1. Introduction
Rice is a fundamental staple food for a substantial portion of the global population and plays a critical role in the economies of many countries. Its cultivation and trade are vital for global food security, necessitating efficient and accurate methods for quality assessment and variety identification. Traditionally, rice classification relies on manual inspection, a process that is inherently subjective, labor-intensive, time-consuming, and prone to human error due to factors such as fatigue, eye-strain, and inconsistent decision-making among technicians. These limitations directly impede effective quality control, accurate market pricing, and the overall efficiency of the agricultural supply chain.
To overcome these challenges, the agricultural sector has increasingly turned to automated, non-destructive and efficient systems utilizing machine vision and deep learning technologies. Convolutional Neural Networks (CNNs) have emerged as a prominent solution, demonstrating considerable success in various image classification tasks, including rice variety identification. For instance, studies have achieved high classification accuracies with CNNs; Murat Koklu et al. reported 100% accuracy in classifying five Turkish rice varieties (Arborio, Basmati, Ipsala, Jasmine, and Karacadag) using a custom CNN on a dataset of 75,000 grain images. Similarly, EfficientNet-b1, a CNN model, achieved 99.87% accuracy for the same five varieties using a dataset of 75,000 images. While CNNs excel at capturing local features through their convolutional layers, they often exhibit limitations in effectively modeling long-range dependencies and global contextual relationships within images, which can be crucial for distinguishing subtle features among visually similar rice varieties.
In recent years, Vision Transformers (ViTs) have revolutionized the field of computer vision, demonstrating competitive, and often superior, performance compared to traditional CNN architectures, especially in complex image classification tasks. Inspired by their success in Natural Language Processing, ViTs process images by dividing them into fixed-size patches, which are then treated as sequences of tokens. This unique architecture, particularly its powerful self-attention mechanism, allows ViTs to inherently capture global dependencies and contextual relationships across the entire image. This capability is particularly advantageous for tasks requiring the discernment of subtle morphological, shape, and color characteristics, making ViTs a promising alternative for precision agriculture applications like plant disease detection and classification.
This paper proposes a novel and robust approach for automated rice-type detection that takes advantage of the advanced capabilities of the Vision Transformer (ViT) model. Specifically, we employ the google/vit-base-patch16-224-in21k model, pre-trained on a large ImageNet-21k dataset, for classifying rice varieties. Our research utilizes a comprehensive dataset consisting of 75,000 RGB images across five distinct rice varieties: Arborio, Basmati, Ipsala, Jasmine, and Karacadag. The dataset was meticulously divided, with 60,000 images allocated for training and 15,000 images for testing. The training set distribution was: Arborio (11988), Basmati (11994), Ipsala (12021), Jasmine (12008), and Karacadag (11989). The test set distribution was: Arborio (3012), Basmati (3006), Ipsala (2979), Jasmine (2992), and Karacadag (3011). The ViT model was trained with the following parameters: output_dir=root_dir, per_device_train_batch_size=16, evaluation_strategy="epoch", save_strategy="epoch", fp16=True, num_train_epochs=20, logging_steps=500, learning_rate=2e-4, save_total_limit=1, remove_unused_columns=False, push_to_hub=False, report_to=’tensorboard’, and load_best_model_at_end=True. Through this method, our proposed model achieved a remarkable classification accuracy of 99.9984%.
The primary contribution of this research is the successful demonstration of a highly accurate and robust system for rice type detection using a Vision Transformer model. This work not only highlights the superior performance of ViTs in handling the intricate visual characteristics of different rice varieties but also provides a scalable and reliable solution for quality control in the agricultural industry. The anticipated impact includes reducing reliance on manual, error-prone methods, enhancing the efficiency of rice sorting and grading, and ultimately contributing to improved food quality and global food security.
2. Literature Review
Recent years have witnessed a remarkable surge in the application of deep learning and Transformer-based architectures in agricultural image analysis, particularly for rice classification and disease detection. Early surveys such as Khan et al. [
1] and Han et al. [
2] provided comprehensive insights into the evolution of Vision Transformers (ViTs), highlighting their ability to model long-range dependencies and deliver competitive performance across image recognition and segmentation tasks. Building upon these foundations, Yao et al. [
3] introduced the Dual-ViT architecture, which leveraged dual semantic and pixel pathways to achieve high accuracy on ImageNet benchmarks with reduced computational overhead. Similarly, Mehdipour et al. [
4] and Zhang et al. [
5] emphasized the growing importance of ViTs in precision agriculture, demonstrating their superiority over CNNs in scalability, interpretability, and accuracy, with results exceeding 98% in crop disease identification.
In rice-focused research, Koklu et al. [
6] and Qadri et al. [
7] showcased the capabilities of CNNs and texture-driven machine vision approaches, attaining near-perfect classification accuracy (reaching 100%) for different rice varieties. Fabiyi et al. [
8] enhanced this by combining RGB and hyperspectral features with Random Forest classifiers, achieving an accuracy of up to 98.17% for subsets, whereas Chatnuntawech et al. [
9] introduced spatio-spectral CNNs that attained over 97% accuracy on processed rice. Jin et al. [
10] also investigated hyperspectral imaging techniques, attaining 99.94% accuracy through the use of near-infrared hyperspectral data paired with deep learning. Simultaneously, Kiratiratanapruk et al. [
11] and Rajalakshmi et al. [
12] developed automated grading and seed classification models, with RiceSeedNet reaching 99% accuracy, underscoring the viability of these techniques for practical uses. Komal et al. [
13] presented an extensive analysis of automatic rice variety identification systems, outlining the application of morphological, color, and textural attributes alongside classifiers like SVM, KNN, and hybrid neural networks. Their results showed accuracies between 92% and 99.73%, with Neuro-Fuzzy Neural Networks attaining the best performance, while also highlighting difficulties concerning dataset quality and feature extraction.
For rice disease identification, Singh et al. [
14] employed CNNs to achieve 96.08% accuracy, while Ulukaya and Deari [
15] applied ViT-based methods under field conditions, obtaining 88.57% accuracy despite environmental challenges. Leu et al. [
16] introduced RiceTalk, an IoT-enabled deep learning system that achieved 91.18% accuracy for rice blast detection, showcasing the potential of integrating AI with IoT. More advanced models include Patil et al. [
17], who proposed a Faster R-CNN + EfficientNet-B0 framework for rice disease severity estimation with 96.43% accuracy, and Verma et al. [
18][
19], who designed a hybrid CNN with transfer learning, achieving 98.6% accuracy.
Beyond rice, similar hybrid CNN–ViT approaches have shown promise in other crops. Shu et al. [
20] reported 99.70% accuracy in maize disease detection, and Bhujel et al. [
21] achieved 99.83% in tomato disease classification. These findings align with Bhujel et al.’s [
21] and Shu et al.’s [
20] conclusions that combining CNN spatial learning with ViT’s global contextual understanding significantly improves classification robustness. Gudipalli et al. [
22] earlier noted that deep learning methods consistently outperform traditional machine vision systems, confirming this trend.
3. Dataset
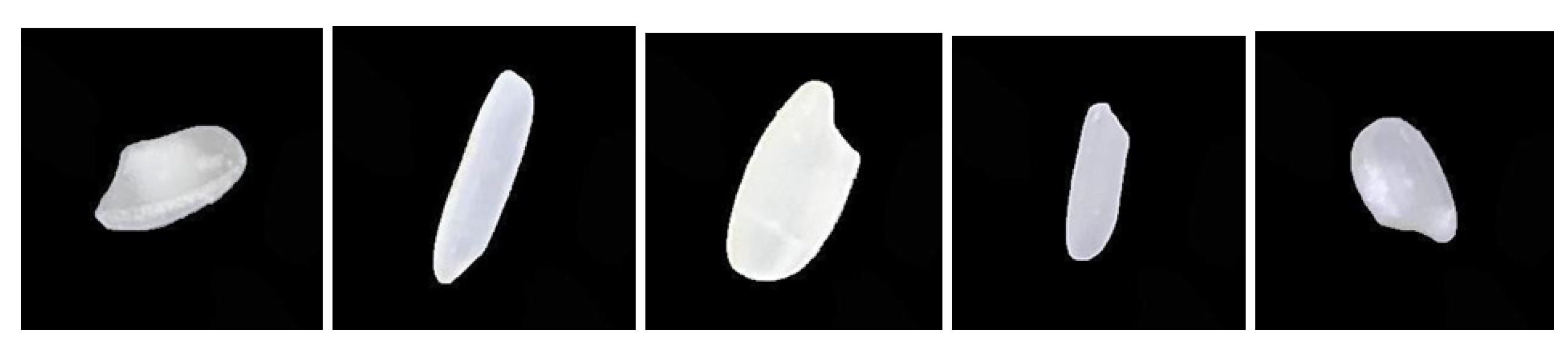
In this study, we utilized a publicly available rice grain image dataset comprising five distinct rice varieties, namely Arborio, Basmati, Ipsala, Jasmine, and Karacadag, which are commonly cultivated in Turkey. The dataset contains a total of 75,000 images, with 15,000 samples from each rice variety, ensuring a balanced representation across classes. Each rice grain is captured individually in an RGB image with a fixed resolution of 250 × 250 pixels, thereby providing sufficient detail for fine-grained classification tasks. Representative samples from each rice variety are illustrated in Fig. 1. For model development, the dataset was partitioned into training and testing sets with an 80:20 split, where the folder structure is organized accordingly.
Figure 1.
Image of the rice grains. (1. Arborio, 2. Basmati, 3. Ipsala, 4. Jasmine, 5. Karacadag).
Figure 1.
Image of the rice grains. (1. Arborio, 2. Basmati, 3. Ipsala, 4. Jasmine, 5. Karacadag).
4. Methods
The present study details the development of an automated and data-driven methodology for the classification of rice varieties from digital imagery, achieved through the employment of a Vision Transformer (ViT) model. This framework was designed to overcome the limitations associated with manual classification, which is often subjective, time-consuming, and error-prone, thereby constraining efficient quality assessment and price standardization. The methodology approach integrates a curated dataset, standardized preprocessing, a fine-tuned ViT architecture, and an optimized training protocol to achieve reliable and reproducible classification performance.
A comprehensive image dataset was used, consisting of 75,000 RGB images representing five rice varieties: Arborio, Basmati, Ipsala, Jasmine, and Karacadag where each one captured at a resolution of 250 × 250 pixels. To ensure balanced learning and unbiased validation, the dataset was divided into training, validation, and testing subsets, adhering to an 80:10:10 ratio. The detailed distribution of samples for each class is presented in
Table 1. Each image underwent resizing to 224 × 224 pixels to match the ViT input specifications, and pixel values were subsequently normalized from the 0–255 range to the [0, 1] interval. This normalization stabilized the optimization process by mitigating large numerical variations across channels. To improve model robustness, a controlled data augmentation comprising horizontal and vertical flips, small rotations (±10°), and moderate brightness and contrast adjustments was applied exclusively to the training images, thereby enhancing dataset diversity while preserving the morphological integrity of each grain.
The Vision Transformer model used in this study follows the ViT-Base-Patch16-224 configuration. Each input image was partitioned into non-overlapping 16 × 16 patches, generating 196 tokens, each of which was flattened and linearly projected into a 768-dimensional embedding space. To preserve spatial relationships, learnable positional embeddings
were added to the patch embeddings, resulting in an input sequence represented as
where
denotes the
flattened image patch,
the trainable projection matrix, and
the resulting sequence of embeddings passed into the Transformer encoder.
The encoder consists of 12 stacked layers, each comprising a Multi-Head Self-Attention (MHSA) module and a Feed-Forward Network (FFN). Within each attention head, relationships between all patches are modeled simultaneously through scaled dot-product attention:
where
Q,
K, and
V are the query, key, and value projections, respectively, and
is the key-vector dimension. Multiple attention heads operate in parallel, enabling the network to capture diverse contextual cues such as texture, curvature, and grain shape. Residual connections and layer normalization follow both the MHSA and FFN blocks to ensure stable gradient propagation and improved convergence.
To perform the final classification, a learnable [CLS] token is prepended to the patch sequence before encoding. As this token passes through the transformer layers, it aggregates contextual information from all patches. After the final encoder layer, its output vector serves as a global image representation, which is fed into a Multi-Layer Perceptron (MLP) head that produces five logits corresponding to the five rice varieties. The Softmax function converts these logits into class probabilities for the final prediction.
The network parameters were optimized using the AdamW optimizer with a learning rate of
, batch size = 32, weight decay = 0.01, and 20 epochs of training. The optimization objective was the Categorical Cross-Entropy loss, defined as
where
represents the number of rice classes,
is the true one-hot label, and
is the predicted probability for class
i. The model was initialized from the
google/vit-base-patch16-224-in21k checkpoint and pre-trained on the large-scale ImageNet-21k dataset. A comprehensive fine-tuning of all layers, encompassing the patch embedding, encoder blocks, and classification, facilitated the efficient transfer of generalized visual features to the rice-classification task, thereby reducing both training time and the risk of overfitting.
Training and evaluation were implemented in Python 3.10 using PyTorch and the Hugging Face Transformers library on an NVIDIA GPU with CUDA support. Mixed precision (FP16) training was employed to accelerate computation and minimize memory usage. Model checkpoints were saved after each epoch, and the configuration achieving the highest validation accuracy was retained for testing. Performance was assessed using standard evaluation metrics—accuracy, precision, recall, and F1-score—derived from the confusion matrix. For each class, precision and recall were calculated as
and the harmonic mean of these two metrics provided the F1-score:
Macro and weighted-average values were computed to evaluate performance across all categories. In addition, Receiver Operating Characteristic (ROC) and Precision–Recall (PR) curves were plotted, and the Area Under the Curve (AUC) was reported to quantify the model’s overall discriminative capability. All preprocessing parameters, random seeds, and hyperparameter settings were fixed to guarantee reproducibility. The experimental pipeline including dataset organization, model fine-tuning, and metric evaluation was executed under identical conditions across all runs. This methodology provides a reliable and scalable strategy for fine-grained classification of rice grain varieties using advanced transformer-based vision models.
5. Proposed Architecture
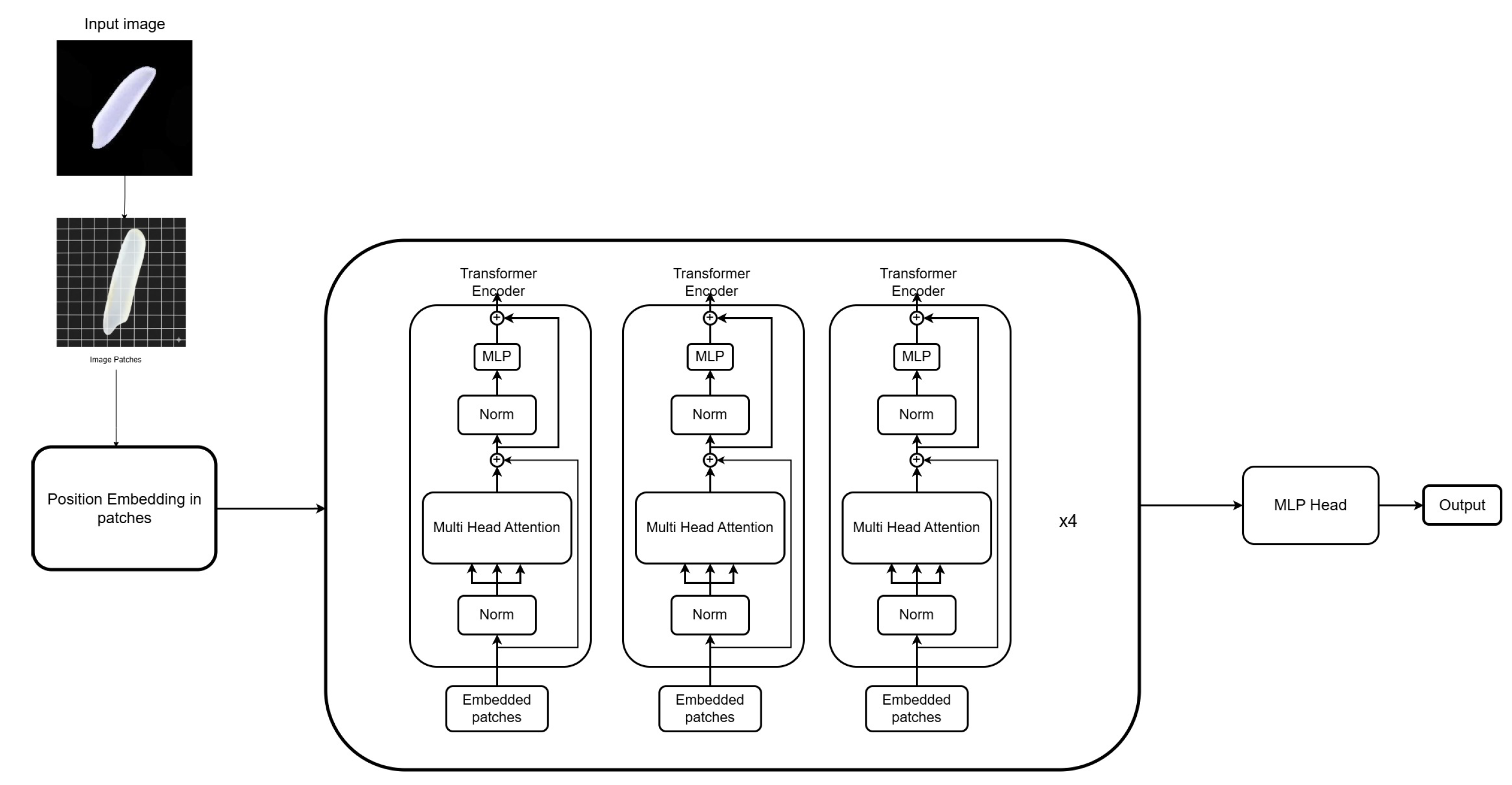
In this work, we propose a transformer-based architecture inspired by the Vision Transformer (ViT) [Dosovitskiy et al., 2020] and adapt it for the classification of rice grain varieties. The backbone of the network is the pre-trained ViT-Base-Patch16-224 model, which was fine-tuned on our custom dataset of five rice categories. The overall pipeline of the proposed approach is illustrated in Fig. 2.
The model accepts input images of size , corresponding to the three RGB channels. Each input image is first partitioned into non-overlapping patches of size 16 × 16, generating a sequence of patches, where each patch is flattened and linearly projected into a fixed-dimensional embedding space. To preserve spatial information, learnable positional embeddings are added to the patch embeddings, resulting in a sequence of image tokens. These tokens are then passed into the transformer encoder block.
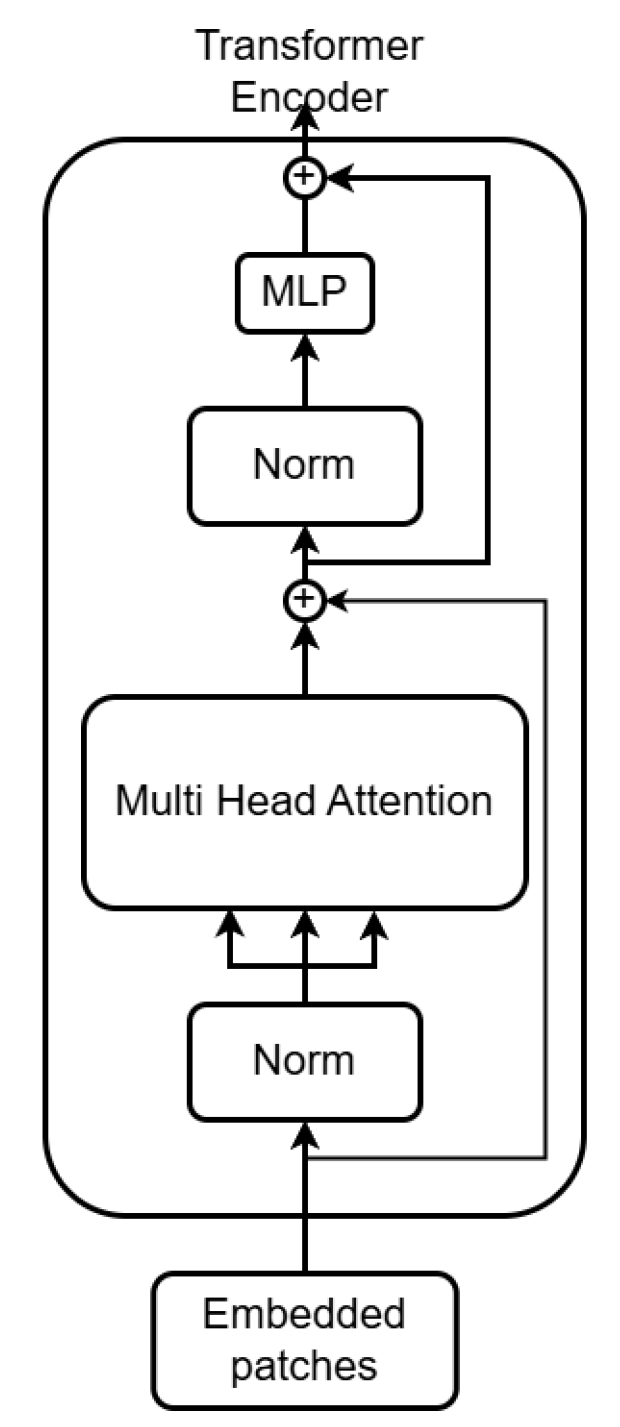
Figure 3.
Transformer encoder.
Figure 3.
Transformer encoder.
The encoder is composed of multiple layers, each consisting of three key operations:
(i) Layer Normalization, (ii) Multi-Head Self-Attention (MHSA), and
(iii) a feed-forward network (FFN). The MHSA mechanism maps each token into
Query (Q), Key (K), and
Value (V) representations via learnable weight matrices
. The attention weights are computed as:
where
denotes the dimension of the key vector. By aggregating information across all patches, the self-attention mechanism enables the model to capture long-range dependencies between different parts of the image. In the multi-head setting, this operation is repeated in parallel with multiple sets of
projections, and the outputs are concatenated before being passed through a linear projection. Finally, the transformer encoder produces a contextualized representation of the input image tokens, which is passed to a classification head (a fully connected layer) to output the class probabilities.
For training, the pre-trained ViT model was fine-tuned on our dataset with five output labels, using the Adam optimizer with a learning rate of 2e-5. We set the batch size to 16 and trained the network for 10 epochs. The experiments were accelerated with mixed precision (FP16) when a compatible GPU was available. The training and evaluation followed an 80:20 split of the dataset. Data augmentation was applied during training to improve generalization. The loss function employed was cross-entropy, defined as:
where
C is the number of classes,
is the true label, and
is the predicted probability for class
i.
Through this design, the transformer-based model leverages both the representational power of pre-trained features and the discriminative capacity of fine-tuning, enabling accurate classification of rice grain varieties from image data.
6. Results And Analysis
The Vision Transformer (ViT) model demonstrated exceptional performance across all five rice varieties, achieving a near-perfect classification. As shown in the classification report, the model attained an overall accuracy of 99.99%, with weighted averages of precision, recall, and F1-score all equal to 0.9999. Class-wise metrics further confirm this consistency, with each rice variety (Arborio, Basmati, Ipsala, Jasmine, and Karacadag) yielding F1-scores of approximately 1.00, highlighting the model’s ability to generalize effectively across categories.
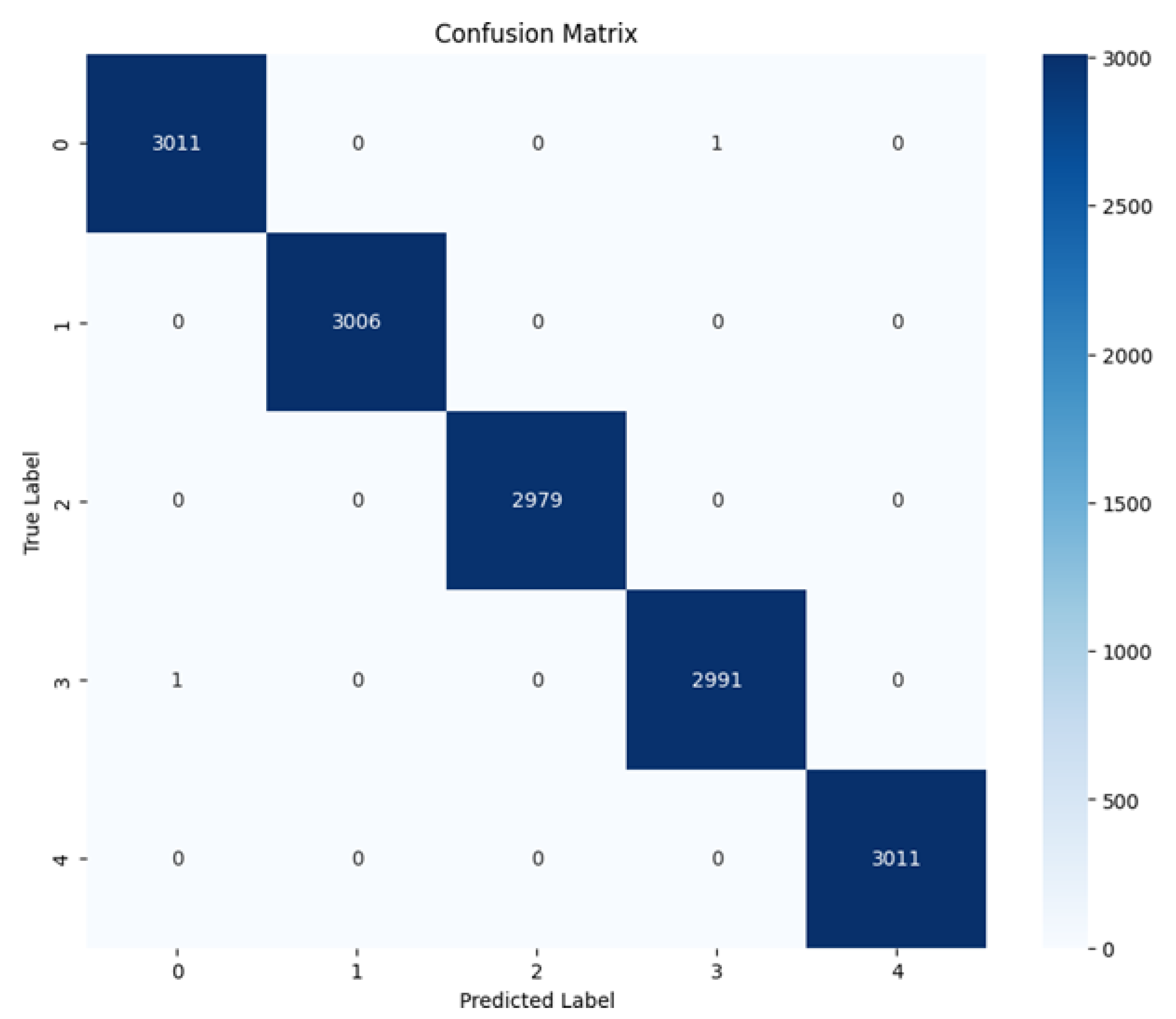
The confusion matrix (Fig. 4) provides a more granular view of the predictions. Almost all instances were correctly classified, with negligible misclassifications. For example, a minimal number of Jasmine grains were predicted as Arborio, likely due to subtle morphological and color similarities between the two varieties. The absence of significant misclassifications underscores the discriminative power of ViT’s attention mechanism.
Figure 4.
Confusion Matrix.
Figure 4.
Confusion Matrix.
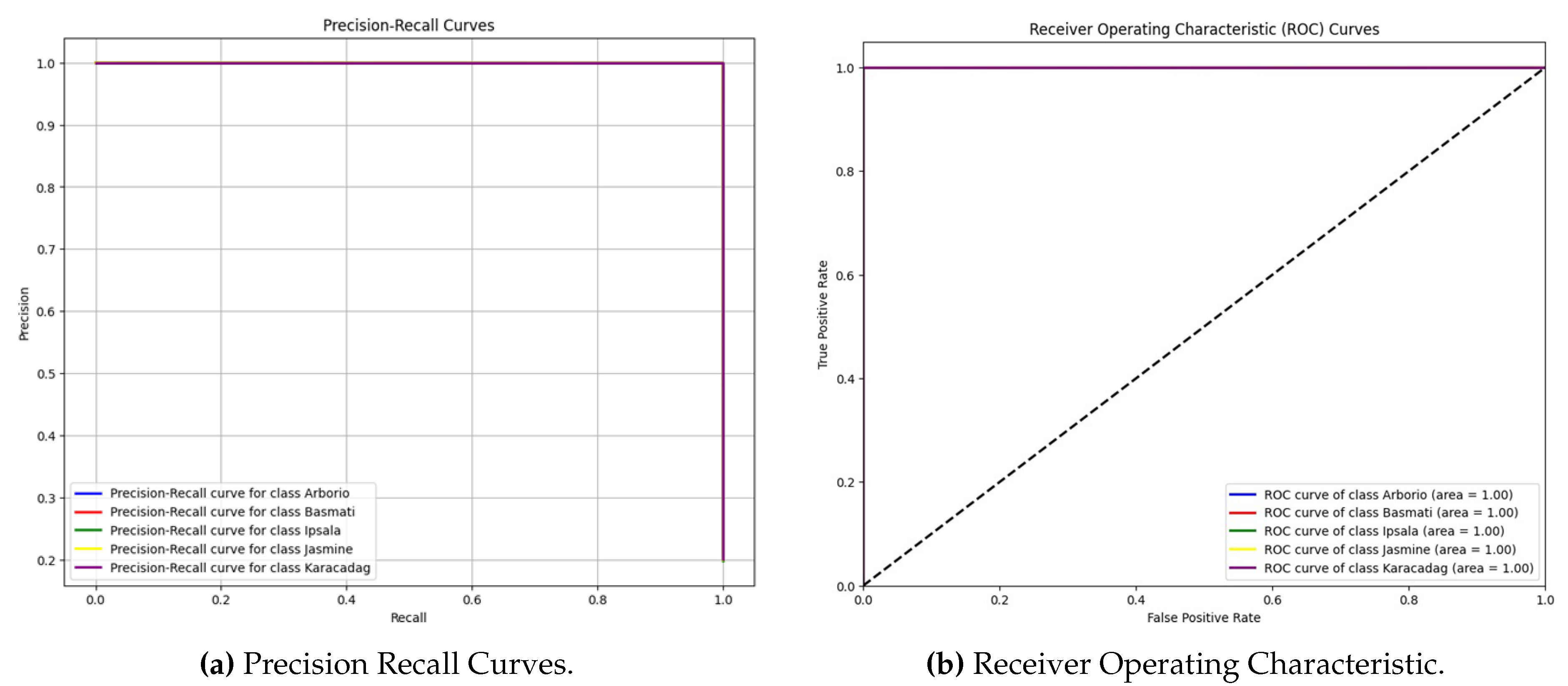
The Precision-Recall (PR) curves (Fig. 5) illustrate consistently high performance across all classes, with curves closely aligned at the top-right corner, reinforcing the reliability of predictions even under class imbalance conditions. Similarly, the Receiver Operating Characteristic (ROC) curves (Fig. 6) demonstrate an AUC of 1.00 for all classes, further confirming the robustness of the classifier and its ability to minimize false positives.
Figure 5.
Comparison of Precision-Recall and ROC Curves.
Figure 5.
Comparison of Precision-Recall and ROC Curves.
A quantitative summary is provided in
Table 2, consolidating accuracy, precision, recall, and F1-score for each class alongside macro and weighted averages. This table facilitates easy comparison and highlights the uniformity of the model’s performance across all rice varieties
Overall, these results validate the efficacy of Vision Transformers in fine-grained grain classification tasks. The global self-attention mechanism likely enabled the model to capture complex dependencies between grain shape and color distributions, which traditional CNN baselines (e.g., ResNet) or feature-based methods (e.g., SVM) typically struggle with. The marginal misclassifications observed can be attributed to intraclass visual similarity and could be further mitigated by integrating domain-specific augmentation strategies or hybrid feature fusion approac
7. Conclusions
This study successfully demonstrated the potential of Vision Transformer (ViT) models for fine-grained rice variety classification. By leveraging the self-attention mechanism, the proposed ViT-based system effectively captured subtle morphological, shape, and color characteristics of rice grains, overcoming the limitations of traditional manual methods and even surpassing conventional CNN architectures. Using a balanced dataset of 75,000 images across five rice varieties—Arborio, Basmati, Ipsala, Jasmine, and Karacadag—the model achieved an exceptional classification accuracy of 99.99%, with precision, recall, and F1-scores consistently near 1.0 for all classes.
These results confirm the robustness and scalability of ViTs in agricultural applications, particularly for enhancing precision, reliability, and efficiency in quality control. The minimal misclassifications observed highlight both the discriminative strength of the model and areas where further refinement, such as domain-specific augmentation or hybrid feature integration, may further optimize performance.
Overall, this research provides a strong foundation for deploying ViT-based solutions in precision agriculture. By reducing reliance on subjective and error-prone manual inspection, the approach contributes to improved sorting, grading, and pricing of rice, ultimately supporting global food security. Future work could extend this framework to other staple crops and explore integration with real-time field and industrial applications, thereby broadening the impact of advanced deep learning models in agricultural innovation.
References
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in Vision: A Survey. ACM Comput. Surv. 2022, 54. [Google Scholar] [CrossRef]
- Han, K.; Wang, Y.; Chen, H.; Chen, X.; Guo, J.; Liu, Z.; Tang, Y.; Xiao, A.; Xu, C.; Xu, Y.; et al. A Survey on Vision Transformer. IEEE Transactions on Pattern Analysis and Machine Intelligence 2023, 45, 87–110. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Li, Y.; Pan, Y.; Wang, Y.; Zhang, X.P.; Mei, T. 2022; arXiv:cs.CV/2207.04976].
- Mehdipour, S.; Mirroshandel, S.A.; Tabatabaei, S.A. A: Transformers in Precision Agriculture, 2025; arXiv:cs.CV/2504.21706].
- Zhang, M.; Liu, C.; Li, Z.; Yin, B. From Convolutional Networks to Vision Transformers: Evolution of Deep Learning in Agricultural Pest and Disease Identification. Agronomy 2025, 15. [Google Scholar] [CrossRef]
- Koklu, M.; Cinar, I.; Taspinar, Y.S. Classification of rice varieties with deep learning methods. Computers and Electronics in Agriculture 2021, 187, 106285. [Google Scholar] [CrossRef]
- Qadri, S.; Aslam, T.; Nawaz, S.A.; Saher, N.; Abdul-Razzaq. ; Rehman, M.U.; Ahmad, N.; Shahzad, F.; Qadri, S.F. Machine Vision Approach for Classification of Rice Varieties Using Texture Features. International Journal of Food Properties 2021, 24, 1615–1630. [Google Scholar] [CrossRef]
- Fabiyi, S.D.; Vu, H.; Tachtatzis, C.; Murray, P.; Harle, D.; Dao, T.K.; Andonovic, I.; Ren, J.; Marshall, S. Varietal Classification of Rice Seeds Using RGB and Hyperspectral Images. IEEE Access 2020, 8, 22493–22505. [Google Scholar] [CrossRef]
- Chatnuntawech, I.; Tantisantisom, K.; Khanchaitit, P.; Boonkoom, T.; Bilgic, B.; Chuangsuwanich, E. Rice Classification Using Spatio-Spectral Deep Convolutional Neural Network, 2019. arXiv:cs.CV/1805.11491].
- Jin, B.; Zhang, C.; Jia, L.; Tang, Q.; Gao, L.; Zhao, G.; Qi, H. Identification of Rice Seed Varieties Based on Near-Infrared Hyperspectral Imaging Technology Combined with Deep Learning. ACS Omega 2022, 7, 4735–4749. [Google Scholar] [CrossRef] [PubMed]
- Kiratiratanapruk, K.; Temniranrat, P.; Sinthupinyo, W.; Prempree, P.; Chaitavon, K.; Porntheeraphat, S.; Prasertsak, A. Development of Paddy Rice Seed Classification Process using Machine Learning Techniques for Automatic Grading Machine. Journal of Sensors 2020, 2020, 1–14. [Google Scholar] [CrossRef]
- Rajalakshmi, R.; Faizal, S.; Sivasankaran, S.; Geetha, R. RiceSeedNet: Rice seed variety identification using deep neural network. Journal of Agriculture and Food Research 2024, 16, 101062. [Google Scholar] [CrossRef]
- Komal. ; Sethi, G.K.; Bawa, R.K. Automatic Rice Variety Identification System: state-of-the-art review, issues, challenges and future directions. Multimedia Tools and Applications 2023, 82, 27305–27336. [Google Scholar] [CrossRef]
- Singh, N.; Kumar, P.; Kumar, A. Rice Leaf Disease Detection and Classification Using Convolutional Neural Network. NA 2022. NA.
- Ulukaya, S.; Deari, S. A robust vision transformer-based approach for classification of labeled rices in the wild. Computers and Electronics in Agriculture 2025, 231, 109950. [Google Scholar] [CrossRef]
- Melo, D.F.Q.; Silva, B.M.C.; Pombo, N.; Xu, L. Internet of Things Assisted Monitoring Using Ultrasound-Based Gesture Recognition Contactless System. IEEE Access 2021, 9, 90185–90194. [Google Scholar] [CrossRef]
- Patil, R.R.; Kumar, S.; Chiwhane, S.; Rani, R.; Pippal, S.K. An Artificial-Intelligence-Based Novel Rice Grade Model for Severity Estimation of Rice Diseases. Agriculture 2023, 13. [Google Scholar] [CrossRef]
- Verma, D.; Dafadar, M.; Mishra, J.; Kumar, A.; Mahato, S. AI-Enable Rice Image Classification Using Hybrid Convolutional Neural Network Models. International Journal of Intelligent Systems 2025, 2025. [Google Scholar] [CrossRef]
- ORCID — orcid.org. Available online: https://orcid.org/0000-0003-0818-6746 (accessed on 18 September 2025).
- Barman, U.; Sarma, P.; Rahman, M.; Deka, V.; Lahkar, S.; Sharma, V.; Saikia, M.J. ViT-SmartAgri: Vision Transformer and Smartphone-Based Plant Disease Detection for Smart Agriculture. Agronomy 2024, 14. [Google Scholar] [CrossRef]
- Bhujel, R.; Li, X.; Baniya, S.; Yin, Z. A Deep Learning Approach for Tomato Disease Detection Using Hybrid Convolutional Neural Network and Vision Transformer. Sensors 2023, 23, 6949. [Google Scholar] [CrossRef]
- Gudipalli, A.; N. , A.; Reddy Ch, P. A review on analysis and grading of rice using image processing. ARPN Journal of Engineering and Applied Sciences 2016, 11, 13550–13555. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).