0,96 | 10,17 | 6,28 | 4,26 | Burotto et. al., 2014 |
| 2 | ERCC1 | 22,36 | 3,72 | 32,01 | 12,6 | 1,43 | Scott & Salgia, 2008; Burotto et. al., 2014 |
| 3 | FGFR2 | 30,91 | 14,78 | 49,14 | 49,37 | 1,59 | Patel et al., 2015; Villalobos & Wistuba, 2017 |
| 4 | AURKA | 4,26 | 1,89 | 43,31 | 22,78 | 10,17 | Yunchu et. al., 2024 |
| 5 | TPX2 | 5 | 4,39 | 127,85 | 87,53 | 25,58 | Wang & Li., 2022 |
| 6 | BIRC5 | 3,22 | 3,25 | 85,75 | 48,09 | 26,65 | Yunchu et. al., 2024 |
| 7 | PIK3CA | 6,47 | 2,06 | 11,97 | 8,38 | 1,85 | Patel et al., 2015; Villalobos & Wistuba, 2017; Zhang et. al., 2021 |
| 8 | KLK10 | 10,62 | 4,75 | 27,39 | 46,8 | 2,58 | Zhang et. al., 2021 |
| 9 | TOP2A | 5,64 | 5,9 | 116,07 | 92,53 | 20,59 | Yunchu et. al., 2024 |
| 10 | PCNA | 102,48 | 28,78 | 404,4 | 200,94 | 3,95 | Yunchu et. al., 2024 |
| 11 | KRAS | 21,86 | 5,41 | 37,03 | 33,92 | 1,69 | Burotto et. al., 2014 |
| 12 | CCNB1 | 7,85 | 5,34 | 101,89 | 51,12 | 12,98 | Cai et. al., 2023 |
| 13 | CEP55 | 2,8 | 2,43 | 49,17 | 29,62 | 17,55 | Wang & Li., 2022 |
| 14 | TP53 | 26,41 | 7,09 | 48,55 | 40,35 | 1,84 | Scott & Salgia, 2008; Burotto et. al., 2014 |
| 15 | EGFR | 35,85 | 12,63 | 113,54 | 225,28 | 3,17 | Scott & Salgia, 2008; Burotto et. al., 2014; Patel et al., 2015; Villalobos & Wistuba, 2017 |
| 16 | CHEK1 | 1,85 | 0,96 | 18,72 | 9,62 | 10,11 | Cai et. al., 2023 |
| 17 | CCNB2 | 3,36 | 2,55 | 60,1 | 30,73 | 17,89 | Cai et. al., 2023 |
| 18 | BRAF | 4,76 | 1,28 | 4,97 | 2,16 | 1,04 | Patel et al., 2015; Villalobos & Wistuba, 2017 |
| 19 | RRM1 | 27,57 | 7,56 | 87,18 | 49,05 | 3,16 | Scott & Salgia, 2008; Burotto et. al., 2014 |
| 20 | CDK1 | 5,52 | 3,16 | 61,52 | 33,84 | 11,15 | Cai et. al., 2023 |
| 21 | MCC | 11,04 | 4,62 | 16,25 | 11,81 | 1,47 | Zhang et. al., 2021 |
| 22 | UBE2C | 6,03 | 5,21 | 180,1 | 108,34 | 29,88 | Yunchu et. al., 2024 |
| 23 | TYMS | 2,69 | 1,22 | 12,67 | 10,37 | 4,71 | Yunchu et. al., 2024 |
| 24 | AURKB | 2,23 | 2,12 | 48,9 | 30,33 | 21,96 | Yunchu et. al., 2024 |
| 25 | EXT1 | 29,82 | 8,98 | 49,45 | 25,92 | 1,66 | Zhang et. al., 2021 |
| 26 | PYCR1 | 8,91 | 5,79 | 71,7 | 56,09 | 8,05 | Wang & Li., 2022 |
| 27 | PDCD1 | 4,07 | 2,99 | 5,57 | 6,34 | 1,37 | Patel et al., 2015 |
| Tumor/normal | Stage I/normal | |||||
|---|---|---|---|---|---|---|
| SYMBOL | log2fc | pvalue | FDR | log2fc | pvalue | FDR |
| CRLF1 | 1,25 | 7,42E-02 | 0,1 | 1,49 | 4,63E-03 | 4,97E-03 |
| LTF | 0,47 | 4,09E-01 | 0,46 | 1,27 | 2,11E-03 | 2,31E-03 |
| RAI14 | 1,14 | 3,28E-05 | 0 | 1,06 | 1,02E-11 | 1,56E-11 |
| SERTAD4 | 1,01 | 3,56E-05 | 0 | 1,1 | 5,37E-16 | 9,63E-16 |
| OLFM4 | 4,1 | 3,04E-03 | 0,01 | 6,63 | 1,15E-02 | 1,21E-02 |
| CCDC91 | 0,87 | 2,35E-04 | 0 | 1,04 | 1,78E-04 | 2,04E-04 |
| NPL | 0,59 | 3,98E-02 | 0,06 | 1,01 | 5,30E-06 | 6,48E-06 |
| GRB7 | 0,88 | 4,08E-06 | 0 | 1,34 | 1,56E-02 | 1,63E-02 |
| NSDHL | 1,08 | 5,75E-08 | 0 | 1,01 | 1,87E-39 | 1,05E-38 |
| TKFC | 0,91 | 4,21E-10 | 0 | 1,02 | 1,36E-40 | 8,06E-40 |
| NADK2 | 1,05 | 1,47E-07 | 0 | 1,03 | 1,29E-21 | 2,92E-21 |
| CXADR | 1 | 4,47E-04 | 0 | 1,01 | 9,61E-17 | 1,78E-16 |
| FABP7 | 9,76 | 3,13E-01 | 0,36 | 7,83 | 4,06E-03 | 4,37E-03 |
| RPUSD2 | 0,92 | 3,69E-09 | 0 | 1,02 | 2,00E-42 | 1,36E-41 |
| KRT4 | 0,93 | 2,98E-01 | 0,35 | 1,59 | 1,69E-02 | 1,76E-02 |
| SNCG | 0,42 | 5,30E-01 | 0,58 | 1,19 | 3,96E-04 | 4,47E-04 |
| DHX36 | 1,05 | 1,08E-08 | 0 | 1,08 | 5,36E-46 | 4,63E-45 |
| COPB2 | 0,99 | 7,04E-09 | 0 | 1,05 | 2,90E-48 | 3,05E-47 |
| KPNA4 | 1,01 | 6,32E-10 | 0 | 1,01 | 4,12E-41 | 2,53E-40 |
| AMTN | 8,69 | 2,79E-02 | 0,04 | 7,97 | 1,50E-03 | 1,64E-03 |
| IGHA2 | 1 | 8,76E-02 | 0,12 | 1,42 | 1,02E-04 | 1,18E-04 |
| PCP4L1 | 0,74 | 2,51E-01 | 0,3 | 1,18 | 1,52E-05 | 1,82E-05 |
| MARCKS | 1,14 | 1,10E-10 | 0 | 1,03 | 2,63E-27 | 7,60E-27 |
Conclusions
Supplementary Materials
Authors' contributions
Funding declaration
Consent for publication
Availability of data and materials
Acknowledgment
Declaration of interest statement
References
- Barta JA, Powell CA, Wisnivesky JP. Global Epidemiology of Lung Cancer. Ann Glob Health. 2019 Jan 22;85(1):8. [CrossRef]
- Mauricio Burotto,Anish Thomas,Deepa Subramaniam,Giuseppe Giaccone and Arun Rajan. Biomarkers in Early-Stage Non-Small Cell Lung Cancer: Current Concepts and Future Directions. J Thorac Oncol. 2014 Nov; 9(11): 1609–1617.
- Kaier Cai, Zhilong Xie, Yingao Liu, Junfeng Wu, Hao Song, Wang Liu, Xinyi Wang, Yinghuan Xiong, Siyuan Gan, and Yanqin. Identification of Potential Key Genes and Prognostic Biomarkers of Lung Cancer Based on Bioinformatics. Biomed Res Int. 2023; 2023: 2152432.
- Qi Huang, Haiming Chen, Dandan Yin, Jie Wang, Shaodong Wang, Feng Yang, Jiawei Li, Teng Mu, Jilun Li, Jia Zhao, Rong Yin, Wei Li, Mantang Qiu, Erbao Zhang & Xiangnan Li. Multi-omics analysis reveals NNMT as a master metabolic regulator of metastasis in esophageal squamous cell carcinoma. npj Precision Oncology volume 8, Article number: 24 (2024).
- Chuantao Zhang, Man Jiang, Na Zhou, Helei Hou, Tianjun Li, Hongsheng Yu, Yuan-De Tan, and Xiaochun Zhang. Use tumor suppressor genes as biomarkers for diagnosis of non-small cell lung cancer. Sci Rep. 2021; 11: 3596.
- Fangwei Wang,Qisheng Su, and Chaoqian Li. Identidication of novel biomarkers in non-small cell lung cancer using machine learning. Sci Rep. 2022; 12: 16693.
- Jai N. Patel, Jennifer L. Ersek, and Edward S. Kim. Lung cancer biomarkers, targeted therapies and clinical assays. Transl Lung Cancer Res. 2015 Oct; 4(5): 503–514.
- Jiyun Zhang, Miru Tang, andJinsai Shang. PPARγ Modulators in Lung Cancer: Molecular Mechanisms, Clinical Prospects, and Challenges. Biomolecules 2024, 14(2), 190.
- Kaier Cai, Zhilong Xie, Yingao Liu, Junfeng Wu, Hao Song, Wang Liu, Xinyi Wang, Yinghuan Xiong, Siyuan Gan, and Yanqin. Identification of Potential Key Genes and Prognostic Biomarkers of Lung Cancer Based on Bioinformatics. Biomed Res Int. 2023; 2023: 2152432.
- Lau SCM, Pan Y, Velcheti V, Wong KK. Squamous cell lung cancer: Current landscape and future therapeutic options. Cancer Cell. 2022 Nov 14;40(11):1279-1293. [CrossRef]
- Li C, Lei S, Ding L, Xu Y, Wu X, Wang H, Zhang Z, Gao T, Zhang Y, Li L. Global burden and trends of lung cancer incidence and mortality. Chin Med J (Engl). 2023 Jul 5;136(13):1583-1590. [CrossRef]
- Li N, Li Y, Zheng P, Zhan X. Cancer Stemness-Based Prognostic Immune-Related Gene Signatures in Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. Front Endocrinol (Lausanne). 2021 Oct 21;12:755805. [CrossRef]
- Liu H, Li T, Dong C, Lyu J. Identification of miRNA signature for predicting the prognostic biomarker of squamous cell lung carcinoma. PloS One (2022) 17(3):e0264645. [CrossRef]
- Mauricio Burotto,Anish Thomas,Deepa Subramaniam,Giuseppe Giaccone and Arun Rajan. Biomarkers in Early-Stage Non-Small Cell Lung Cancer: Current Concepts and Future Directions. J Thorac Oncol. 2014 Nov; 9(11): 1609–1617.
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5: 621–628. pmid:18516045.
- O'Keeffe LM, Taylor G, Huxley RR, et al. Smoking as a risk factor for lung cancer in women and men: a systematic review and meta-analysis. BMJ Open 2018;8:e021611. [CrossRef]
- Pamela Villalobos, Ignacio I. Wistuba. Lung Cancer Biomarkers. Hematol Oncol Clin North Am. 2017 Feb; 31(1): 13–29.
- Jai N. Patel, Jennifer L. Ersek, and Edward S. Kim. Lung cancer biomarkers, targeted therapies and clinical assays. Transl Lung Cancer Res. 2015 Oct; 4(5): 503–514.
- Jai N. Patel, Jennifer L. Ersek, and Edward S. Kim. Lung cancer biomarkers, targeted therapies and clinical assays. Transl Lung Cancer Res. 2015 Oct; 4(5): 503–514.
- Relli V, Trerotola M, Guerra E, Alberti S. Abandoning the notion of non-small cell lung cancer. Trends Mol Med (2019) 25(7):585–94. [CrossRef]
- Scott, A.; Salgia, R. Biomarkers in lung cancer: from early detection to novel therapeutics and decision making. Biomark Med. 2008 Dec 1; 2(6): 577–586.
- Pamela Villalobos, Ignacio I. Wistuba. Lung Cancer Biomarkers. Hematol Oncol Clin North Am. 2017 Feb; 31(1): 13–29.
- Wang, J., Liu, Q., Yuan, S. et al. Genetic predisposition to lung cancer: comprehensive literature integration, meta-analysis, and multiple evidence assessment of candidate-gene association studies. Sci Rep 7, 8371 (2017). [CrossRef]
- Wang X, Su R, Guo Q, Liu J, Ruan B, Wang G. Competing endogenous RNA (ceRNA) hypothetic model based on comprehensive analysis of long non-coding RNA expression in lung adenocarcinoma. PeerJ (2019) 7:e8024. [CrossRef]
- Wu R, Ma R, Duan X, Zhang J, Li K, Yu L, Zhang M, Liu P, Wang C. Identification of specific prognostic markers for lung squamous cell carcinoma based on tumor progression, immune infiltration, and stem index. Front Immunol. 2023 Sep 29;14:1236444. [CrossRef]
- Yang Yunchu, Akihiko Miyanaga,corresponding author Kuniko Matsuda, Koichiro Kamio, and Masahiro Seike. Exploring effective biomarkers and potential immune related gene in small cell lung cancer. Sci Rep. 2024; 14: 7604.
- Yang Yunchu, Akihiko Miyanaga,corresponding author Kuniko Matsuda, Koichiro Kamio, and Masahiro Seike. Exploring effective biomarkers and potential immune related gene in small cell lung cancer. Sci Rep. 2024; 14: 7604.
- Chuantao Zhang, Man Jiang, Na Zhou, Helei Hou, Tianjun Li, Hongsheng Yu, Yuan-De Tan, and Xiaochun Zhang. Use tumor suppressor genes as biomarkers for diagnosis of non-small cell lung cancer. Sci Rep. 2021; 11: 3596.
- Zhou, Y., Tao, L., Qiu, J. et al. Tumor biomarkers for diagnosis, prognosis and targeted therapy. Sig Transduct Target Ther 9, 132 (2024). [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





